“Soft Protein Corona” as the Stabilizer of the Methionine-Coated Silver Nanoparticles in the Physiological Environment: Insights into the Mechanism of the Interaction
Abstract
:1. Introduction
2. Results and Discussion
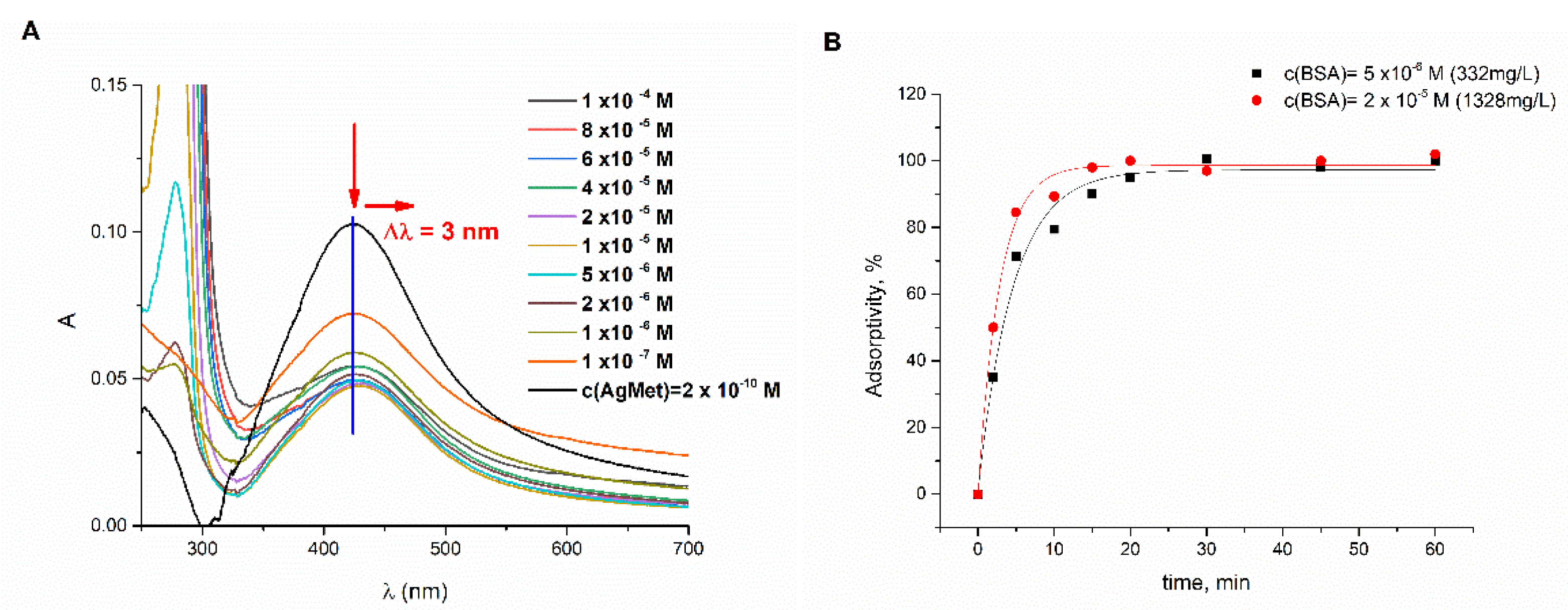
2.1. UV/Vis Spectrophotometric Analysis of the Interaction of BSA–L-Met-Coated AgNPs under Physiological-like Conditions
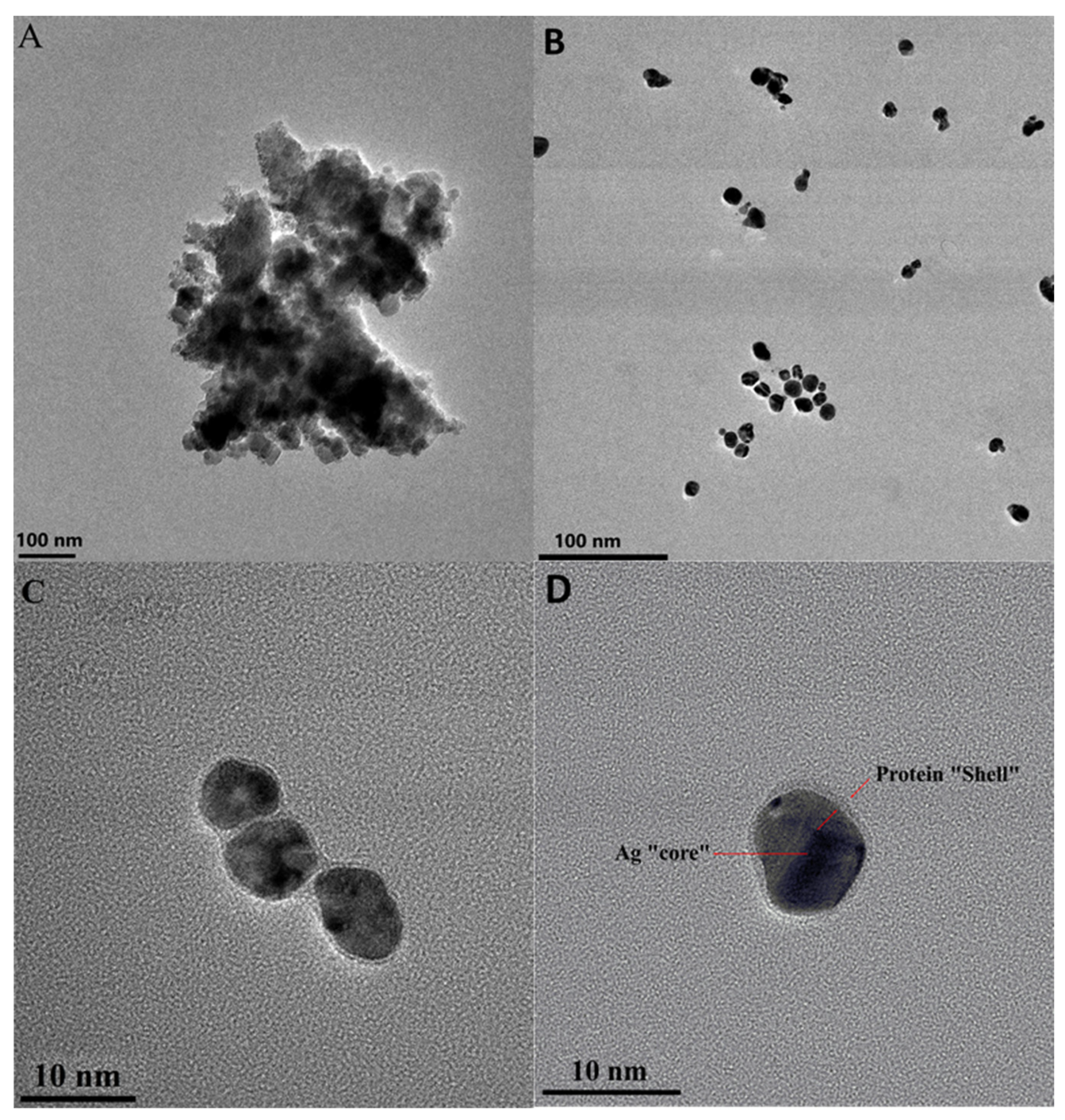
2.2. Characterization of Formed BSA/AgMet Conjugates by TEM, Dynamic Light Scattering, and Zeta Potential Measurements
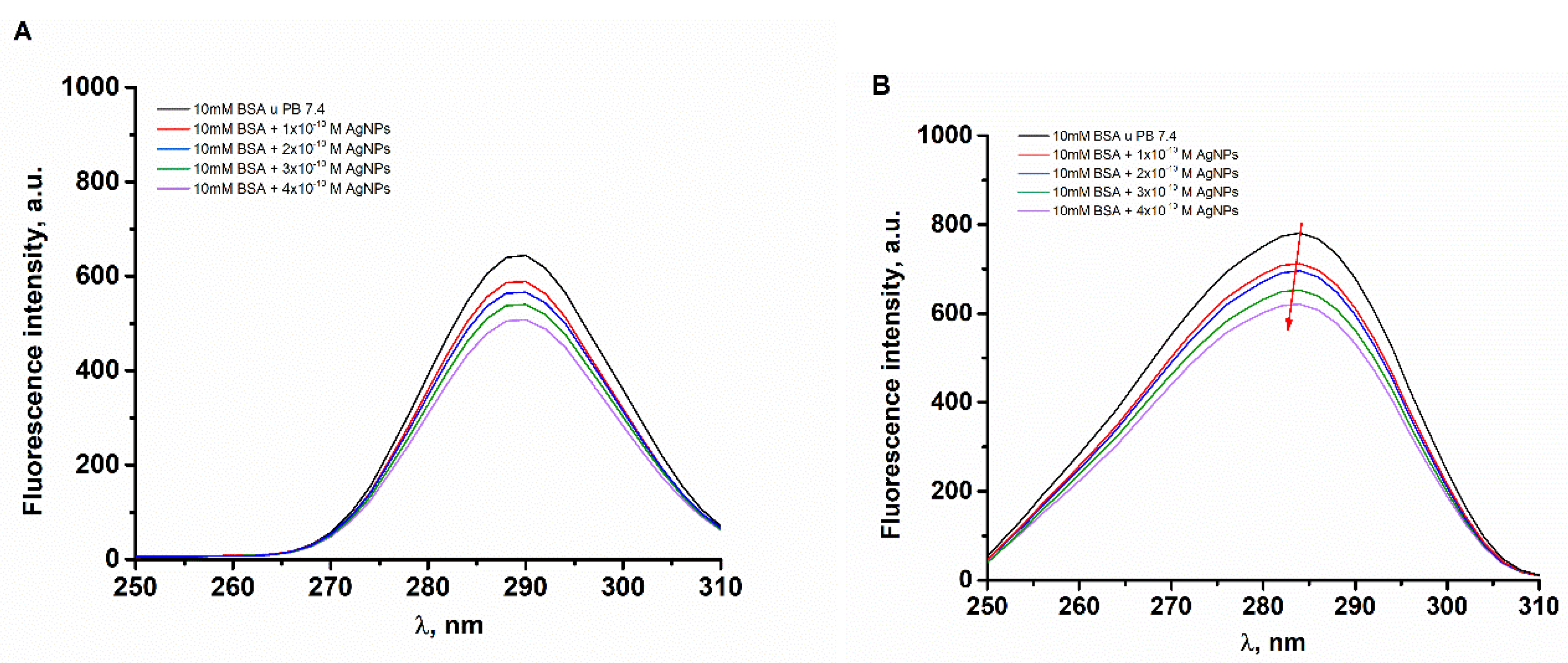
2.3. Quenching of Tryptophan Fluorescence of BSA by MET-Coated AgNPs
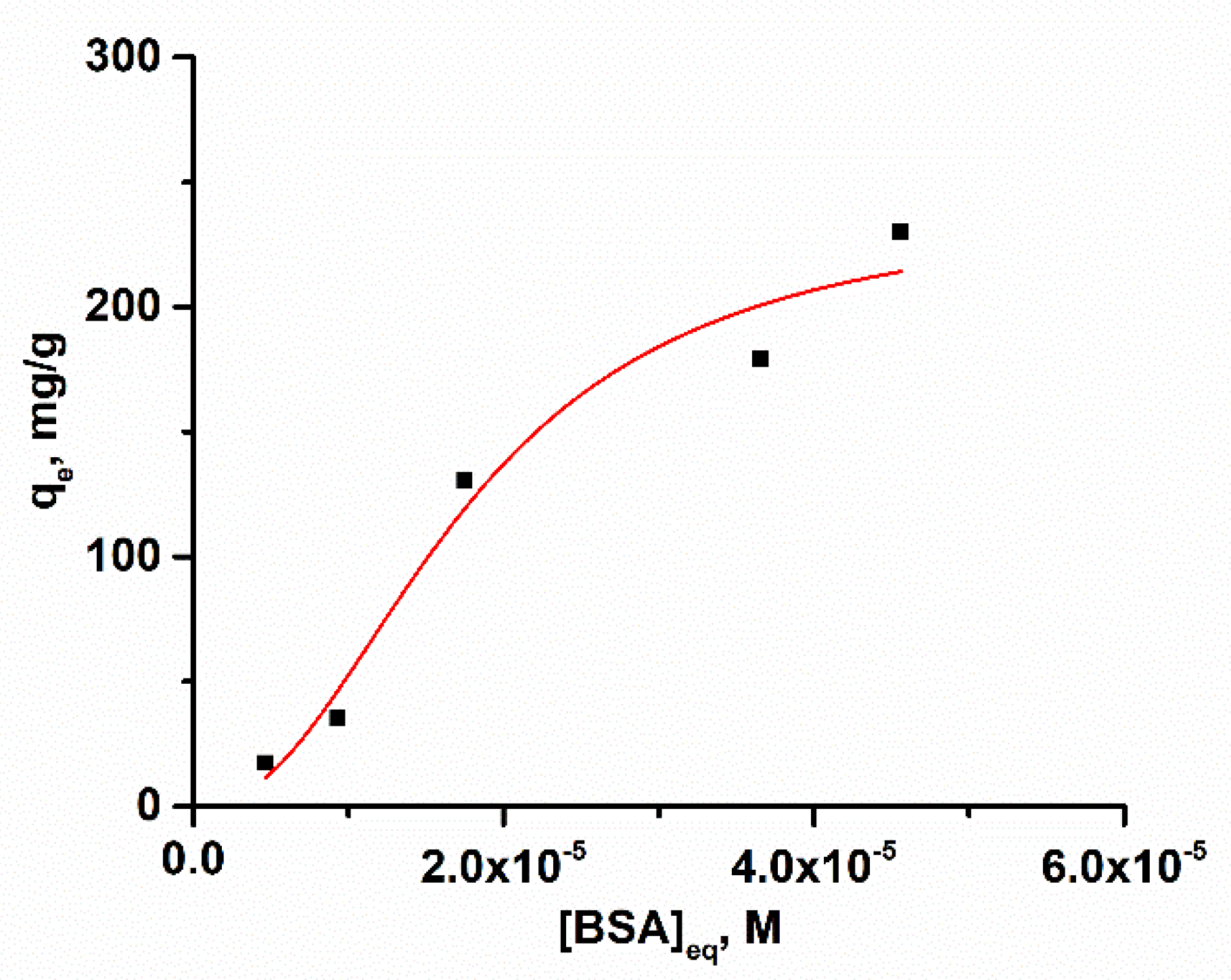
2.4. Adsorption Isotherm and Kinetic Analysis
3. Materials and Methods
3.1. Synthesis of Silver Nanoparticles
3.2. Batch Mode Adsorption Studies
3.3. UV/Vis Measurements
3.4. Transmission Electron Microscopy
3.5. Dynamic Light Scattering and Zeta Potential Measurements
3.6. Fluorescence Measurements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- García-Álvarez, R.; Vallet-Regí, M. Hard and Soft Protein Corona of Nanomaterials: Analysis and Relevance. Nanomaterials 2021, 11, 888. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef]
- Pisani, C.; Gaillard, J.-C.; Odorico, M.; Nyalosaso, J.L.; Charnay, C.; Guari, Y.; Chopineau, J.; Devoisselle, J.-M.; Armengaud, J.; Prat, O. The timeline of corona formation around silica nanocarriers highlights the role of the protein interactome. Nanoscale 2017, 9, 1840–1851. [Google Scholar] [CrossRef]
