New Insights into Neutrophil Extracellular Trap (NETs) Formation from Porcine Neutrophils in Response to Bacterial Infections
Abstract
:1. Introduction
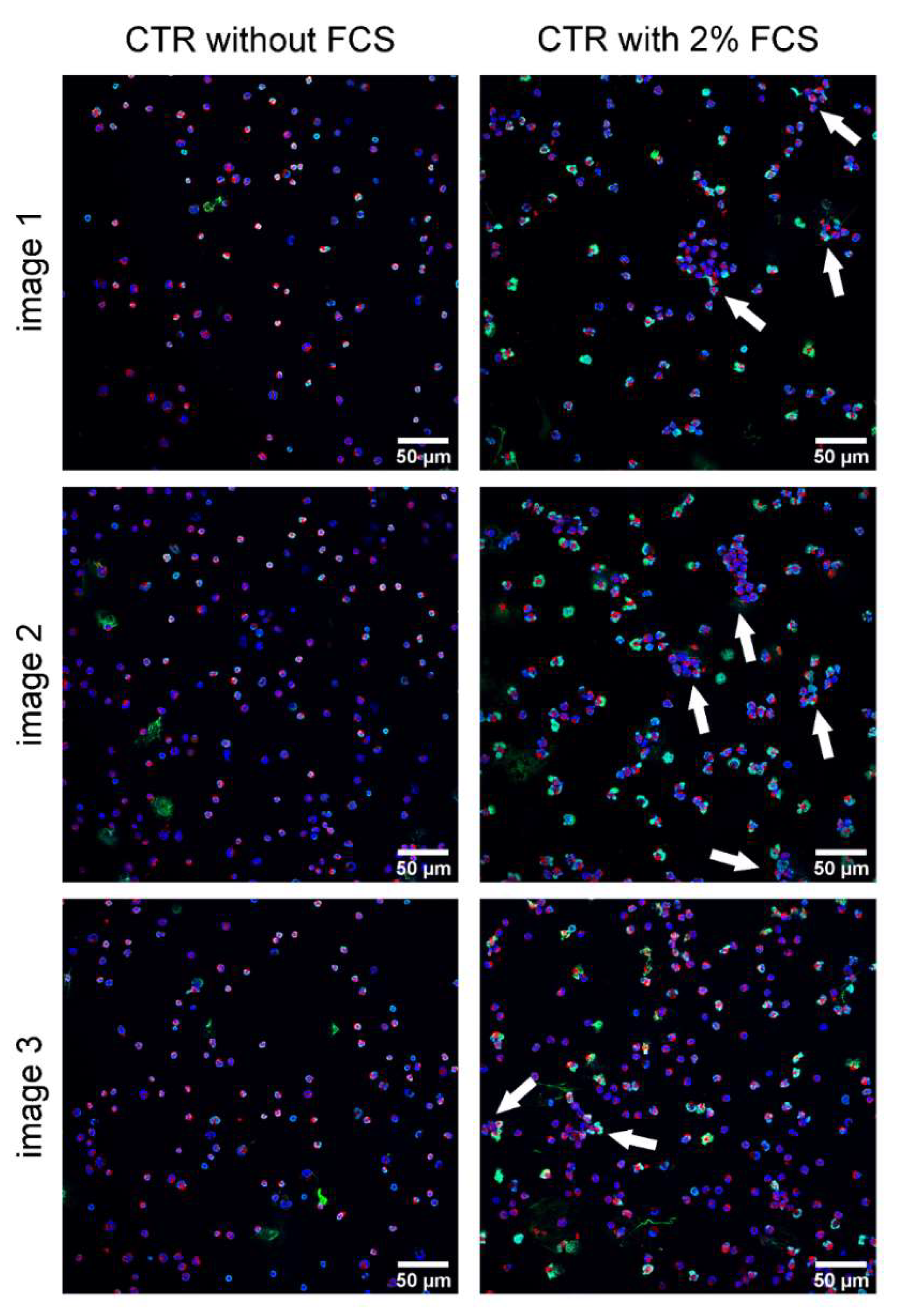
2. Results
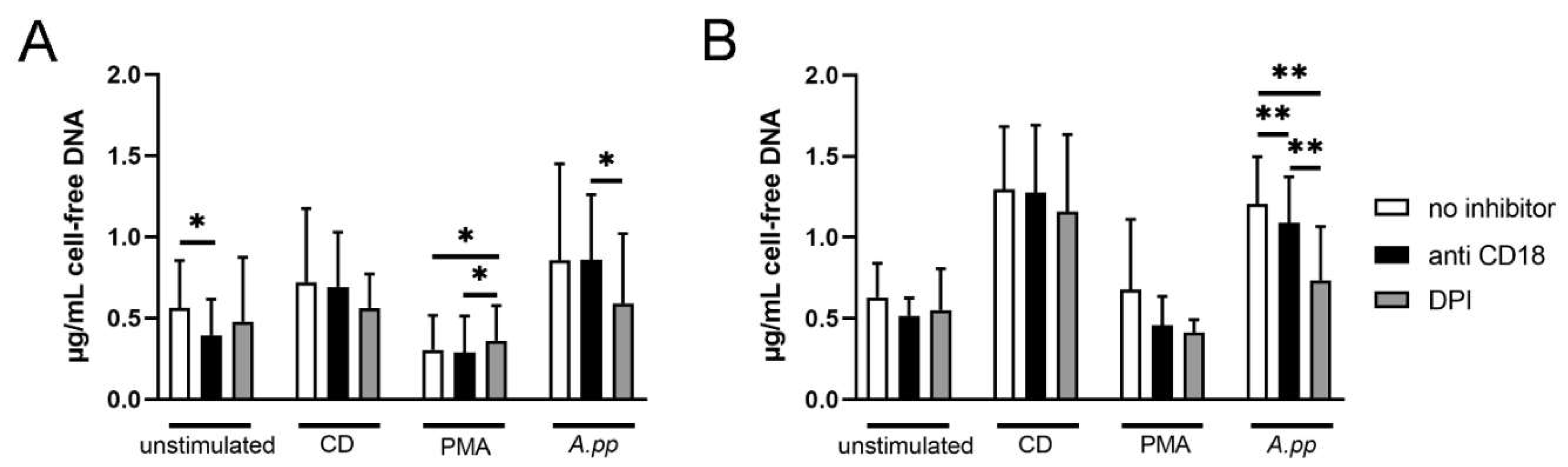
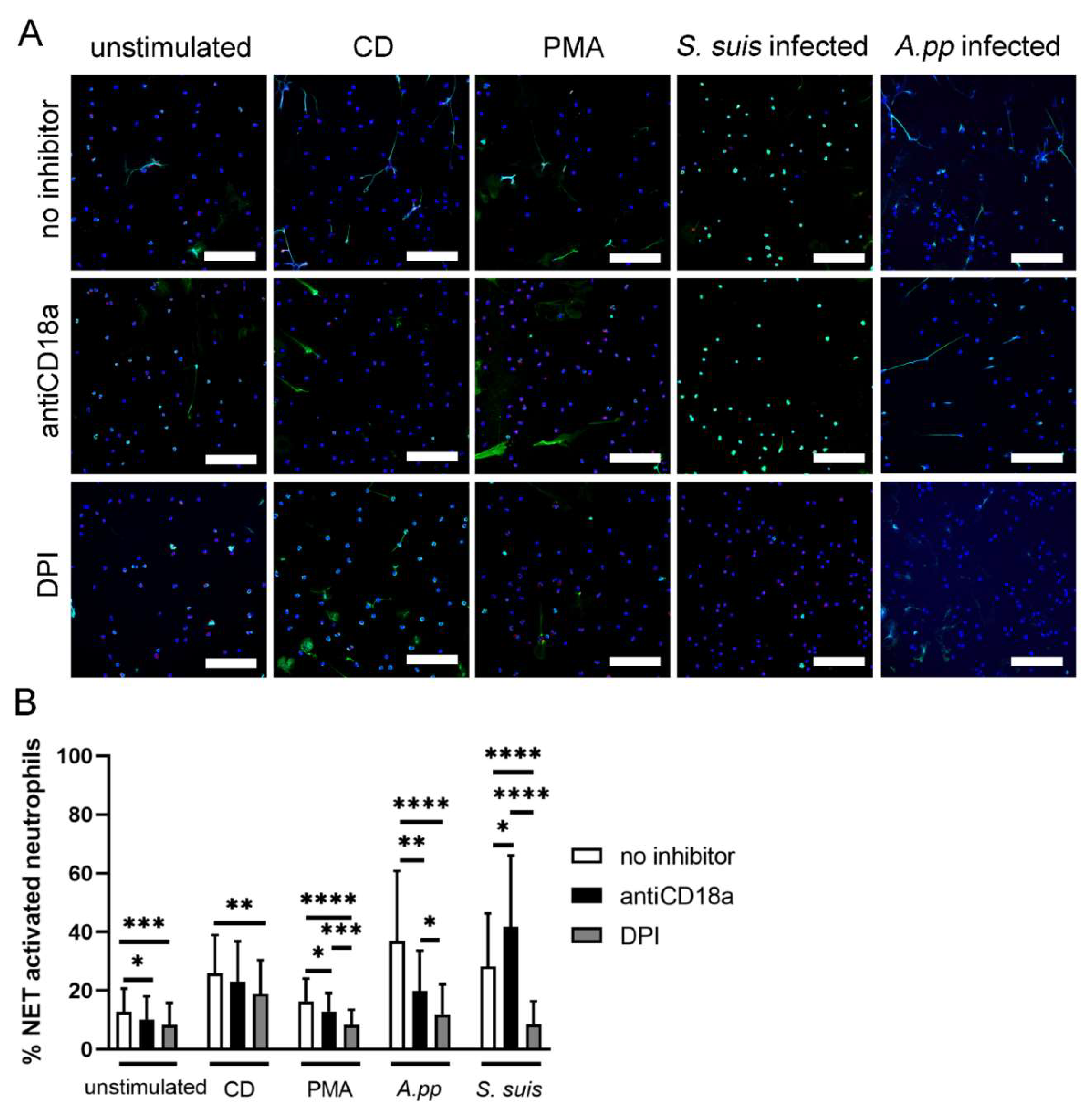
- A.pp and S. suis induce NADPH oxidase-dependent NETs and A.pp also via CD18 activation
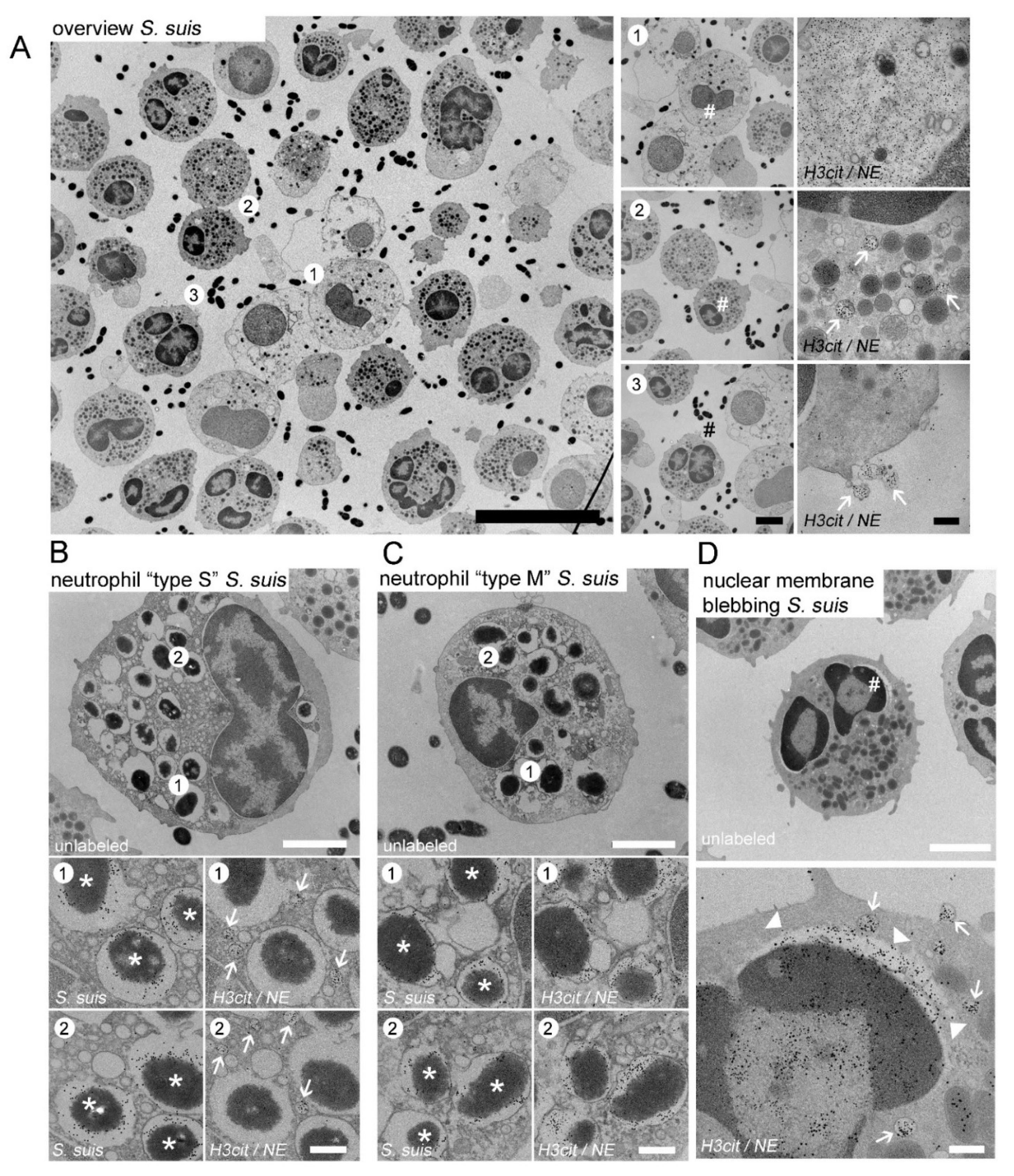
- A.pp and S. suis infected porcine neutrophils release vesicular NETs and undergo phagocytosis
3. Discussion
4. Materials and Methods
4.1. Collection of Porcine Blood
4.2. Growth Conditions of A.pp and S. suis
4.3. Neutrophil Isolation
4.4. NET Inhibition Assay in Presence of A.pp and S. suis
4.5. NET Staining
4.6. Staining of Bound Anti CD18a on Neutrophils
4.7. NET Quantification in Immunofluorescence Images
4.8. Electron Microscopy of Neutrophils Infected with A.pp and S. suis
4.9. NET Quantification in EM Images
- NET release from the nuclei: loss of intact cytoplasmatic structure with release of nuclear DNA was counted as positive. The percentage of positive cells was calculated in relation to all of the observed cells investigated;
- NET surface coverage: the area covered with NET structures that was found in the extracellular environment of neutrophils was measured. The percentage indicated the fraction of the area covered by NETs in relation to the total area of the respective field;
- Nuclear vesicles per neutrophil: the number of the intracellular nuclear vesicles (H3cit and NE positive) were counted per neutrophil in the randomly selected fields. The analyzed neutrophils show an intact cytoplasmic structure with nucleus, double nuclear membrane, and intact granules.
