The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes
Abstract
1. Introduction
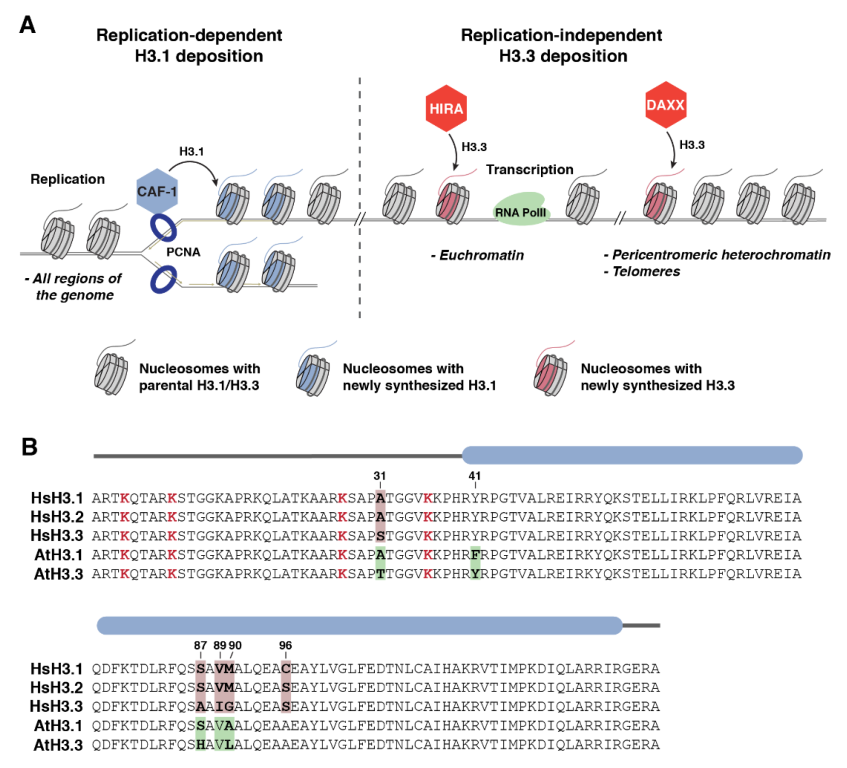
2. Discovery of the Replication-Dependent H3.1 Variant
3. Identification of Readers and Writers of H3 Variants
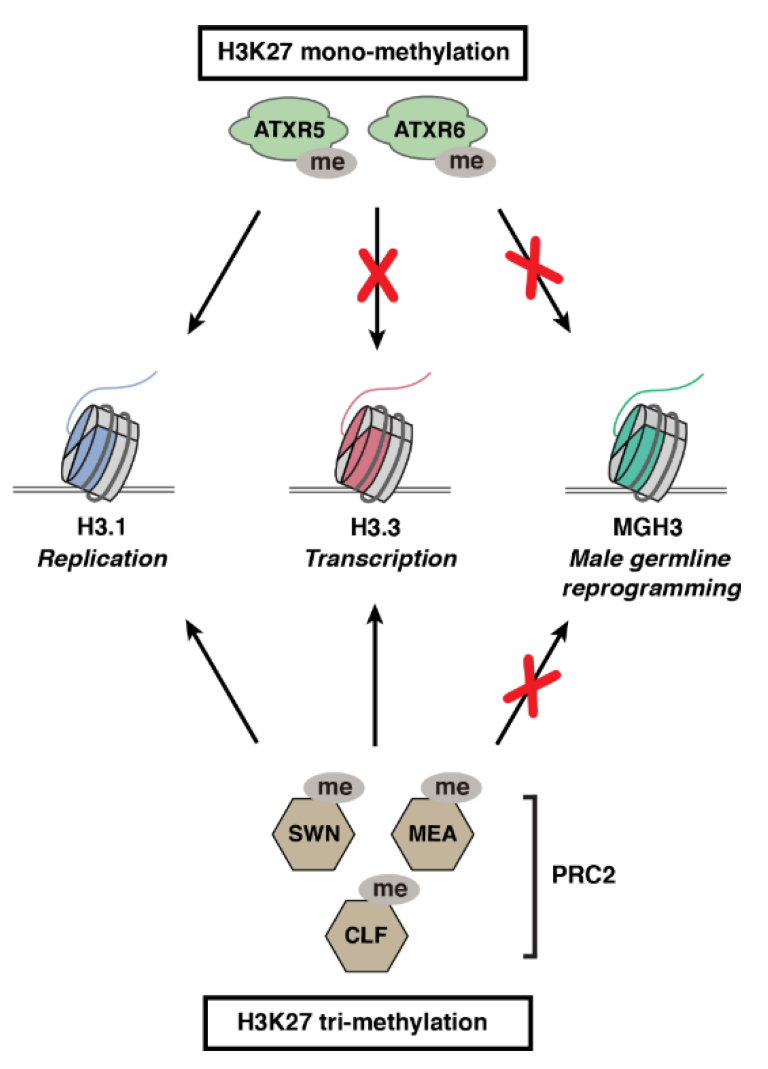
3.1. H3.1-Specific Deposition of H3K27me1 by ATXR5 and ATXR6 in Plants
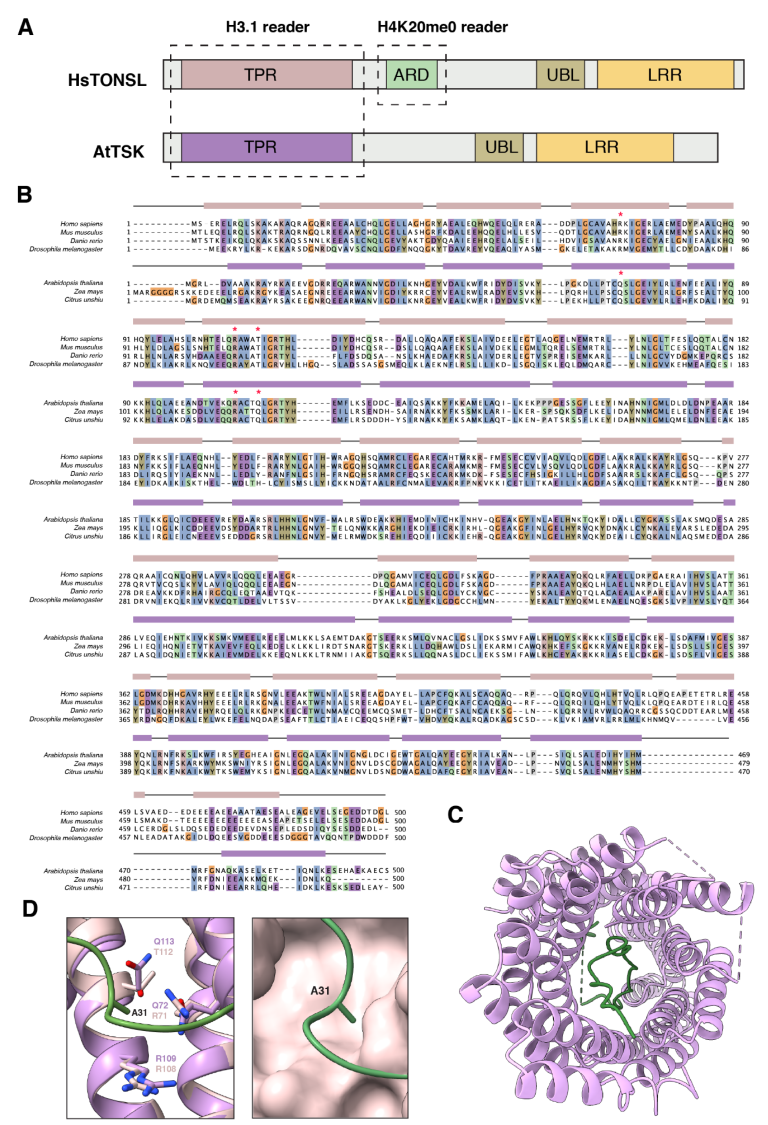
3.2. TSK/TONSL Acts as an H3.1 Reader via its TPR Domain
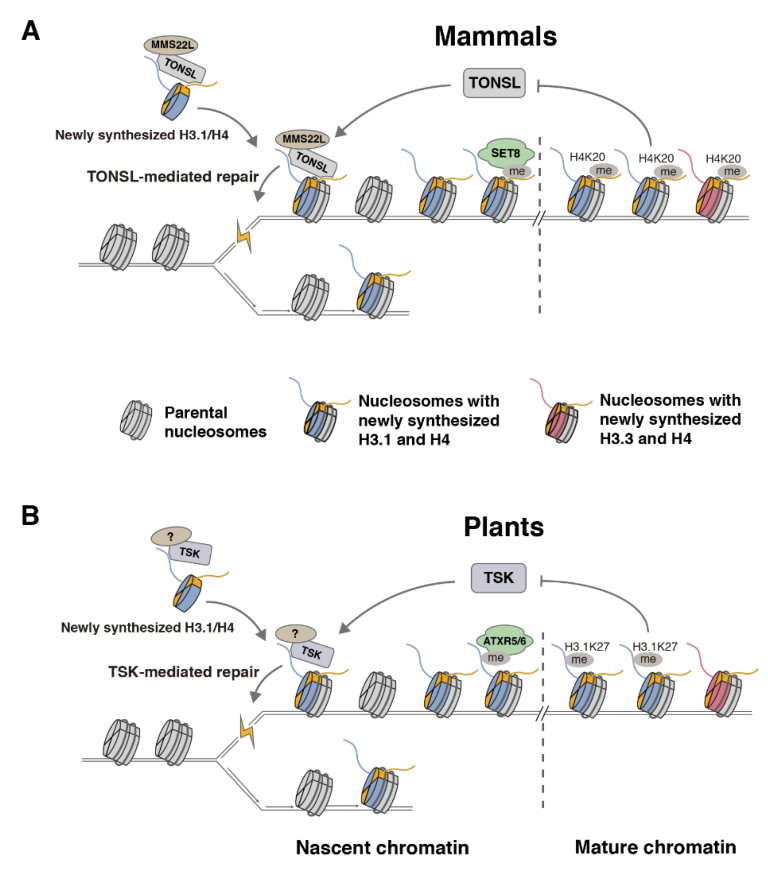
4. Role of the TSK/TONSL-H3.1 Pathway in DNA Repair
5. Differences between Plants and Animals in the TSK/TONSL-H3.1 Pathway
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef]
- Jiang, D.; Borg, M.; Lorkovic, Z.J.; Montgomery, S.A.; Osakabe, A.; Yelagandula, R.; Axelsson, E.; Berger, F. The evolution and functional divergence of the histone H2B family in plants. PLoS Genet. 2020, 16, e1008964. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Ahmad, K.; Almouzni, G.; Ausio, J.; Berger, F.; Bhalla, P.L.; Bonner, W.M.; Cande, W.Z.; Chadwick, B.P.; Chan, S.W.; et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics. Chromatin. 2012, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.B.; Allis, C.D. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 2006, 103, 6428–6435. [Google Scholar] [CrossRef] [PubMed]
- Franklin, S.G.; Zweidler, A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature 1977, 266, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.; Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 2002, 9, 1191–1200. [Google Scholar] [CrossRef]
- Ray-Gallet, D.; Woolfe, A.; Vassias, I.; Pellentz, C.; Lacoste, N.; Puri, A.; Schultz, D.C.; Pchelintsev, N.A.; Adams, P.D.; Jansen, L.E.; et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 2011, 44, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Roche, D.; Almouzni, G. New histone incorporation marks sites of UV repair in human cells. Cell 2006, 127, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Postberg, J.; Forcob, S.; Chang, W.J.; Lipps, H.J. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 2010, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Endo, M.; Singh, M.B.; Bhalla, P.L. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005, 44, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Bonner, W.M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell 1981, 27, 321–330. [Google Scholar] [CrossRef]
- Ray-Gallet, D.; Almouzni, G. The Histone H3 Family and Its Deposition Pathways. Adv. Exp. Med. Biol. 2021, 1283, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar] [CrossRef]
- Shibahara, K.; Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 1999, 96, 575–585. [Google Scholar] [CrossRef]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes. Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Ray-Gallet, D.; Quivy, J.P.; Scamps, C.; Martini, E.M.; Lipinski, M.; Almouzni, G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 2002, 9, 1091–1100. [Google Scholar] [CrossRef]
- Sarai, N.; Nimura, K.; Tamura, T.; Kanno, T.; Patel, M.C.; Heightman, T.D.; Ura, K.; Ozato, K. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. Embo. J. 2013, 32, 2392–2406. [Google Scholar] [CrossRef]
