Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease
Abstract
:1. Introduction
2. Results and Discussion
2.1. Experimental Strategy and Recombinant Expression of Tagged Csy4* Protein Variants
2.2. Functional Characterization of Different Tagged Csy4* Proteins
2.3. Preparation of Nucleoplasmic-Enriched Extracts
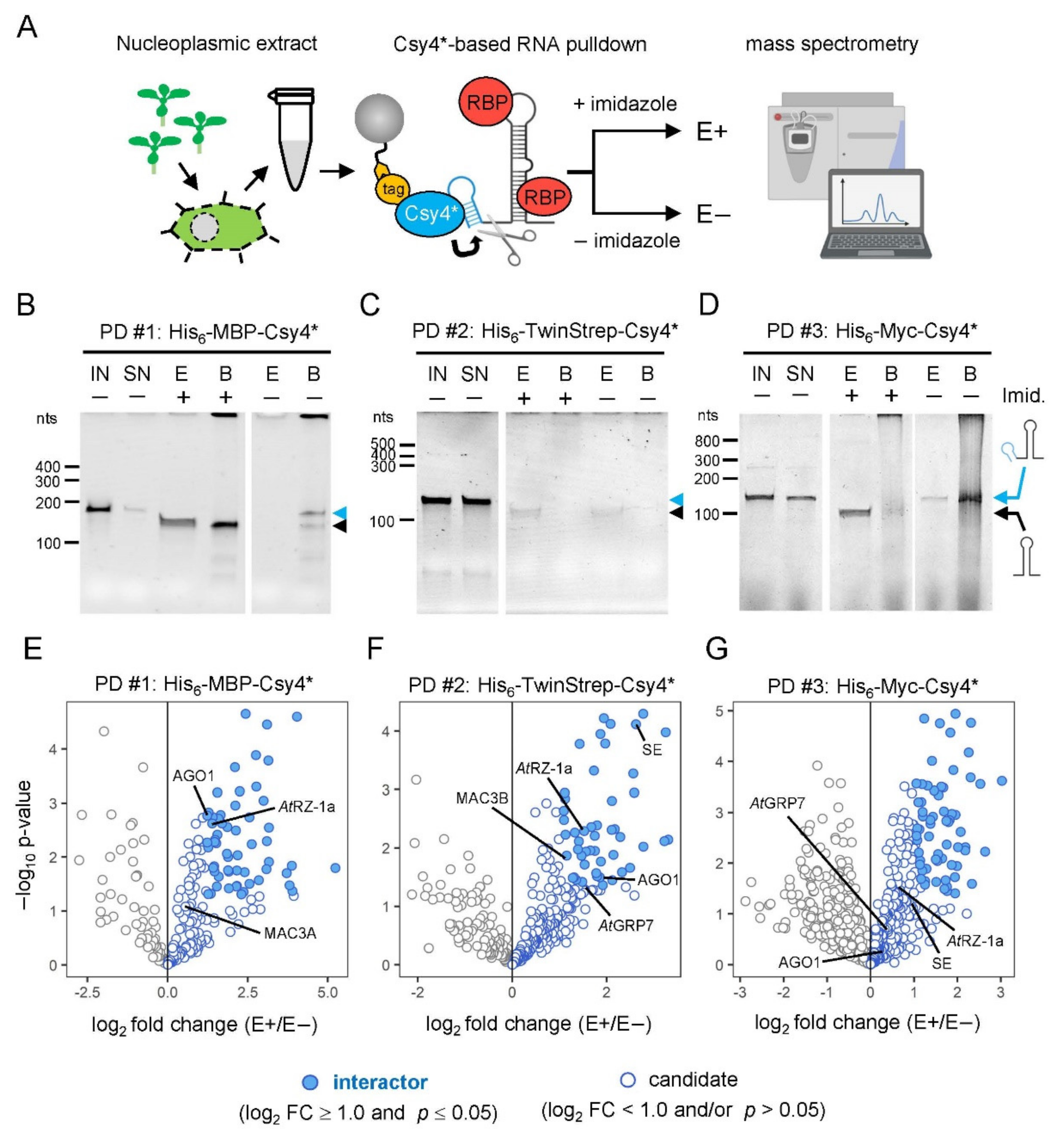
2.4. Proof-of-Principle: Comparative Analysis of Csy4*-Based RNA Pulldown
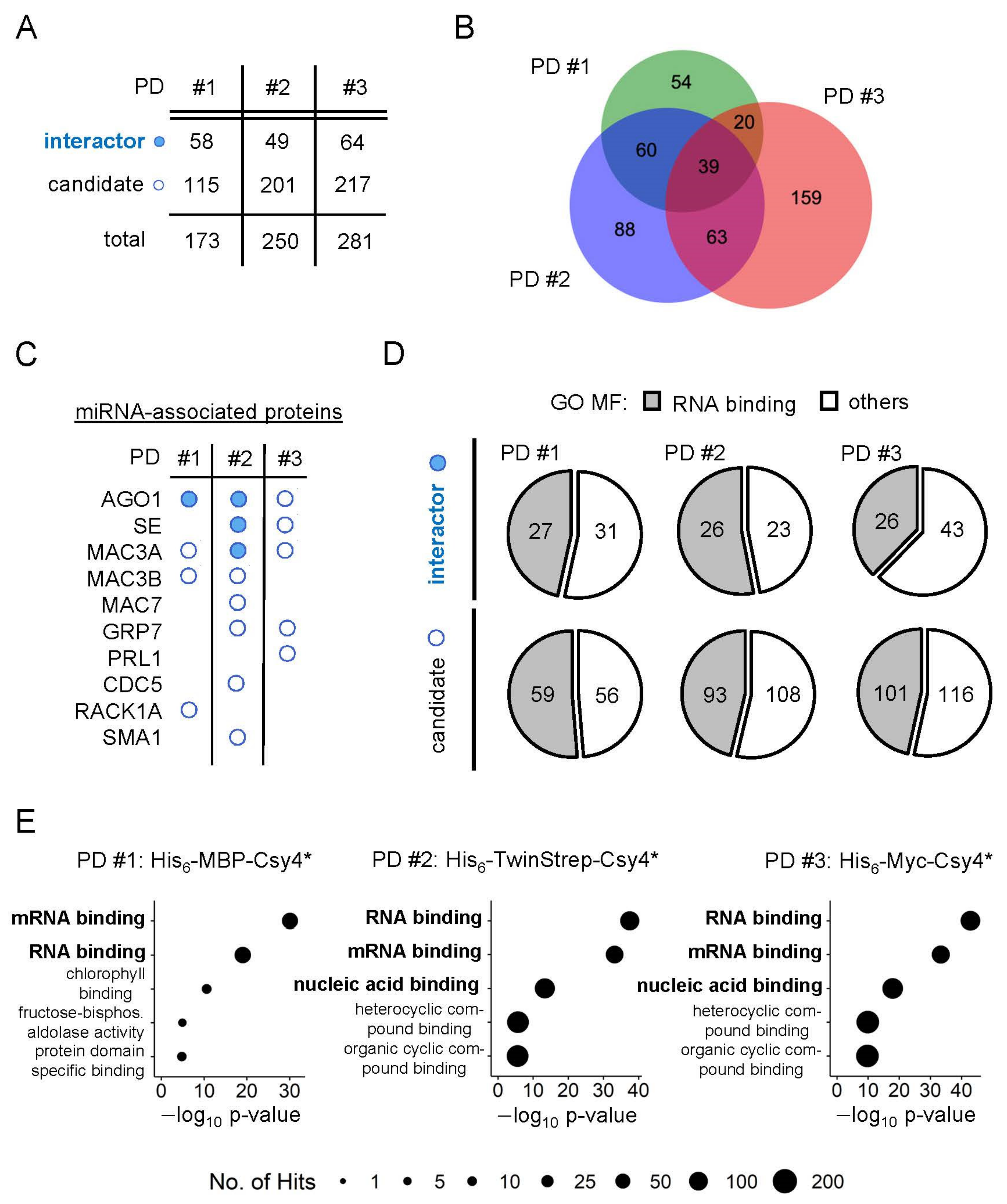
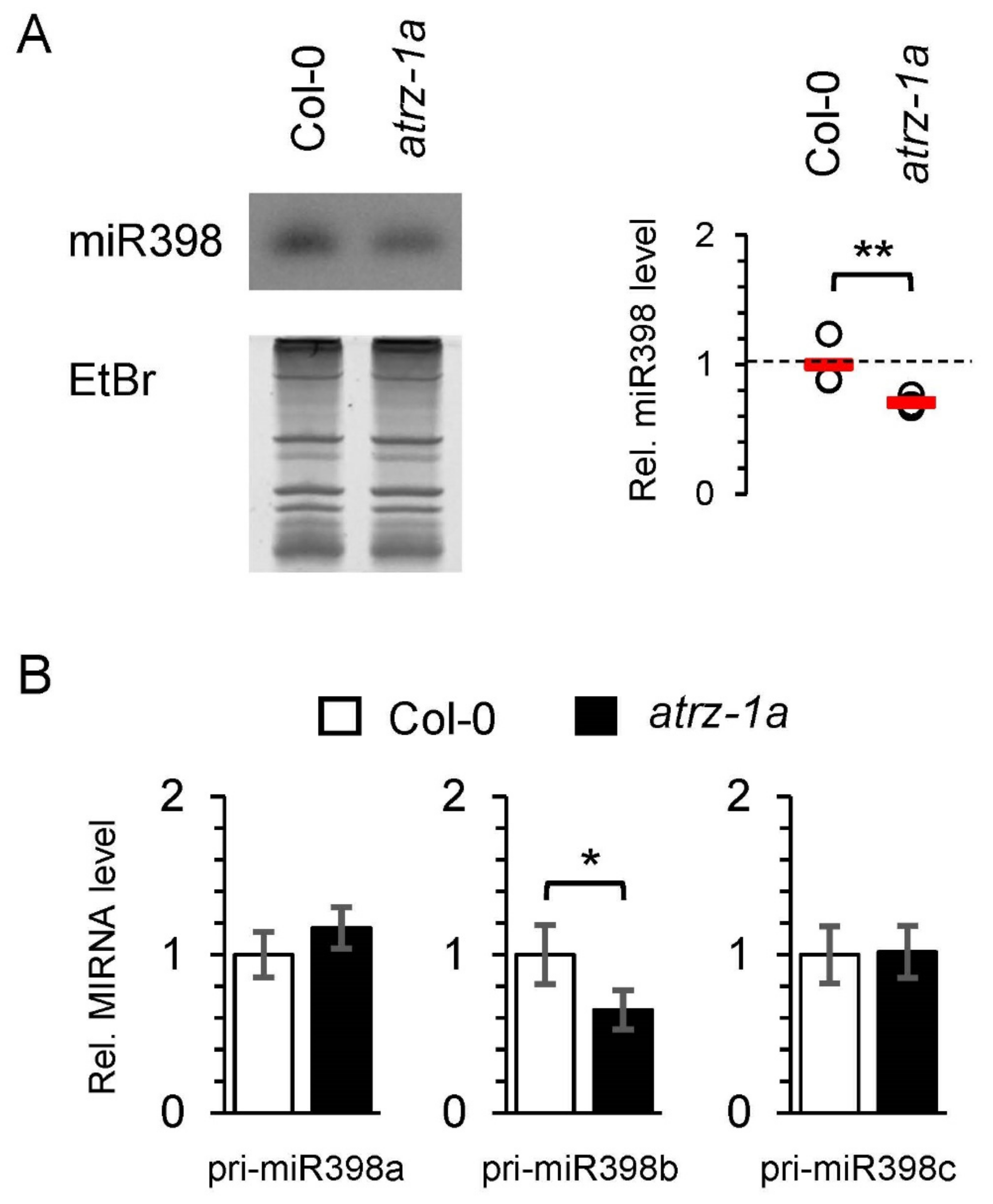
2.5. Identification of New Potential Regulators of miRNA Biogenesis
3. Materials and Methods
3.1. Cloning and In Vitro Transcription
3.2. Cloning and Recombinant Protein Expression of Tagged Csy4* Protein Variants
3.3. Nucleoplasmic Extract Preparation
3.4. Immunoblot Analysis
3.5. Csy4*-Based RNA Pulldowns
3.6. Mass Spectrometry and Data Analysis
3.7. Analysis of GO Terms and Protein Domains
3.8. Plant Material and Growth Conditions
3.9. Detection of Small RNAs by Northern Blotting
3.10. RT-qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogers, K.; Chen, X. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Allen, E.; Fahlgren, N.; Calamar, A.; Givan, S.A.; Carrington, J.C. Expression of Arabidopsis MIRNA Genes. Plant Physiol. 2005, 138, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Gonzalez, N.; Inzé, D.; Dubois, M. Emerging Connections between Small RNAs and Phytohormones. Trends Plant Sci. 2020, 25, 912–929. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of MicroRNAs in Plant Stress Responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant MiRNAs and SiRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Dong, Z.; Han, M.-H.; Fedoroff, N. The RNA-Binding Proteins HYL1 and SE Promote Accurate in Vitro Processing of Pri-MiRNA by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 9970–9975. [Google Scholar] [CrossRef]
- Kurihara, Y.; Takashi, Y.; Watanabe, Y. The Interaction between DCL1 and HYL1 Is Important for Efficient and Precise Processing of Pri-MiRNA in Plant MicroRNA Biogenesis. RNA 2006, 12, 206–212. [Google Scholar] [CrossRef]
- Ren, G.; Xie, M.; Dou, Y.; Zhang, S.; Zhang, C.; Yu, B. Regulation of MiRNA Abundance by RNA Binding Protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 12817–12821. [Google Scholar] [CrossRef]
- Yu, B.; Bi, L.; Zheng, B.; Ji, L.; Chevalier, D.; Agarwal, M.; Ramachandran, V.; Li, W.; Lagrange, T.; Walker, J.C.; et al. The FHA Domain Proteins DAWDLE in Arabidopsis and SNIP1 in Humans Act in Small RNA Biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 10073–10078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dou, Y.; Li, S.; Ren, G.; Chevalier, D.; Zhang, C.; Yu, B. DAWDLE Interacts with DICER-LIKE Proteins to Mediate Small RNA Biogenesis. Plant Physiol. 2018, 177, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Gregory, B. A Link between RNA Metabolism and Silencing Affecting Arabidopsis Development. Dev. Cell 2008, 14, 854–866. [Google Scholar] [CrossRef]