- Espeche Turbay, M.B.; Rey, V.; Dorado, R.D.; Sosa, M.C.; Borsarelli, C.D. Silver nanoparticle-protein interactions and the role of lysozyme as an antagonistic antibacterial agent. Colloids Surf. B 2021, 208, 112030. [Google Scholar] [CrossRef] [PubMed]
- Shannahan, J.H.; Lai, X.; Ke, P.C.; Podila, R.; Brown, J.M.; Witzmann, F.A. Silver Nanoparticle Protein Corona Composition in Cell Culture Media. PLoS ONE 2013, 8, e74001. [Google Scholar] [CrossRef]
- Winzen, S.; Schoettler, S.; Baier, G.; Rosenauer, C.; Mailaender, V.; Landfester, K.; Mohr, K. Complementary analysis of the hard and soft protein corona: Sample preparation critically effects corona composition. Nanoscale 2015, 7, 2992–3001. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Chan, W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef]
- Mohammad-Beigi, H.; Hayashi, Y.; Zeuthen, C.M.; Eskandari, H.; Scavenius, C.; Juul-Madsen, K.; Vorup-Jensen, T.; Enghild, J.J.; Sutherland, D.S. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 2020, 11, 4535. [Google Scholar] [CrossRef] [PubMed]
- Burdușel, A.-C.; Gherasim, O.; Grumezescu, A.M.; Mogoantă, L.; Ficai, A.; Andronescu, E. Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview. Nanomaterials 2018, 8, 681. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Korbekandi, H.; Mirmohammadi, S.V.; Zolfaghari, B. Synthesis of silver nanoparticles: Chemical, physical and biological methods. Res. Pharm. Sci. 2014, 9, 385–406. [Google Scholar] [PubMed]