- Soni, S.; Pchelintsev, N.; Adams, P.D.; Bieker, J.J. Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for beta-globin gene expression. Proc. Natl. Acad. Sci. USA 2014, 111, 13337–13342. [Google Scholar] [CrossRef]
- Zhang, H.; Gan, H.; Wang, Z.; Lee, J.H.; Zhou, H.; Ordog, T.; Wold, M.S.; Ljungman, M.; Zhang, Z. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol. Cell 2017, 65, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Maze, I.; Wenderski, W.; Noh, K.M.; Bagot, R.C.; Tzavaras, N.; Purushothaman, I.; Elsasser, S.J.; Guo, Y.; Ionete, C.; Hurd, Y.L.; et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 2015, 87, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Pina, B.; Suau, P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 1987, 123, 51–58. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Sekeri-Pataryas, K.E. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 1999, 34, 741–754. [Google Scholar] [CrossRef]
- Urban, M.K.; Zweidler, A. Changes in nucleosomal core histone variants during chicken development and maturation. Dev. Biol. 1983, 95, 421–428. [Google Scholar] [CrossRef]
- Ingouff, M.; Rademacher, S.; Holec, S.; Soljic, L.; Xin, N.; Readshaw, A.; Foo, S.H.; Lahouze, B.; Sprunck, S.; Berger, F. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 2010, 20, 2137–2143. [Google Scholar] [CrossRef]
- Loppin, B.; Bonnefoy, E.; Anselme, C.; Laurencon, A.; Karr, T.L.; Couble, P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 2005, 437, 1386–1390. [Google Scholar] [CrossRef]
- Torres-Padilla, M.E.; Bannister, A.J.; Hurd, P.J.; Kouzarides, T.; Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 2006, 50, 455–461. [Google Scholar] [CrossRef]
- Van der Heijden, G.W.; Dieker, J.W.; Derijck, A.A.; Muller, S.; Berden, J.H.; Braat, D.D.; van der Vlag, J.; de Boer, P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005, 122, 1008–1022. [Google Scholar] [CrossRef]
- Stroud, H.; Otero, S.; Desvoyes, B.; Ramirez-Parra, E.; Jacobsen, S.E.; Gutierrez, C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 5370–5375. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, H.; Holec, S.; Alden, K.; Clarke, N.D.; Jacques, P.E.; Berger, F. Dynamic deposition of histone variant h3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012, 8, e1002658. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, J.; Hong, F.; Spector, D.L.; Fang, Y. Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes. Proc. Natl. Acad. Sci. USA 2011, 108, 10574–10578. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Chen, X.; Qian, S.; Zhong, X. The plant-specific histone residue Phe41 is important for genome-wide H3.1 distribution. Nat. Commun. 2018, 9, 630. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.J.; Huang, H.; Lewis, P.W.; Chin, J.W.; Allis, C.D.; Patel, D.J. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 2012, 491, 560–565. [Google Scholar] [CrossRef]
- Liu, C.P.; Xiong, C.; Wang, M.; Yu, Z.; Yang, N.; Chen, P.; Zhang, Z.; Li, G.; Xu, R.M. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 2012, 19, 1287–1292. [Google Scholar] [CrossRef]
- Ricketts, M.D.; Frederick, B.; Hoff, H.; Tang, Y.; Schultz, D.C.; Singh Rai, T.; Grazia Vizioli, M.; Adams, P.D.; Marmorstein, R. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 2015, 6, 7711. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Benson, L.J.; Gu, Y.; Yakovleva, T.; Tong, K.; Barrows, C.; Strack, C.L.; Cook, R.G.; Mizzen, C.A.; Annunziato, A.T. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 2006, 281, 9287–9296. [Google Scholar] [CrossRef]