- Kim, S.; Yang, J.-Y.; Xu, J.; Jang, I.-C.; Prigge, M.J.; Chua, N.-H. Two Cap-Binding Proteins CBP20 and CBP80 Are Involved in Processing Primary MicroRNAs. Plant Cell Physiol. 2008, 49, 1634–1644. [Google Scholar] [CrossRef]
- Laubinger, S.; Sachsenberg, T.; Zeller, G.; Busch, W.; Lohmann, J.U.; Rätsch, G.; Weigel, D. Dual Roles of the Nuclear Cap-Binding Complex and SERRATE in Pre-MRNA Splicing and MicroRNA Processing in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 8795–8800. [Google Scholar] [CrossRef] [PubMed]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A.; et al. Arabidopsis MicroRNA Expression Regulation in a Wide Range of Abiotic Stress Responses. Front. Plant Sci 2015, 6, 410. [Google Scholar] [CrossRef] [PubMed]
- Jodder, J. Regulation of Pri-MIRNA Processing: Mechanistic Insights into the MiRNA Homeostasis in Plant. Plant Cell Rep. 2021, 40, 783–798. [Google Scholar] [CrossRef]
- Hafner, M.; Katsantoni, M.; Köster, T.; Marks, J.; Mukherjee, J.; Staiger, D.; Ule, J.; Zavolan, M. CLIP and Complementary Methods. Nat. Rev. Methods Primers 2021, 1, 20. [Google Scholar] [CrossRef]
- Hogg, J.R.; Collins, K. RNA-Based Affinity Purification Reveals 7SK RNPs with Distinct Composition and Regulation. RNA 2007, 13, 868–880. [Google Scholar] [CrossRef]
- Slobodin, B.; Gerst, J.E. A Novel MRNA Affinity Purification Technique for the Identification of Interacting Proteins and Transcripts in Ribonucleoprotein Complexes. RNA 2010, 16, 2277–2290. [Google Scholar] [CrossRef]
- Lee, H.Y.; Haurwitz, R.E.; Apffel, A.; Zhou, K.; Smart, B.; Wenger, C.D.; Laderman, S.; Bruhn, L.; Doudna, J.A. RNA–Protein Analysis Using a Conditional CRISPR Nuclease. Proc. Natl. Acad. Sci. USA 2013, 110, 5416–5421. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Haurwitz, R.E.; Doudna, J.A. Mechanism of Substrate Selection by a Highly Specific CRISPR Endoribonuclease. RNA 2012, 18, 661–672. [Google Scholar] [CrossRef]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science 2010, 329, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant Protein Expression in Escherichia Coli: Advances and Challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Terpe, K. Overview of Tag Protein Fusions: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Rennie, S.; Köster, T.; Porcelli, C.; Lewinski, M.; Staiger, D.; Andersson, R.; Brodersen, P. Principles of MRNA Targeting via the Arabidopsis M6A-Binding Protein ECT2. eLife 2021, 10, e72375. [Google Scholar] [CrossRef]
