Modulation of KV4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2
Abstract
:1. Introduction
2. Results
2.1. IQM-266 Binding to KChIP2
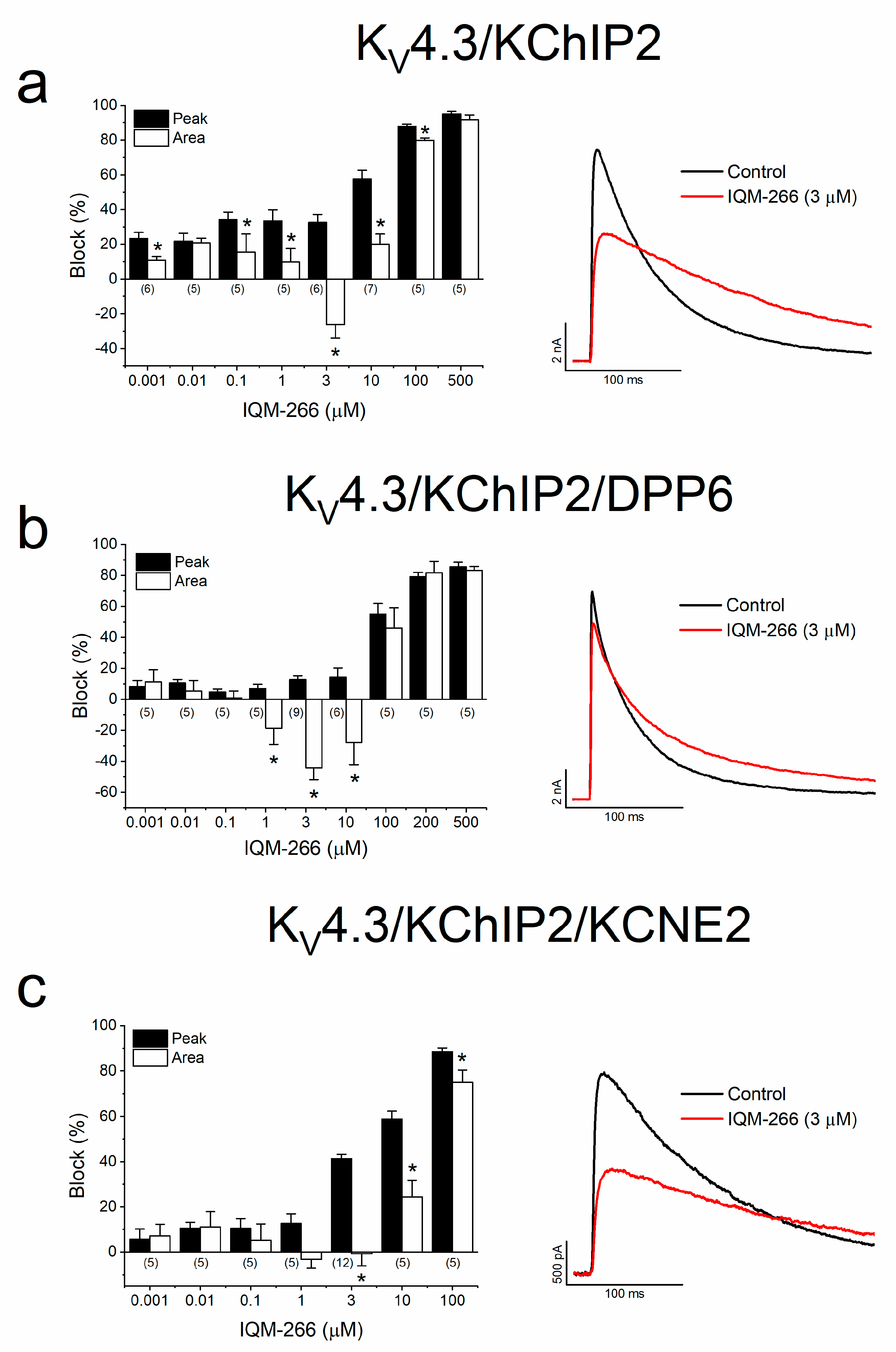
2.2. Effects of IQM-266 on KV4.3/KChIP2, KV4.3/KChIP2/DPP6 and KV4.3/KChIP2/KCNE2 Channels
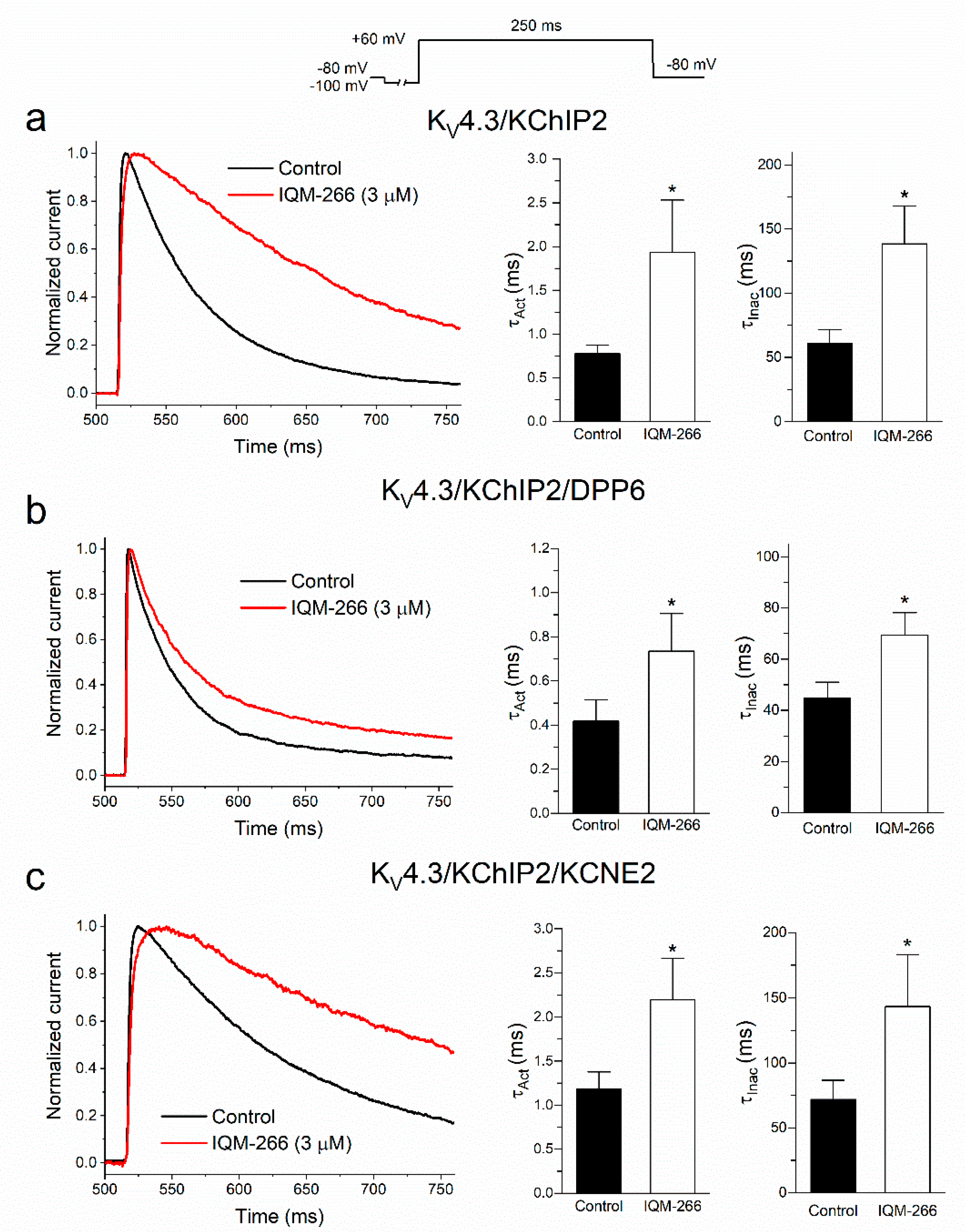
2.3. Time-Dependent Effects of IQM-266 on KV4.3/KChIP2, KV4.3/KChIP2/DPP6 and KV4.3/KChIP2/KCNE2 Channels
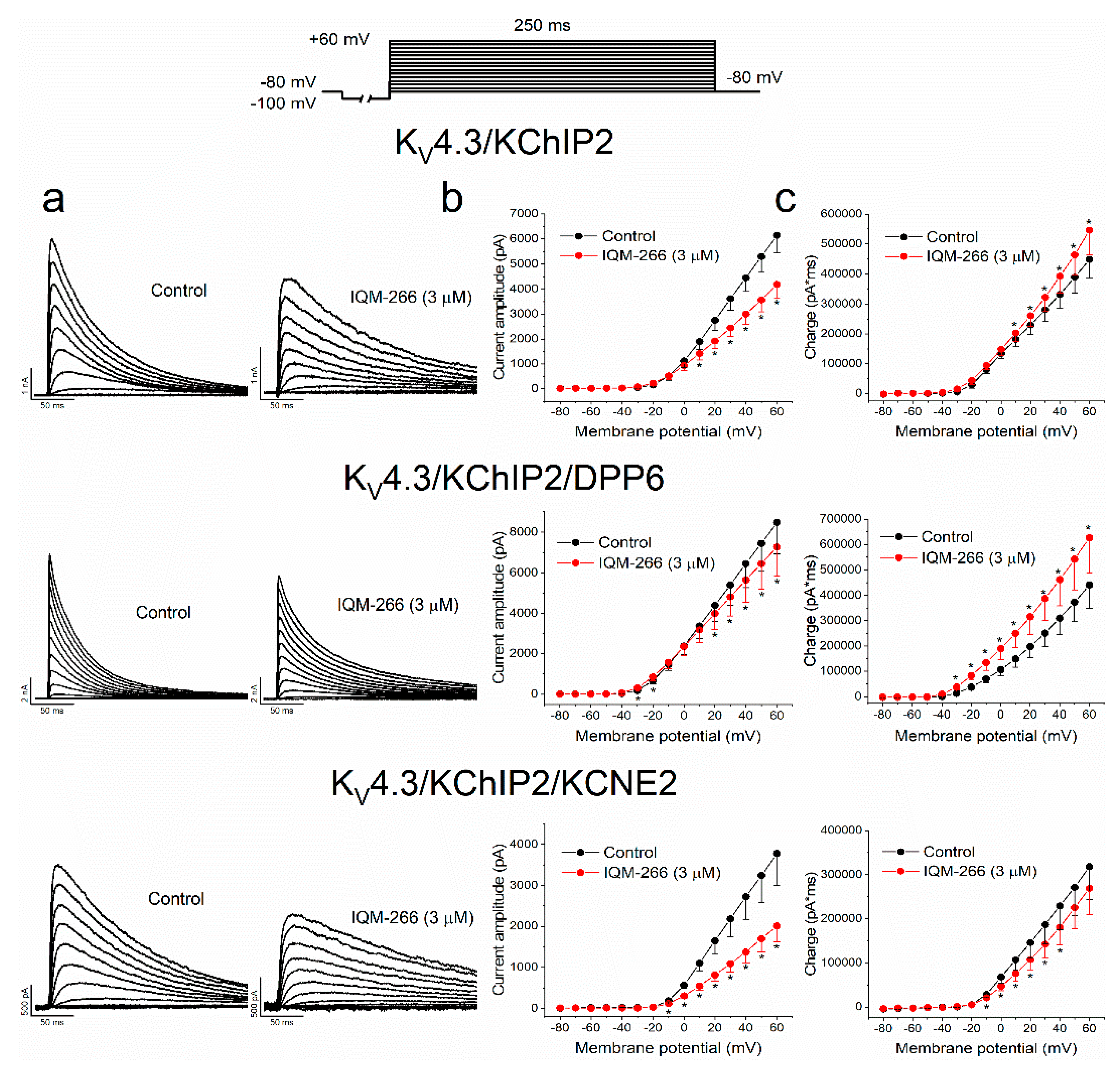
2.4. Voltage Dependence of the Block Produced by IQM-266 on KV4.3/KChIP2, KV4.3/KChIP2/DPP6 and KV4.3/KChIP2/KCNE2 Channels
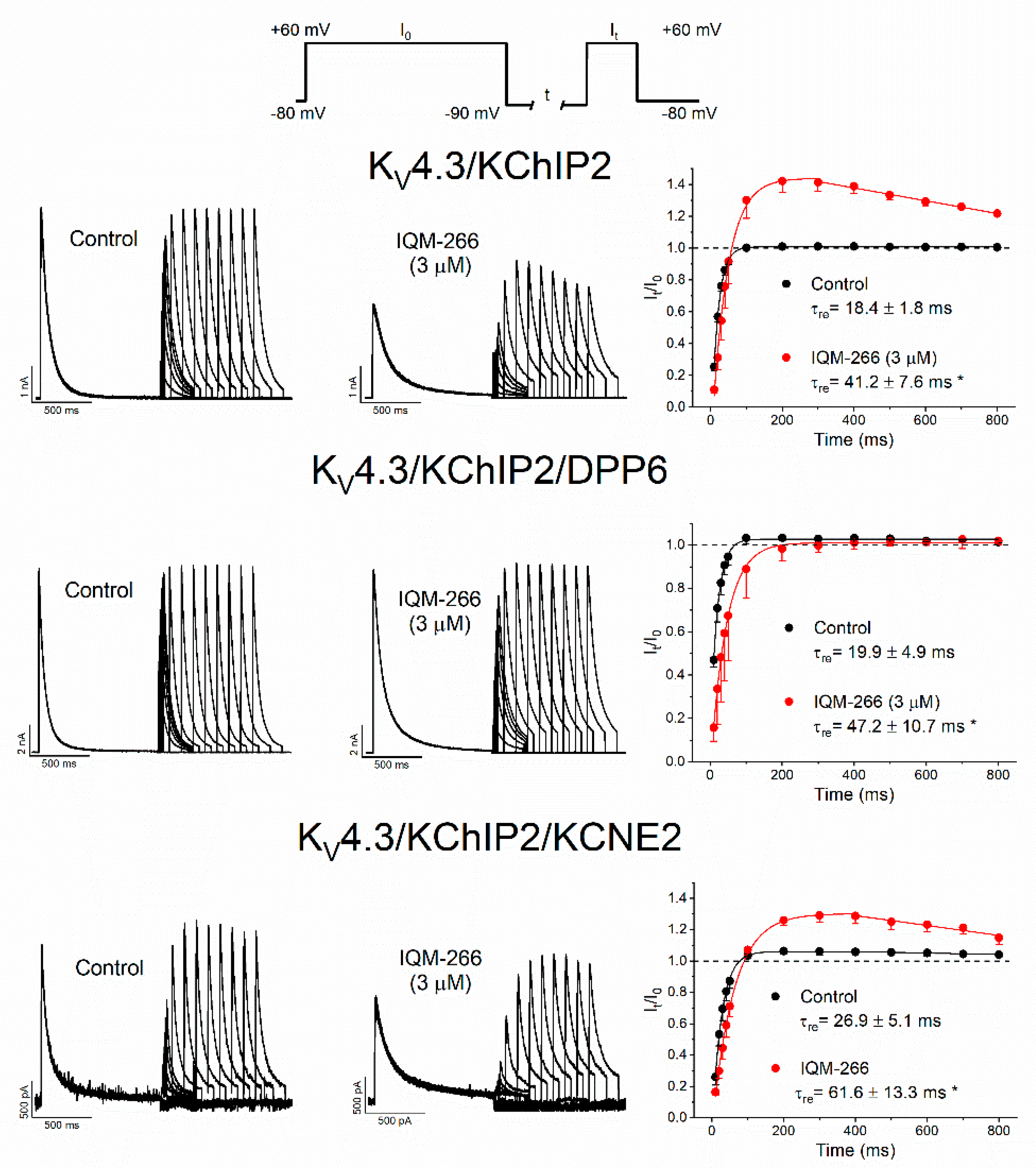
2.5. Use-Dependent Effects of IQM-266 on KV4.3/KChIP2, KV4.3/KChIP2/DPP6 and KV4.3/KChIP2/KCNE2 Channels
3. Discussion
4. Materials and Methods
4.1. Cellular Cultures and Transient Transfection
4.2. Purification Protocol
4.3. Concentration Determination
4.4. Chemistry
4.5. Fluorescence Binding Assay
4.6. Homology Modeling
4.7. Ligand Preparation
4.8. Docking Studies
4.9. Electrophysiology Methods
4.10. Statistics
4.11. Materials
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, W.; Po, S.; Yamagishi, T.; Ashen, M.D.; Stetten, G.; Tomaselli, G.F. Isolation and characterization of the human gene encoding Ito: Further diversity by alternative mRNA splicing. Am. J. Physiol. 1998, 275, H1963–H1970. [Google Scholar] [PubMed]
- Nerbonne, J.M.; Kass, R.S. Molecular physiology of cardiac repolarization. Physiol. Rev. 2005, 85, 1205–1253. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.E.; Shi, W.; Wang, H.S.; McDonald, C.; Yu, H.; Wymore, R.S.; Cohen, I.S.; McKinnon, D. Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circ. Res. 1996, 79, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Franqueza, L.; Valenzuela, C.; Eck, J.; Tamkun, M.M.; Tamargo, J.; Snyders, D.J. Functional expression of an inactivating potassium channel (Kv4.3) in a mammalian cell line. Cardiovasc. Res. 1999, 41, 212–219. [Google Scholar] [CrossRef]