- Hake, S.B.; Garcia, B.A.; Duncan, E.M.; Kauer, M.; Dellaire, G.; Shabanowitz, J.; Bazett-Jones, D.P.; Allis, C.D.; Hunt, D.F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006, 281, 559–568. [Google Scholar] [CrossRef]
- Johnson, L.; Mollah, S.; Garcia, B.A.; Muratore, T.L.; Shabanowitz, J.; Hunt, D.F.; Jacobsen, S.E. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic. Acids. Res. 2004, 32, 6511–6518. [Google Scholar] [CrossRef] [PubMed]
- Loyola, A.; Bonaldi, T.; Roche, D.; Imhof, A.; Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 2006, 24, 309–316. [Google Scholar] [CrossRef] [PubMed]
- McKittrick, E.; Gafken, P.R.; Ahmad, K.; Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 2004, 101, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Waterborg, J.H. Sequence analysis of acetylation and methylation in two histone H3 variants of alfalfa. J. Biol. Chem. 1990, 265, 17157–17161. [Google Scholar] [CrossRef]
- Hake, S.B.; Garcia, B.A.; Kauer, M.; Baker, S.P.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. USA 2005, 102, 6344–6349. [Google Scholar] [CrossRef]
- Schulmeister, A.; Schmid, M.; Thompson, E.M. Phosphorylation of the histone H3.3 variant in mitosis and meiosis of the urochordate Oikopleura dioica. Chromosome. Res. 2007, 15, 189–201. [Google Scholar] [CrossRef]
- Van der Heijden, G.W.; Derijck, A.A.; Posfai, E.; Giele, M.; Pelczar, P.; Ramos, L.; Wansink, D.G.; van der Vlag, J.; Peters, A.H.; de Boer, P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007, 39, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Ren, H.; Williams, E.; McGhie, J.; Ahn, S.; Sim, M.; Tam, A.; Earle, E.; Anderson, M.A.; Mann, J.; et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome. Res. 2009, 19, 404–414. [Google Scholar] [CrossRef]
- Sitbon, D.; Boyarchuk, E.; Dingli, F.; Loew, D.; Almouzni, G. Histone variant H3.3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway. Nat. Commun. 2020, 11, 1256. [Google Scholar] [CrossRef]
- Martire, S.; Gogate, A.A.; Whitmill, A.; Tafessu, A.; Nguyen, J.; Teng, Y.C.; Tastemel, M.; Banaszynski, L.A. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat. Genet. 2019, 51, 941–946. [Google Scholar] [CrossRef]
- Armache, A.; Yang, S.; Martinez de Paz, A.; Robbins, L.E.; Durmaz, C.; Cheong, J.Q.; Ravishankar, A.; Daman, A.W.; Ahimovic, D.J.; Klevorn, T.; et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 2020, 583, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Andrews, F.H.; Gatchalian, J.; Krajewski, K.; Strahl, B.D.; Kutateladze, T.G. Regulation of Methyllysine Readers through Phosphorylation. ACS Chem. Biol. 2016, 11, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zheng, L.; Park, J.W.; Lv, R.; Chen, H.; Jiao, F.; Xu, W.; Mu, S.; Wen, H.; Qiu, J.; et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 2014, 56, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Li, Y.; Xi, Y.; Jiang, S.; Stratton, S.; Peng, D.; Tanaka, K.; Ren, Y.; Xia, Z.; Wu, J.; et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 2014, 508, 263–268. [Google Scholar] [CrossRef]