- Köster, T.; Staiger, D. Plant Individual Nucleotide Resolution Cross-Linking and Immunoprecipitation to Characterize RNA-Protein Complexes. In RNA Tagging: Methods in Molecular Biology; Heinlein, M., Ed.; Springer: New York, NY, USA, 2020; Volume 2166, pp. 255–267. ISBN 978-1-07-160711-4. [Google Scholar]
- Meyer, K.; Köster, T.; Nolte, C.; Weinholdt, C.; Lewinski, M.; Grosse, I.; Staiger, D. Adaptation of ICLIP to Plants Determines the Binding Landscape of the Clock-Regulated RNA-Binding Protein AtGRP7. Genome Biol. 2017, 18, 204. [Google Scholar] [CrossRef]
- Evan, G.I.; Lewis, G.K.; Ramsay, G.; Bishop, J.M. Isolation of Monoclonal Antibodies Specific for Human C-Myc Proto-Oncogene Product. Mol. Cell. Biol. 1985, 5, 3610–3616. [Google Scholar] [CrossRef]
- Korndörfer, I.P.; Skerra, A. Improved Affinity of Engineered Streptavidin for the Strep-Tag II Peptide Is Due to a Fixed Open Conformation of the Lid-like Loop at the Binding Site. Protein Sci. 2002, 11, 883–893. [Google Scholar] [CrossRef]
- Voss, S.; Skerra, A. Mutagenesis of a Flexible Loop in Streptavidin Leads to Higher Affinity for the Strep-Tag II Peptide and Improved Performance in Recombinant Protein Purification. Protein Eng. 1997, 10, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Busso, D.; Delagoutte-Busso, B.; Moras, D. Construction of a Set Gateway-Based Destination Vectors for High-Throughput Cloning and Expression Screening in Escherichia Coli. Anal. Biochem. 2005, 343, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions That Promote Protein Stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of MiR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Song, L.; Axtell, M.J.; Fedoroff, N.V. RNA Secondary Structural Determinants of MiRNA Precursor Processing in Arabidopsis. Curr. Biol. 2010, 20, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Papp, I.; Mette, M.F.; Aufsatz, W.; Daxinger, L.; Schauer, S.E.; Ray, A.; van der Winden, J.; Matzke, M.; Matzke, A.J.M. Evidence for Nuclear Processing of Plant Micro RNA and Short Interfering RNA Precursors. Plant Physiol. 2003, 132, 1382–1390. [Google Scholar] [CrossRef]
- Gonzalo, L.; Tossolini, I.; Gulanicz, T.; Cambiagno, D.A.; Kasprowicz-Maluski, A.; Smolinski, D.J.; Mammarella, M.F.; Ariel, F.D.; Marquardt, S.; Szweykowska-Kulinska, Z.; et al. R-Loops at MicroRNA Encoding Loci Promote Co-Transcriptional Processing of Pri-MiRNAs in Plants. Nat. Plants 2022, 8, 402–418. [Google Scholar] [CrossRef]
- Köster, T.; Meyer, K.; Weinholdt, C.; Smith, L.M.; Lummer, M.; Speth, C.; Grosse, I.; Weigel, D.; Staiger, D. Regulation of Pri-MiRNA Processing by the HnRNP-like Protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014, 42, 9925–9936. [Google Scholar] [CrossRef]