- Patel, S.P.; Campbell, D.L. Transient outward potassium current, ‘Ito’, phenotypes in the mammalian left ventricle: Underlying molecular, cellular and biophysical mechanisms. J. Physiol. 2005, 569, 7–39. [Google Scholar] [CrossRef] [PubMed]
- Nattel, S.; Maguy, A.; Le Bouter, S.; Yeh, Y.H. Arrhythmogenic ion-channel remodeling in the heart: Heart failure, myocardial infarction, and atrial fibrillation. Physiol. Rev. 2007, 87, 425–456. [Google Scholar] [CrossRef]
- Huo, R.; Sheng, Y.; Guo, W.T.; Dong, D.L. The potential role of Kv4.3 K+ channel in heart hypertrophy. Channels 2014, 8, 203–209. [Google Scholar] [CrossRef]
- Caballero, R.; de la Fuente, M.G.; Gomez, R.; Barana, A.; Amoros, I.; Dolz-Gaiton, P.; Osuna, L.; Almendral, J.; Atienza, F.; Fernandez-Aviles, F.; et al. In humans, chronic atrial fibrillation decreases the transient outward current and ultrarapid component of the delayed rectifier current differentially on each atria and increases the slow component of the delayed rectifier current in both. J. Am. Coll. Cardiol. 2010, 55, 2346–2354. [Google Scholar] [CrossRef]
- Dilks, D.; Ling, H.P.; Cockett, M.; Sokol, P.; Numann, R. Cloning and expression of the human kv4.3 potassium channel. J. Neurophysiol. 1999, 81, 1974–1977. [Google Scholar] [CrossRef]
- Ohya, S.; Tanaka, M.; Oku, T.; Asai, Y.; Watanabe, M.; Giles, W.R.; Imaizumi, Y. Molecular cloning and tissue distribution of an alternatively spliced variant of an A-type K+ channel alpha-subunit, Kv4.3 in the rat. FEBS Lett. 1997, 420, 47–53. [Google Scholar] [CrossRef]