- Tai, J.T.; Lai, C.S.; Ho, H.C.; Yeh, Y.S.; Wang, H.F.; Ho, R.M.; Tsai, D.H. Protein-silver nanoparticle interactions to colloidal stability in acidic environments. Langmuir 2014, 30, 12755–12764. [Google Scholar] [CrossRef]
- Boehmler, D.J.; O’Dell, Z.J.; Chung, C.; Riley, K.R. Bovine Serum Albumin Enhances Silver Nanoparticle Dissolution Kinetics in a Size- and Concentration-Dependent Manner. Langmuir 2020, 36, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.C.; Qian, H.; Gies, V.; Yang, L. Human serum albumin stabilizes aqueous silver nanoparticle suspensions and inhibits particle uptake by cells. Environ. Sci. Nano 2018, 5, 863–867. [Google Scholar] [CrossRef]
- Laban, B.; Ralević, U.; Petrović, S.; Leskovac, A.; Vasić-Anićijević, D.; Marković, M.; Vasić, V. Green synthesis and characterization of nontoxic L-methionine capped silver and gold nanoparticles. J. Inorg. Biochem. 2020, 204, 110958. [Google Scholar] [CrossRef]
- Wang, G.; Lu, Y.; Hou, H.; Liu, Y. Probing the binding behavior and kinetics of silver nanoparticles with bovine serum albumin. RSC Adv. 2017, 7, 9393–9401. [Google Scholar] [CrossRef]
- Yoo, E.J.; Li, T.; Park, H.G.; Chang, Y.K. Size-dependent flocculation behavior of colloidal Au nanoparticles modified with various biomolecules. Ultramicroscopy 2008, 108, 1273–1277. [Google Scholar] [CrossRef]
- White, P.; Hjortkjaer, J. Preparation and characterisation of a stable silver colloid for SER(R)S spectroscopy. J. Raman Spectrosc. 2014, 45, 32–40. [Google Scholar] [CrossRef]