- Schöning, J.C.; Streitner, C.; Page, D.R.; Hennig, S.; Uchida, K.; Wolf, E.; Furuya, M.; Staiger, D. Auto-regulation of the Circadian Slave Oscillator Component AtGRP7 and Regulation of Its Targets Is Impaired by a Single RNA Recognition Motif Point Mutation. Plant J. 2007, 52, 1119–1130. [Google Scholar] [CrossRef]
- Machida, S.; Chen, H.-Y.; Adam Yuan, Y. Molecular Insights into MiRNA Processing by Arabidopsis Thaliana SERRATE. Nucleic Acids Res. 2011, 39, 7828–7836. [Google Scholar] [CrossRef] [PubMed]
- Lobbes, D.; Rallapalli, G.; Schmidt, D.D.; Martin, C.; Clarke, J. SERRATE: A New Player on the Plant MicroRNA Scene. EMBO Rep. 2006, 7, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Z.; Lu, F.; Dong, A.; Huang, H. SERRATE Is a Novel Nuclear Regulator in Primary MicroRNA Processing in Arabidopsis. Plant J. 2006, 47, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, K.; Zhou, B.; Li, M.; Zhang, S.; Zeng, L.; Zhang, C.; Yu, B. MAC3A and MAC3B, Two Core Subunits of the MOS4-Associated Complex, Positively Influence MiRNA Biogenesis. Plant Cell 2018, 30, 481–494. [Google Scholar] [CrossRef]
- Jia, T.; Zhang, B.; You, C.; Zhang, Y.; Zeng, L.; Li, S.; Johnson, K.C.M.; Yu, B.; Li, X.; Chen, X. The Arabidopsis MOS4-Associated Complex Promotes MicroRNA Biogenesis and Precursor Messenger RNA Splicing. Plant Cell 2017, 29, 2626–2643. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Yu, B. PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of MiRNAs and SiRNAs in Arabidopsis. PLoS Genet. 2014, 10, e1004841. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, M.; Ren, G.; Yu, B. CDC5, a DNA Binding Protein, Positively Regulates Posttranscriptional Processing and/or Transcription of Primary MicroRNA Transcripts. Proc. Natl. Acad. Sci. USA 2013, 110, 17588–17593. [Google Scholar] [CrossRef]
- Li, S.; Xu, R.; Li, A.; Liu, K.; Gu, L.; Li, M.; Zhang, H.; Zhang, Y.; Zhuang, S.; Wang, Q.; et al. SMA1, a Homolog of the Splicing Factor Prp28, Has a Multifaceted Role in MiRNA Biogenesis in Arabidopsis. Nucleic Acids Res. 2018, 46, 9148–9159. [Google Scholar] [CrossRef]
- Speth, C.; Willing, E.-M.; Rausch, S.; Schneeberger, K.; Laubinger, S. RACK1 Scaffold Proteins Influence MiRNA Abundance in Arabidopsis. Plant J. 2013, 76, 433–445. [Google Scholar] [CrossRef]
- Jannot, G.; Bajan, S.; Giguère, N.J.; Bouasker, S.; Banville, I.H.; Piquet, S.; Hutvagner, G.; Simard, M.J. The Ribosomal Protein RACK1 Is Required for MicroRNA Function in Both C. Elegans and Humans. EMBO Rep. 2011, 12, 581–586. [Google Scholar] [CrossRef]