- Serodio, P.; Kentros, C.; Rudy, B. Identification of molecular components of A-type channels activating at subthreshold potentials. J. Neurophysiol. 1994, 72, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Serodio, P.; Vega-Saenz de Miera, E.; Rudy, B. Cloning of a novel component of A-type K+ channels operating at subthreshold potentials with unique expression in heart and brain. J. Neurophysiol. 1996, 75, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Abbott, G.W. beta Subunits Functionally Differentiate Human Kv4.3 Potassium Channel Splice Variants. Front. Physiol. 2017, 8, 66. [Google Scholar] [PubMed]
- Birnbaum, S.G.; Varga, A.W.; Yuan, L.L.; Anderson, A.E.; Sweatt, J.D.; Schrader, L.A. Structure and function of Kv4-family transient potassium channels. Physiol. Rev. 2004, 84, 803–833. [Google Scholar] [CrossRef]
- Nerbonne, J.M. Molecular basis of functional voltage-gated K+ channel diversity in the mammalian myocardium. J. Physiol. 2000, 525, 285–298. [Google Scholar] [CrossRef]
- Radicke, S.; Cotella, D.; Graf, E.M.; Banse, U.; Jost, N.; Varro, A.; Tseng, G.N.; Ravens, U.; Wettwer, E. Functional modulation of the transient outward current Ito by KCNE beta-subunits and regional distribution in human non-failing and failing hearts. Cardiovasc. Res. 2006, 71, 695–703. [Google Scholar] [CrossRef]
- Radicke, S.; Cotella, D.; Graf, E.M.; Ravens, U.; Wettwer, E. Expression and function of dipeptidyl-aminopeptidase-like protein 6 as a putative beta-subunit of human cardiac transient outward current encoded by Kv4.3. J. Physiol. 2005, 565, 751–756. [Google Scholar] [CrossRef]
- Capera, J.; Serrano-Novillo, C.; Navarro-Perez, M.; Cassinelli, S.; Felipe, A. The Potassium Channel Odyssey: Mechanisms of Traffic and Membrane Arrangement. Int. J. Mol. Sci. 2019, 20, 734. [Google Scholar] [CrossRef]
- Cercos, P.; Peraza, D.A.; Benito-Bueno, A.; Socuellamos, P.G.; Aziz-Nignan, A.; Arrechaga-Estevez, D.; Beato, E.; Pena-Acevedo, E.; Albert, A.; Gonzalez-Vera, J.A.; et al. Pharmacological Approaches for the Modulation of the Potassium Channel KV4.x and KChIPs. Int. J. Mol. Sci. 2021, 22, 1419. [Google Scholar] [CrossRef]