- Gao, S.; Xiong, J.; Zhang, C.; Berquist, B.R.; Yang, R.; Zhao, M.; Molascon, A.J.; Kwiatkowski, S.Y.; Yuan, D.; Qin, Z.; et al. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes. Dev. 2013, 27, 1662–1679. [Google Scholar] [CrossRef]
- Jacob, Y.; Bergamin, E.; Donoghue, M.T.; Mongeon, V.; LeBlanc, C.; Voigt, P.; Underwood, C.J.; Brunzelle, J.S.; Michaels, S.D.; Reinberg, D.; et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 2014, 343, 1249–1253. [Google Scholar] [CrossRef]
- Davarinejad, H.; Huang, Y.C.; Mermaz, B.; LeBlanc, C.; Poulet, A.; Thomson, G.; Joly, V.; Munoz, M.; Arvanitis-Vigneault, A.; Valsakumar, D.; et al. The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 2022, 375, 1281–1286. [Google Scholar] [CrossRef]
- Boehm, E.M.; Gildenberg, M.S.; Washington, M.T. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 2016, 39, 231–254. [Google Scholar] [CrossRef]
- Raynaud, C.; Sozzani, R.; Glab, N.; Domenichini, S.; Perennes, C.; Cella, R.; Kondorosi, E.; Bergounioux, C. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006, 47, 395–407. [Google Scholar] [CrossRef]
- Baumbusch, L.O.; Thorstensen, T.; Krauss, V.; Fischer, A.; Naumann, K.; Assalkhou, R.; Schulz, I.; Reuter, G.; Aalen, R.B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic. Acids Res. 2001, 29, 4319–4333. [Google Scholar] [CrossRef]
- Springer, N.M.; Napoli, C.A.; Selinger, D.A.; Pandey, R.; Cone, K.C.; Chandler, V.L.; Kaeppler, H.F.; Kaeppler, S.M. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003, 132, 907–925. [Google Scholar] [CrossRef] [PubMed]
- Jacob, Y.; Feng, S.; LeBlanc, C.A.; Bernatavichute, Y.V.; Stroud, H.; Cokus, S.; Johnson, L.M.; Pellegrini, M.; Jacobsen, S.E.; Michaels, S.D. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Jacob, Y.; Michaels, S.D. H3K27me1 is E(z) in animals, but not in plants. Epigenetics 2009, 4, 366–369. [Google Scholar] [CrossRef]
- Jacob, Y.; Stroud, H.; Leblanc, C.; Feng, S.; Zhuo, L.; Caro, E.; Hassel, C.; Gutierrez, C.; Michaels, S.D.; Jacobsen, S.E. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 2010, 466, 987–991. [Google Scholar] [CrossRef]
- Lindroth, A.M.; Shultis, D.; Jasencakova, Z.; Fuchs, J.; Johnson, L.; Schubert, D.; Patnaik, D.; Pradhan, S.; Goodrich, J.; Schubert, I.; et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. Embo. J. 2004, 23, 4286–4296. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendia, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Jacob, Y.; Voigt, P. In Vitro Assays to Measure Histone Methyltransferase Activity Using Different Chromatin Substrates. Methods Mol. Biol. 2018, 1675, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Wang, Y.; Liu, Y.; Gao, S. Histone methyltransferase TXR1 is required for both H3 and H3.3 lysine 27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci. China Life Sci. 2017, 60, 264–270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cui, B.; Liu, Y.; Gorovsky, M.A. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell Biol. 2006, 26, 7719–7730. [Google Scholar] [CrossRef]