- Afshinnia, K.; Baalousha, M. Effect of phosphate buffer on aggregation kinetics of citrate-coated silver nanoparticles induced by monovalent and divalent electrolytes. Sci. Total Environ. 2017, 581–582, 268–276. [Google Scholar] [CrossRef]
- Woods, K.E.; Perera, Y.R.; Davidson, M.B.; Wilks, C.A.; Yadav, D.K.; Fitzkee, N.C. Understanding Protein Structure Deformation on the Surface of Gold Nanoparticles of Varying Size. J. Phys. Chem. C 2016, 120, 27944–27953. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Mulenos, M.R.; Steele, L.R.; Sayes, C.M. Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics. Appl. Sci. 2018, 8, 2669. [Google Scholar] [CrossRef]
- Kokkinopoulou, M.; Simon, J.; Landfester, K.; Mailänder, V.; Lieberwirth, I. Visualization of the protein corona: Towards a biomolecular understanding of nanoparticle-cell-interactions. Nanoscale 2017, 9, 8858–8870. [Google Scholar] [CrossRef] [PubMed]
- Eby, D.M.; Schaeublin, N.M.; Farrington, K.E.; Hussain, S.M.; Johnson, G.R. Lysozyme Catalyzes the Formation of Antimicrobial Silver Nanoparticles. ACS Nano 2009, 3, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Mukherjee, S. Reversibility in protein folding: Effect of β-cyclodextrin on bovine serum albumin unfolded by sodium dodecyl sulphate. Phys. Chem. Chem. Phys. 2013, 15, 9375–9383. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston, MA, USA, 2016; pp. 529–575. [Google Scholar]
- Mariam, J.; Dongre, P.M.; Kothari, D.C. Study of Interaction of Silver Nanoparticles with Bovine Serum Albumin Using Fluorescence Spectroscopy. J. Fluoresc. 2011, 21, 2193. [Google Scholar] [CrossRef]