- Otsuka, M.; Takata, A.; Yoshikawa, T.; Kojima, K.; Kishikawa, T.; Shibata, C.; Takekawa, M.; Yoshida, H.; Omata, M.; Koike, K. Receptor for Activated Protein Kinase C: Requirement for Efficient MicroRNA Function and Reduced Expression in Hepatocellular Carcinoma. PLoS ONE 2011, 6, e24359. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Spector, D.L. Identification of Nuclear Dicing Bodies Containing Proteins for MicroRNA Biogenesis in Living Arabidopsis Plants. Curr. Biol. 2007, 17, 818–823. [Google Scholar] [CrossRef]
- Bologna, N.G.; Iselin, R.; Abriata, L.A.; Sarazin, A.; Pumplin, N.; Jay, F.; Grentzinger, T.; Dal Peraro, M.; Voinnet, O. Nucleo-Cytosolic Shuttling of ARGONAUTE1 Prompts a Revised Model of the Plant MicroRNA Pathway. Mol. Cell 2018, 69, 709–719.e5. [Google Scholar] [CrossRef] [PubMed]
- Eamens, A.L.; Smith, N.A.; Curtin, S.J.; Wang, M.-B.; Waterhouse, P.M. The Arabidopsis Thaliana Double-Stranded RNA Binding Protein DRB1 Directs Guide Strand Selection from MicroRNA Duplexes. RNA 2009, 15, 2219–2235. [Google Scholar] [CrossRef] [PubMed]
- Bach-Pages, M.; Homma, F.; Kourelis, J.; Kaschani, F.; Mohammed, S.; Kaiser, M.; van der Hoorn, R.A.L.; Castello, A.; Preston, G.M. Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules 2020, 10, 661. [Google Scholar] [CrossRef]
- Marondedze, C.; Thomas, L.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-Binding Protein Repertoire of Arabidopsis Thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.-M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In Planta Determination of the MRNA-Binding Proteome of Arabidopsis Etiolated Seedlings. Plant Cell Online 2016, 28, 2435–2452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Boonen, K.; Ferrari, P.; Schoofs, L.; Janssens, E.; van Noort, V.; Rolland, F.; Geuten, K. UV Crosslinked MRNA-Binding Proteins Captured from Leaf Mesophyll Protoplasts. Plant Methods 2016, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Silverman, I.M.; Li, F.; Gregory, B.D. Genomic Era Analyses of RNA Secondary Structure and RNA-Binding Proteins Reveal Their Significance to Post-Transcriptional Regulation in Plants. Plant Sci. 2013, 205–206, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD Repeat: A Common Architecture for Diverse Functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, S.; Huang, J.; Zheng, C. Genome-Wide Comparative In Silico Analysis of the RNA Helicase Gene Family in Zea Mays and Glycine Max: A Comparison with Arabidopsis and Oryza Sativa. PLoS ONE 2013, 8, e78982. [Google Scholar] [CrossRef]
- Bycroft, M.; Hubbard, T.J.P.; Proctor, M.; Freund, S.M.V.; Murzin, A.G. The Solution Structure of the S1 RNA Binding Domain: A Member of an Ancient Nucleic Acid–Binding Fold. Cell 1997, 88, 235–242. [Google Scholar] [CrossRef]