- Takeda, S.; Tadele, Z.; Hofmann, I.; Probst, A.V.; Angelis, K.J.; Kaya, H.; Araki, T.; Mengiste, T.; Mittelsten Scheid, O.; Shibahara, K.; et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes. Dev. 2004, 18, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Guyomarc’h, S.; Vernoux, T.; Traas, J.; Zhou, D.X.; Delarue, M. MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J. Exp. Bot. 2004, 55, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Inagaki, S.; Nakajima, S.; Akashi, T.; Ohto, M.A.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 2004, 38, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Duro, E.; Lundin, C.; Ask, K.; Sanchez-Pulido, L.; MacArtney, T.J.; Toth, R.; Ponting, C.P.; Groth, A.; Helleday, T.; Rouse, J. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 2010, 40, 632–644. [Google Scholar] [CrossRef]
- O’Connell, B.C.; Adamson, B.; Lydeard, J.R.; Sowa, M.E.; Ciccia, A.; Bredemeyer, A.L.; Schlabach, M.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol. Cell 2010, 40, 645–657. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.; Panier, S.; Wildenhain, J.; Tkach, J.M.; Al-Hakim, A.; Landry, M.C.; Escribano-Diaz, C.; Szilard, R.K.; Young, J.T.; Munro, M.; et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 2010, 40, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Piwko, W.; Olma, M.H.; Held, M.; Bianco, J.N.; Pedrioli, P.G.; Hofmann, K.; Pasero, P.; Gerlich, D.W.; Peter, M. RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. Embo. J. 2010, 29, 4210–4222. [Google Scholar] [CrossRef]
- Campos, E.I.; Smits, A.H.; Kang, Y.H.; Landry, S.; Escobar, T.M.; Nayak, S.; Ueberheide, B.M.; Durocher, D.; Vermeulen, M.; Hurwitz, J.; et al. Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol. Cell 2015, 60, 697–709. [Google Scholar] [CrossRef]
- Saredi, G.; Huang, H.; Hammond, C.M.; Alabert, C.; Bekker-Jensen, S.; Forne, I.; Reveron-Gomez, N.; Foster, B.M.; Mlejnkova, L.; Bartke, T.; et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature 2016, 534, 714–718. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Piwko, W.; Mlejnkova, L.J.; Mutreja, K.; Ranjha, L.; Stafa, D.; Smirnov, A.; Brodersen, M.M.; Zellweger, R.; Sturzenegger, A.; Janscak, P.; et al. The MMS22L-TONSL heterodimer directly promotes RAD51-dependent recombination upon replication stress. Embo. J. 2016, 35, 2584–2601. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.H.; Fowler, F.; Chen, C.C.; Shen, Z.J.; Sleckman, B.; Tyler, J.K. The Histone Chaperones ASF1 and CAF-1 Promote MMS22L-TONSL-Mediated Rad51 Loading onto ssDNA during Homologous Recombination in Human Cells. Mol. Cell 2018, 69, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; LeBlanc, C.; Poulet, A.; Mermaz, B.; Villarino, G.; Webb, K.M.; Joly, V.; Mendez, J.; Voigt, P.; Jacob, Y. H3.1K27me1 maintains transcriptional silencing and genome stability by preventing GCN5-mediated histone acetylation. Plant Cell 2021, 33, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Houston, S.I.; McManus, K.J.; Adams, M.M.; Sims, J.K.; Carpenter, P.B.; Hendzel, M.J.; Rice, J.C. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 2008, 283, 19478–19488. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.; Elvers, I.; Trelle, M.B.; Menzel, T.; Eskildsen, M.; Jensen, O.N.; Helleday, T.; Helin, K.; Sorensen, C.S. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 2007, 179, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; Okamoto, I.; Murphy, N.; Chu, J.; Price, S.M.; Shen, M.M.; Torres-Padilla, M.E.; Heard, E.; Reinberg, D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell Biol. 2009, 29, 2278–2295. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Murr, R.; Herceg, Z.; Sardet, C.; Julien, E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 2007, 179, 1413–1426. [Google Scholar] [CrossRef]