- An, W.F.; Bowlby, M.R.; Betty, M.; Cao, J.; Ling, H.P.; Mendoza, G.; Hinson, J.W.; Mattsson, K.I.; Strassle, B.W.; Trimmer, J.S.; et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature 2000, 403, 553–556. [Google Scholar] [CrossRef]
- Kuo, H.C.; Cheng, C.F.; Clark, R.B.; Lin, J.J.; Lin, J.L.; Hoshijima, M.; Nguyen-Tran, V.T.; Gu, Y.; Ikeda, Y.; Chu, P.H.; et al. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell 2001, 107, 801–813. [Google Scholar] [CrossRef]
- Benitah, J.P.; Gomez, A.M.; Bailly, P.; Da Ponte, J.P.; Berson, G.; Delgado, C.; Lorente, P. Heterogeneity of the early outward current in ventricular cells isolated from normal and hypertrophied rat hearts. J. Physiol. 1993, 469, 111–138. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, K.; Covarrubias, M. A dipeptidyl aminopeptidase-like protein remodels gating charge dynamics in Kv4.2 channels. J. Gen. Physiol. 2006, 128, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Jerng, H.H.; Pfaffinger, P.J.; Covarrubias, M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol. Cell. Neurosci. 2004, 27, 343–369. [Google Scholar] [CrossRef]
- Kise, Y.; Kasuya, G.; Okamoto, H.H.; Yamanouchi, D.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Nakajo, K.; Nureki, O. Structural basis of gating modulation of Kv4 channel complexes. Nature 2021, 599, 158–164. [Google Scholar] [CrossRef]
- Zhang, M.; Jiang, M.; Tseng, G.N. minK-related peptide 1 associates with Kv4.2 and modulates its gating function: Potential role as beta subunit of cardiac transient outward channel? Circ. Res. 2001, 88, 1012–1019. [Google Scholar] [CrossRef]