- Kim, W.Y.; Kim, J.Y.; Jung, H.J.; Oh, S.H.; Han, Y.S.; Kang, H. Comparative Analysis of Arabidopsis Zinc Finger-Containing Glycine-Rich RNA-Binding Proteins during Cold Adaptation. Plant Physiol. Biochem. 2010, 48, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-O.; Kim, J.S.; Kang, H. Cold-Inducible Zinc Finger-Containing Glycine-Rich RNA-Binding Protein Contributes to the Enhancement of Freezing Tolerance in Arabidopsis Thaliana. Plant J. 2005, 42, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and Plant Stress Responses. Physiol. Plant 2011, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-O.; Pan, S.; Jung, C.-H.; Kang, H. A Zinc Finger-Containing Glycine-Rich RNA-Binding Protein, AtRZ-1a, Has a Negative Impact on Seed Germination and Seedling Growth of Arabidopsis Thaliana Under Salt or Drought Stress Conditions. Plant Cell Physiol. 2007, 48, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Umate, P.; Tuteja, R.; Tuteja, N. Genome-Wide Analysis of Helicase Gene Family from Rice and Arabidopsis: A Comparison with Yeast and Human. Plant Mol Biol 2010, 73, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Bologna, N.G.; Chorostecki, U.; Palatnik, J.F. Identification of MicroRNA Processing Determinants by Random Mutagenesis of Arabidopsis MIR172a Precursor. Curr. Biol. 2010, 20, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Wollmann, H.; Schneeberger, K.; Weigel, D. Structure Determinants for Accurate Processing of MiR172a in Arabidopsis Thaliana. Curr. Biol. 2010, 20, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, Z.; Castillo-González, C.; Sun, D.; Li, Y.; Yu, B.; Zhao, B.; Li, P.; Zhang, X. SWI2/SNF2 ATPase CHR2 Remodels Pri-MiRNAs via Serrate to Impede MiRNA Production. Nature 2018, 557, 516–521. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.-Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-Binding Proteins Contribute to Small RNA Loading in Plant Extracellular Vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants Send Small RNAs in Extracellular Vesicles to Fungal Pathogen to Silence Virulence Genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-L.; Chen, W.-Q.; Hou, Y.; Gong, H.-Q.; Sun, J.; Wang, Z.; Zhao, H.; Cao, X.; Song, X.-F.; Liu, C.-M. DEAD-BOX RNA HELICASE 27 Regulates MicroRNA Biogenesis, Zygote Division, and Stem Cell Homeostasis. Plant Cell 2021, 33, 66–84. [Google Scholar] [CrossRef] [PubMed]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. MiRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef]