- Ravindran, A.; Singh, A.; Raichur, A.M.; Chandrasekaran, N.; Mukherjee, A. Studies on interaction of colloidal Ag nanoparticles with Bovine Serum Albumin (BSA). Colloids Surf. B 2010, 76, 32–37. [Google Scholar] [CrossRef]
- Waghmare, M.; Khade, B.; Chaudhari, P.; Dongre, P. Multiple layer formation of bovine serum albumin on silver nanoparticles revealed by dynamic light scattering and spectroscopic techniques. J. Nanopart. Res. 2018, 20, 185. [Google Scholar] [CrossRef]
- Jiang, S.; Liu, H.-Z.; Cai, W.-L.; Bai, A.-M.; Ouyang, Y.; Hu, Y.-J. Quasi-spherical silver nanoparticles with high dispersity and uniform sizes: Preparation, characterization and bioactivity in their interaction with bovine serum albumin. Luminescence 2016, 31, 1146–1151. [Google Scholar] [CrossRef]
- Al-Thabaiti, N.S.; Malik, M.A.; Khan, Z. Protein interactions with silver nanoparticles: Green synthesis, and biophysical approach. Int. J. Biol. Macromol. 2017, 95, 421–428. [Google Scholar] [CrossRef]
- Wang, G.; Liu, X.; Yan, C.; Bai, G.; Lu, Y. Probing the binding of trypsin to glutathione-stabilized gold nanoparticles in aqueous solution. Colloids Surf. B 2015, 135, 261–266. [Google Scholar] [CrossRef]
- Mozaffari Majd, M.; Kordzadeh-Kermani, V.; Ghalandari, V.; Askari, A.; Sillanpää, M. Adsorption isotherm models: A comprehensive and systematic review (2010–2020). Sci. Total Environ. 2022, 812, 151334. [Google Scholar] [CrossRef]