- Wettwer, E.; Amos, G.J.; Posival, H.; Ravens, U. Transient outward current in human ventricular myocytes of subepicardial and subendocardial origin. Circ. Res. 1994, 75, 473–482. [Google Scholar] [CrossRef]
- Peraza, D.A.; Cercos, P.; Miaja, P.; Merinero, Y.G.; Lagartera, L.; Socuellamos, P.G.; Izquierdo Garcia, C.; Sanchez, S.A.; Lopez-Hurtado, A.; Martin-Martinez, M.; et al. Identification of IQM-266, a Novel DREAM Ligand That Modulates KV4 Currents. Front. Mol. Neurosci. 2019, 12, 11. [Google Scholar] [CrossRef]
- Gonzalez, W.G.; Pham, K.; Miksovska, J. Modulation of the voltage-gated potassium channel (Kv4.3) and the auxiliary protein (KChIP3) interactions by the current activator NS5806. J. Biol. Chem. 2014, 289, 32201–32213. [Google Scholar] [CrossRef]
- Lusin, J.D.; Vanarotti, M.; Li, C.; Valiveti, A.; Ames, J.B. NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization. Biochemistry 2008, 47, 2252–2264. [Google Scholar] [CrossRef]
- Pioletti, M.; Findeisen, F.; Hura, G.L.; Minor, D.L., Jr. Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 2006, 13, 987–995. [Google Scholar] [CrossRef]
- Jerng, H.H.; Qian, Y.; Pfaffinger, P.J. Modulation of Kv4.2 channel expression and gating by dipeptidyl peptidase 10 (DPP10). Biophys. J. 2004, 87, 2380–2396. [Google Scholar] [CrossRef] [PubMed]
- Anno, T.; Hondeghem, L.M. Interactions of flecainide with guinea pig cardiac sodium channels. Importance of activation unblocking to the voltage dependence of recovery. Circ. Res. 1990, 66, 789–803. [Google Scholar] [CrossRef]
- Snyders, D.J.; Hondeghem, L.M. Effects of quinidine on the sodium current of guinea pig ventricular myocytes. Evidence for a drug-associated rested state with altered kinetics. Circ. Res. 1990, 66, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Jespersen, T.; Schmitt, N.; Grunnet, M.; Olesen, S.P.; Cordeiro, J.M.; Calloe, K. Effect of the I(to) activator NS5806 on cloned K(V)4 channels depends on the accessory protein KChIP2. Br. J. Pharmacol. 