- Streitner, C.; Köster, T.; Simpson, C.G.; Shaw, P.; Danisman, S.; Brown, J.W.S.; Staiger, D. An HnRNP-like RNA-Binding Protein Affects Alternative Splicing by in Vivo Interaction with Transcripts in Arabidopsis Thaliana. Nucleic Acids Res. 2012, 40, 11240–11255. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Koester, T.; Staiger, D. Pre-MRNA Splicing in Plants: In Vivo Functions of RNA-Binding Proteins Implicated in the Splicing Process. Biomolecules 2015, 5, 1717–1740. [Google Scholar] [CrossRef] [PubMed]
- Bielewicz, D.; Kalak, M.; Kalyna, M.; Windels, D.; Barta, A.; Vazquez, F.; Szweykowska-Kulinska, Z.; Jarmolowski, A. Introns of Plant Pri-MiRNAs Enhance MiRNA Biogenesis. EMBO Rep. 2013, 14, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Speth, C.; Laubinger, S.; Voinnet, O. Enhanced MicroRNA Accumulation through Stemloop-Adjacent Introns. EMBO Rep. 2013, 14, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, O.A.; Calder, G.; Pendle, A.F.; Kim, S.H.; Lewandowska, D.; Simpson, C.G.; Jones, I.M.; Brown, J.W.S.; Shaw, P.J. Dynamic Behavior of Arabidopsis EIF4A-III, Putative Core Protein of Exon Junction Complex: Fast Relocation to Nucleolus and Splicing Speckles under Hypoxia. Plant Cell 2009, 21, 1592–1606. [Google Scholar] [CrossRef]
- Pascuan, C.; Frare, R.; Alleva, K.; Ayub, N.D.; Soto, G. MRNA Biogenesis-Related Helicase EIF4AIII from Arabidopsis Thaliana Is an Important Factor for Abiotic Stress Adaptation. Plant Cell Rep. 2016, 35, 1205–1208. [Google Scholar] [CrossRef]
- Capovilla, G.; Delhomme, N.; Collani, S.; Shutava, I.; Bezrukov, I.; Symeonidi, E.; de Francisco Amorim, M.; Laubinger, S.; Schmid, M. PORCUPINE Regulates Development in Response to Temperature through Alternative Splicing. Nat. Plants 2018, 4, 534–539. [Google Scholar] [CrossRef]
- Huertas, R.; Catalá, R.; Jiménez-Gómez, J.M.; Castellano, M.M.; Crevillén, P.; Piñeiro, M.; Jarillo, J.A.; Salinas, J. Arabidopsis SME1 Regulates Plant Development and Response to Abiotic Stress by Determining Spliceosome Activity Specificity. Plant Cell 2019, 31, 537–554. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Wang, D.; Kim, C.-S.; Yadegari, R.; Larkins, B.A. Plant SMU-1 and SMU-2 Homologues Regulate Pre-MRNA Splicing and Multiple Aspects of Development. Plant Physiol. 2009, 151, 1498–1512. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Lin, W.-D.; Fu, J.L.; Matzke, A.J.M.; Matzke, M. A Genetic Screen Implicates a CWC16/Yju2/CCDC130 Protein and SMU1 in Alternative Splicing in Arabidopsis Thaliana. RNA 2017, 23, 1068–1079. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C. Identification and Characterization of Fructose 1,6-Bisphosphate Aldolase Genes in Arabidopsis Reveal a Gene Family with Diverse Responses to Abiotic Stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.-L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins That Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef]