- Fang, J.; Feng, Q.; Ketel, C.S.; Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Simon, J.A.; Zhang, Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002, 12, 1086–1099. [Google Scholar] [CrossRef]
- Nishioka, K.; Rice, J.C.; Sarma, K.; Erdjument-Bromage, H.; Werner, J.; Wang, Y.; Chuikov, S.; Valenzuela, P.; Tempst, P.; Steward, R.; et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 2002, 9, 1201–1213. [Google Scholar] [CrossRef]
- Rice, J.C.; Nishioka, K.; Sarma, K.; Steward, R.; Reinberg, D.; Allis, C.D. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes. Dev. 2002, 16, 2225–2230. [Google Scholar] [CrossRef] [PubMed]
- Kuo, A.J.; Song, J.; Cheung, P.; Ishibe-Murakami, S.; Yamazoe, S.; Chen, J.K.; Patel, D.J.; Gozani, O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 2012, 484, 115–119. [Google Scholar] [CrossRef]
- Li, S.; Yang, Z.; Du, X.; Liu, R.; Wilkinson, A.W.; Gozani, O.; Jacobsen, S.E.; Patel, D.J.; Du, J. Structural Basis for the Unique Multivalent Readout of Unmodified H3 Tail by Arabidopsis ORC1b BAH-PHD Cassette. Structure 2016, 24, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Saredi, G.; Becker, J.R.; Foster, B.M.; Nguyen, N.V.; Beyer, T.E.; Cesa, L.C.; Faull, P.A.; Lukauskas, S.; Frimurer, T.; et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019, 21, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Bukowski-Wills, J.C.; Lee, S.B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stutzer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 2014, 53, 49–62. [Google Scholar] [CrossRef]
- Jiang, D.; Berger, F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 2017, 357, 1146–1149. [Google Scholar] [CrossRef]
- Pesavento, J.J.; Yang, H.; Kelleher, N.L.; Mizzen, C.A. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell Biol. 2008, 28, 468–486. [Google Scholar] [CrossRef]
- Huen, M.S.; Sy, S.M.; van Deursen, J.M.; Chen, J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J. Biol. Chem. 2008, 283, 11073–11077. [Google Scholar] [CrossRef]
- Abbas, T.; Shibata, E.; Park, J.; Jha, S.; Karnani, N.; Dutta, A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 2010, 40, 9–21. [Google Scholar] [CrossRef]
- Centore, R.C.; Havens, C.G.; Manning, A.L.; Li, J.M.; Flynn, R.L.; Tse, A.; Jin, J.; Dyson, N.J.; Walter, J.C.; Zou, L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 2010, 40, 22–33. [Google Scholar] [CrossRef]
- Jorgensen, S.; Eskildsen, M.; Fugger, K.; Hansen, L.; Larsen, M.S.; Kousholt, A.N.; Syljuasen, R.G.; Trelle, M.B.; Jensen, O.N.; Helin, K.; et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 2011, 192, 43–54. [Google Scholar] [CrossRef]