2010, 160, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Witzel, K.; Fischer, P.; Bahring, R. Hippocampal A-type current and Kv4.2 channel modulation by the sulfonylurea compound NS5806. Neuropharmacology 2012, 63, 1389–1403. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Wang, C.; Wang, Y.; Zou, R.; Shi, C.; Guan, B.; Gamper, N.; Xu, Y. Auxiliary subunits control biophysical properties and response to compound NS5806 of the Kv4 potassium channel complex. FASEB J. 2020, 34, 807–821. [Google Scholar] [CrossRef]
- McCrossan, Z.A.; Abbott, G.W. The MinK-related peptides. Neuropharmacology 2004, 47, 787–821. [Google Scholar] [CrossRef]
- Abbott, G.W. beta Subunits Control the Effects of Human Kv4.3 Potassium Channel Phosphorylation. Front. Physiol. 2017, 8, 646. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Schrödinger. Protein Preparation Wizard. Available online: https://www.schrodinger.com/science-articles/protein-preparation-wizard (accessed on 12 August 2022).
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK-a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Schrödinger. LigPrep. Available online: https://www.schrodinger.com/products/ligprep (accessed on 12 August 2022).
- Schrödinger. Induced Fit Docking Protocol. Available online: https://www.schrodinger.com/kb/1352 (accessed on 12 August 2022).
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg. Med. Chem. 2006, 14, 3160–3173. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef]
- Longobardo, M.; Delpon, E.; Caballero, R.; Tamargo, J.; Valenzuela, C. Structural determinants of potency and stereoselective block of hKv1.5 channels induced by local anesthetics. Mol. Pharmacol. 1998, 54, 162–169. [Google Scholar] [CrossRef]