- Barden, S.; Lange, S.; Tegtmeyer, N.; Conradi, J.; Sewald, N.; Backert, S.; Niemann, H.H. A Helical RGD Motif Promoting Cell Adhesion: Crystal Structures of the Helicobacter Pylori Type IV Secretion System Pilus Protein CagL. Structure 2013, 21, 1931–1941. [Google Scholar] [CrossRef]
- Staiger, D.; Kaulen, H.; Schell, J. A CACGTG Motif of the Antirrhinum Majus Chalcone Synthase Promoter Is Recognized by an Evolutionarily Conserved Nuclear Protein. Proc. Natl. Acad. Sci. USA 1989, 86, 6930–6934. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Krishnakumar, V.; Contrino, S.; Cheng, C.-Y.; Belyaeva, I.; Ferlanti, E.S.; Miller, J.R.; Vaughn, M.W.; Micklem, G.; Town, C.D.; Chan, A.P. ThaleMine: A Warehouse for Arabidopsis Data Integration and Discovery. Plant Cell Physiol. 2017, 58, e4. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Apel, K.; Trepp, G. The Atger3 Promoter Confers Circadian Clock-Regulated Transcription with Peak Expression at the Beginning of the Night. Plant Mol. Biol. 1999, 40, 873–882. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 2 March 2022).
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.-K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist LncRNA Interacts Directly with SHARP to Silence Transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Rogell, B.M.; Fischer, B.; Rettel, M.; Krijgsveld, J.; Castello, A.; Hentze, M.W. Specific RNP Capture with Antisense LNA/DNA Mixmers. RNA 2017, 23, 1290–1302. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüders, J.; Winkel, A.R.; Reichel, M.; Bitterer, V.W.; Scheibe, M.; Widmann, C.; Butter, F.; Köster, T. Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease. Int. J. Mol. Sci. 2022, 23, 8961. https://doi.org/10.3390/ijms23168961
Lüders J, Winkel AR, Reichel M, Bitterer VW, Scheibe M, Widmann C, Butter F, Köster T. Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease. International Journal of Molecular Sciences. 2022; 23(16):8961. https://doi.org/10.3390/ijms23168961
Chicago/Turabian StyleLüders, Janina, Andreas R. Winkel, Marlene Reichel, Valentin W. Bitterer, Marion Scheibe, Christiane Widmann, Falk Butter, and Tino Köster. 2022. "Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease" International Journal of Molecular Sciences 23, no. 16: 8961. https://doi.org/10.3390/ijms23168961
APA StyleLüders, J., Winkel, A. R., Reichel, M., Bitterer, V. W., Scheibe, M., Widmann, C., Butter, F., & Köster, T. (2022). Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease. International Journal of Molecular Sciences, 23(16), 8961. https://doi.org/10.3390/ijms23168961