- Oda, H.; Hubner, M.R.; Beck, D.B.; Vermeulen, M.; Hurwitz, J.; Spector, D.L.; Reinberg, D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 2010, 40, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Brustel, J.; Kirsh, O.; Lefevbre, C.; Callanan, M.; Sardet, C.; Julien, E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010, 12, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Davarinejad, H.; Joshi, M.; Ait-Hamou, N.; Munro, K.; Couture, J.F. ATXR5/6 Forms Alternative Protein Complexes with PCNA and the Nucleosome Core Particle. J. Mol. Biol. 2019, 431, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Brambati, A.; Barry, R.M.; Sfeir, A. DNA polymerase theta (Poltheta)—An error-prone polymerase necessary for genome stability. Curr. Opin. Genet. Dev. 2020, 60, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Goldstein, M.; Alexander, P.; Wakeman, T.P.; Sun, T.; Feng, J.; Lou, Z.; Kastan, M.B.; Wang, X.F. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. Embo. J. 2014, 33, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Heitzeberg, F.; Chen, I.P.; Hartung, F.; Orel, N.; Angelis, K.J.; Puchta, H. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 2004, 38, 954–968. [Google Scholar] [CrossRef]
- Ingouff, M.; Berger, F. Histone3 variants in plants. Chromosoma 2010, 119, 27–33. [Google Scholar] [CrossRef]
- Filipescu, D.; Muller, S.; Almouzni, G. Histone H3 variants and their chaperones during development and disease: Contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 2014, 30, 615–646. [Google Scholar] [CrossRef]
- Cheng, Z.; Cheung, P.; Kuo, A.J.; Yukl, E.T.; Wilmot, C.M.; Gozani, O.; Patel, D.J. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes. Dev. 2014, 28, 1758–1771. [Google Scholar] [CrossRef]
- Bowman, A.; Lercher, L.; Singh, H.R.; Zinne, D.; Timinszky, G.; Carlomagno, T.; Ladurner, A.G. The histone chaperone sNASP binds a conserved peptide motif within the globular core of histone H3 through its TPR repeats. Nucleic. Acids Res. 2016, 44, 3105–3117. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, H.; Gao, Y.; Zhu, Z.; Chen, Z.; Zheng, P.; Xue, L.; Li, J.; Teng, M.; Niu, L. Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer. Structure 2016, 24, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Cho, Y.W.; Yu, L.R.; Yu, H.; Veenstra, T.D.; Ge, K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA 2007, 104, 18439–18444. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wu, J.; Sun, B.; Zhong, C.; Ding, J. Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding. Nucleic. Acids. Res. 2011, 39, 4438–4449. [Google Scholar] [CrossRef] [PubMed]
- Burrage, L.C.; Reynolds, J.J.; Baratang, N.V.; Phillips, J.B.; Wegner, J.; McFarquhar, A.; Higgs, M.R.; Christiansen, A.E.; Lanza, D.G.; Seavitt, J.R.; et al. Bi-allelic Variants in TONSL Cause SPONASTRIME Dysplasia and a Spectrum of Skeletal Dysplasia Phenotypes. Am. J. Hum. Genet. 2019, 104, 422–438. [Google Scholar] [CrossRef]
- Chang, H.R.; Cho, S.Y.; Lee, J.H.; Lee, E.; Seo, J.; Lee, H.R.; Cavalcanti, D.P.; Makitie, O.; Valta, H.; Girisha, K.M.; et al. Hypomorphic Mutations in TONSL Cause SPONASTRIME Dysplasia. Am. J. Hum. Genet. 2019, 104, 439–453. [Google Scholar] [CrossRef]
- Micale, L.; Cialfi, S.; Fusco, C.; Cinque, L.; Castellana, S.; Biagini, T.; Talora, C.; Notarangelo, A.; Bisceglia, L.; Taruscio, D.; et al. Novel TONSL variants cause SPONASTRIME dysplasia and associate with spontaneous chromosome breaks, defective cell proliferation and apoptosis. Hum. Mol. Genet. 2020, 29, 3122–3131. [Google Scholar] [CrossRef]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.E.; Michaels, S.D. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar] [CrossRef]