- David, M.; Macias, A.; Moreno, C.; Prieto, A.; Martinez-Marmol, R.; Vicente, R.; Gonzalez, T.; Felipe, A.; Tamkun, M.M.; Valenzuela, C. Protein kinase C (PKC) activity regulates functional effects of Kvbeta1.3 subunit on KV1.5 channels: Identification of a cardiac Kv1.5 channelosome. J. Biol. Chem. 2012, 287, 21416–21428. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Benito-Bueno, A.; Socuellamos, P.G.; Merinero, Y.G.; Cercos, P.; Izquierdo, C.; Daniel-Mozo, M.; Marín-Olivero, I.; Perez-Lara, A.; Gonzalez-Vera, J.A.; Orte, A.; et al. Modulation of KV4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2. Int. J. Mol. Sci. 2022, 23, 9170. https://doi.org/10.3390/ijms23169170
de Benito-Bueno A, Socuellamos PG, Merinero YG, Cercos P, Izquierdo C, Daniel-Mozo M, Marín-Olivero I, Perez-Lara A, Gonzalez-Vera JA, Orte A, et al. Modulation of KV4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2. International Journal of Molecular Sciences. 2022; 23(16):9170. https://doi.org/10.3390/ijms23169170
Chicago/Turabian Stylede Benito-Bueno, Angela, Paula G. Socuellamos, Yaiza G. Merinero, Pilar Cercos, Carolina Izquierdo, Miguel Daniel-Mozo, Irene Marín-Olivero, Angel Perez-Lara, Juan A. Gonzalez-Vera, Angel Orte, and et al. 2022. "Modulation of KV4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2" International Journal of Molecular Sciences 23, no. 16: 9170. https://doi.org/10.3390/ijms23169170
APA Stylede Benito-Bueno, A., Socuellamos, P. G., Merinero, Y. G., Cercos, P., Izquierdo, C., Daniel-Mozo, M., Marín-Olivero, I., Perez-Lara, A., Gonzalez-Vera, J. A., Orte, A., Albert, A., Martin-Martinez, M., Gutierrez-Rodriguez, M., & Valenzuela, C. (2022). Modulation of KV4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2. International Journal of Molecular Sciences, 23(16), 9170. https://doi.org/10.3390/ijms23169170