- Dasgupta, N.; Ranjan, S.; Patra, D.; Srivastava, P.; Kumar, A.; Ramalingam, C. Bovine serum albumin interacts with silver nanoparticles with a “side-on” or “end on” conformation. Chem. Biol. Interact. 2016, 253, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.S.; Tripathy, D.R.; Chatterjee, A.; Dasgupta, S. A spectroscopic study of the interaction of the antioxidant naringin with bovine serum albumin. J. Biophys. Chem. 2010, 1, 141–152. [Google Scholar] [CrossRef]

| dsr, nm | ζ, mV | Mobility, μm cm/Vs | Conductivity, mS/cm | PDI Index | |
|---|---|---|---|---|---|
| BSA | 7.93 ± 0.09 | −20.7 ± 1.3 | −1.62 ± 0.06 | 1.180 ± 0.095 | 0.10 |
| AgNPs | 57.08 ± 0.15 | −25.8 ± 0.8 | −2.03 ± 0.07 | 0.068 ± 0.001 | 0.89 |
| AgMet/BSA | 68.44 ± 0.20 | −31.1 ± 2.7 | −2.44 ± 0.21 | 2.580 ± 0.168 | 0.25 |
| Temperature (K) | 300 | 310 | 315 |
|---|---|---|---|
| KSV (M) | (9.28 ± 0.61) × 108 | (8.79 ± 0.81) × 108 | (8.46 ± 0.62) × 108 |
| kq (M−1 s−1) | (8.7 ± 0.6) × 1016 | (8.9 ± 0.8) × 1016 | (9.3 ± 0.6) × 1016 |
| K (M−1) | (5.13 ± 0.20) × 104 | (1.17 ± 0.10) × 105 | (3.80 ± 0.21) × 105 |
| n | 0.55 ± 0.06 | 0.59 ± 0.08 | 0.65 ± 0.03 |
| ΔH° (kJ mol−1) | 98.64 | ||
| ΔS° (JK−1 mol−1) | 418.11 | ||
| ΔG° (kJ mol−1) | −26.75 | −30.93 | −33.03 |
| Pseudo-first-order model | k1, min−1 | 0.0734 ± 0.0094 |
| qe, mgg−1 | 1023 ± 25 | |
| R2 | 0.2598 | |
| Pseudo-second-order model | k2, min−1 | (1.95 ± 0.01) × 10−4 |
| qe, mgg−1 | 250 ± 4 | |
| R2 | 0.9999 | |
| Intraparticle diffusion model | kI, min−1 | 0.0019 ± 0.0006 |
| I, mgg−1 | 303 ± 19 | |