- Hale, C.J.; Potok, M.E.; Lopez, J.; Do, T.; Liu, A.; Gallego-Bartolome, J.; Michaels, S.D.; Jacobsen, S.E. Identification of Multiple Proteins Coupling Transcriptional Gene Silencing to Genome Stability in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006092. [Google Scholar] [CrossRef]
- Ma, Z.; Castillo-Gonzalez, C.; Wang, Z.; Sun, D.; Hu, X.; Shen, X.; Potok, M.E.; Zhang, X. Arabidopsis Serrate Coordinates Histone Methyltransferases ATXR5/6 and RNA Processing Factor RDR6 to Regulate Transposon Expression. Dev. Cell 2018, 45, 769–784. [Google Scholar] [CrossRef]
- Stroud, H.; Hale, C.J.; Feng, S.; Caro, E.; Jacob, Y.; Michaels, S.D.; Jacobsen, S.E. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet. 2012, 8, e1002808. [Google Scholar] [CrossRef]
- Potok, M.E.; Zhong, Z.; Picard, C.L.; Liu, Q.; Do, T.; Jacobsen, C.E.; Sakr, O.; Naranbaatar, B.; Thilakaratne, R.; Khnkoyan, Z.; et al. The role of ATXR6 expression in modulating genome stability and transposable element repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2115570119. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Narangajavana, J.; Yamamoto, A.; Hattori, T.; Kagaya, Y.; Paszkowski, J.; Gruissem, W.; Hennig, L.; Takeda, S. Ectopic gene expression and organogenesis in Arabidopsis mutants missing BRU1 required for genome maintenance. Genetics 2011, 189, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Picart-Picolo, A.; Grob, S.; Picault, N.; Franek, M.; Llauro, C.; Halter, T.; Maier, T.R.; Jobet, E.; Descombin, J.; Zhang, P.; et al. Large tandem duplications affect gene expression, 3D organization, and plant-pathogen response. Genome. Res. 2020, 30, 1583–1592. [Google Scholar] [CrossRef]
- Volk, A.; Crispino, J.D. The role of the chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in homeostasis and disease. Biochim. Biophys. Acta 2015, 1849, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tonjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [CrossRef]
- Sarthy, J.F.; Meers, M.P.; Janssens, D.H.; Henikoff, J.G.; Feldman, H.; Paddison, P.J.; Lockwood, C.M.; Vitanza, N.A.; Olson, J.M.; Ahmad, K.; et al. Histone deposition pathways determine the chromatin landscapes of H3.1 and H3.3 K27M oncohistones. eLife 2020, 9, e61090. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-C.; Yuan, W.; Jacob, Y. The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes. Int. J. Mol. Sci. 2022, 23, 9029. https://doi.org/10.3390/ijms23169029
Huang Y-C, Yuan W, Jacob Y. The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes. International Journal of Molecular Sciences. 2022; 23(16):9029. https://doi.org/10.3390/ijms23169029
Chicago/Turabian StyleHuang, Yi-Chun, Wenxin Yuan, and Yannick Jacob. 2022. "The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes" International Journal of Molecular Sciences 23, no. 16: 9029. https://doi.org/10.3390/ijms23169029
APA StyleHuang, Y.-C., Yuan, W., & Jacob, Y. (2022). The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes. International Journal of Molecular Sciences, 23(16), 9029. https://doi.org/10.3390/ijms23169029