| R2 | 0.9496 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bondžić, A.M.; Jovanović, D.; Arsenijević, N.; Laban, B.; Lazarević Pašti, T.; Klekotka, U.; Bondžić, B.P. “Soft Protein Corona” as the Stabilizer of the Methionine-Coated Silver Nanoparticles in the Physiological Environment: Insights into the Mechanism of the Interaction. Int. J. Mol. Sci. 2022, 23, 8985. https://doi.org/10.3390/ijms23168985
Bondžić AM, Jovanović D, Arsenijević N, Laban B, Lazarević Pašti T, Klekotka U, Bondžić BP. “Soft Protein Corona” as the Stabilizer of the Methionine-Coated Silver Nanoparticles in the Physiological Environment: Insights into the Mechanism of the Interaction. International Journal of Molecular Sciences. 2022; 23(16):8985. https://doi.org/10.3390/ijms23168985
Chicago/Turabian StyleBondžić, Aleksandra M., Dunja Jovanović, Nevena Arsenijević, Bojana Laban, Tamara Lazarević Pašti, Urszula Klekotka, and Bojan P. Bondžić. 2022. "“Soft Protein Corona” as the Stabilizer of the Methionine-Coated Silver Nanoparticles in the Physiological Environment: Insights into the Mechanism of the Interaction" International Journal of Molecular Sciences 23, no. 16: 8985. https://doi.org/10.3390/ijms23168985
APA StyleBondžić, A. M., Jovanović, D., Arsenijević, N., Laban, B., Lazarević Pašti, T., Klekotka, U., & Bondžić, B. P. (2022). “Soft Protein Corona” as the Stabilizer of the Methionine-Coated Silver Nanoparticles in the Physiological Environment: Insights into the Mechanism of the Interaction. International Journal of Molecular Sciences, 23(16), 8985. https://doi.org/10.3390/ijms23168985