4.10. Measurement of Cell-Free DNA in Supernatants of A.pp NET Assays
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Summers, C.; Rankin, S.M.; Condliffe, A.M.; Singh, N.; Peters, A.M.; Chilvers, E.R. Neutrophil kinetics in health and disease. Trends Immunol. 2010, 31, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Malech, H.L.; DeLeo, F.R.; Quinn, M.T. The Role of Neutrophils in the Immune System: An Overview. Neutrophil 2020, 2087, 3–10. [Google Scholar]
- Rosales, C. Neutrophils at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 2020, 108, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.-S.; Ji, A.; Ji, X.-Y.; Li, Y.-Z. Neutrophils and Immunity: From Bactericidal Action to Being Conquered. J. Immunol. Res. 2017, 2017, 9671604. [Google Scholar] [CrossRef]
- Ravindran, M.; Khan, M.A.; Palaniyar, N. Neutrophil extracellular trap formation: Physiology, pathology, and pharmacology. Biomolecules 2019, 9, 365. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Buck, A.; Sanchez Klose, F.P.; Venkatakrishnan, V.; Khamzeh, A.; Dahlgren, C.; Christenson, K.; Bylund, J. DPI Selectively Inhibits Intracellular NADPH Oxidase Activity in Human Neutrophils. ImmunoHorizons 2019, 3, 488–497. [Google Scholar] [CrossRef]
- Nunes, P.; Demaurex, N.; Dinauer, M.C. Regulation of The NADPH Oxidase And Associated Ion Fluxes During Phagocytosis. Traffic 2013, 14, 1118–1131. [Google Scholar] [CrossRef]
- Li, P.; Li, M.; Lindberg, M.R.; Kennett, M.J.; Xiong, N.; Wang, Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010, 207, 1853–1862. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef]
- De Buhr, N.; von Köckritz-Blickwede, M. How Neutrophil Extracellular Traps Become Visible. J. Immunol. Res. 2016, 2016, 4604713. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yipp, B.G.; Kubes, P. NETosis: How vital is it? Blood 2013, 122, 2784–2794. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Biermann, M.H.; Brauner, J.M.; Liu, Y.; Zhao, Y.; Herrmann, M. New Insights into Neutrophil Extracellular Traps: Mechanisms of Formation and Role in Inflammation. Front. Immunol. 2016, 7, 302. [Google Scholar] [CrossRef] [PubMed]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef]
- Jorch, S.K.; Kubes, P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017, 23, 279–287. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, X.; Chatterjee, V.; Meegan, J.E.; Beard, R.S., Jr.; Yuan, S.Y. Role of Neutrophil Extracellular Traps and Vesicles in Regulating Vascular Endothelial Permeability. Front. Immunol. 2019, 10, 1037. [Google Scholar] [CrossRef]
- Von Köckritz-Blickwede, M.; Winstel, V. Molecular Prerequisites for Neutrophil Extracellular Trap Formation and Evasion Mechanisms of Staphylococcus aureus. Front. Immunol. 2022, 13, 836278. [Google Scholar] [CrossRef]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A Novel Mechanism of Rapid Nuclear Neutrophil Extracellular Trap Formation in Response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef]
- Yousefi, S.; Mihalache, C.; Kozlowski, E.; Schmid, I.; Simon, H.U. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009, 16, 1438–1444. [Google Scholar] [CrossRef]
- Remijsen, Q.; Kuijpers, T.W.; Wirawan, E.; Lippens, S.; Vandenabeele, P.; Vanden Berghe, T. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011, 18, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Hilda, J.N.; Das, S.; Tripathy, S.P.; Hanna, L.E. Role of neutrophils in tuberculosis: A bird’s eye view. Innate Immun. 2020, 26, 240–247. [Google Scholar] [CrossRef] [PubMed]
- De Buhr, N.; Stehr, M.; Neumann, A.; Naim, H.Y.; Valentin-Weigand, P.; von Köckritz-Blickwede, M.; Baums, C.G. Identification of a novel DNase of Streptococcus suis (EndAsuis) important for neutrophil extracellular trap degradation during exponential growth. Microbiology 2015, 161, 838–850. [Google Scholar] [CrossRef] [PubMed]
- De Buhr, N.; Neumann, A.; Jerjomiceva, N.; von Köckritz-Blickwede, M.; Baums, C.G. Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 2014, 160, 385–395. [Google Scholar] [CrossRef] [PubMed]
- De Buhr, N.; Bonilla, M.C.; Pfeiffer, J.; Akhdar, S.; Schwennen, C.; Kahl, B.C.; Waldmann, K.; Valentin-Weigand, P.; Hennig-Pauka, I.; von Köckritz-Blickwede, M. Degraded neutrophil extracellular traps promote the growth of Actinobacillus pleuropneumoniae. Cell Death Dis. 2019, 10, 657. [Google Scholar] [CrossRef]
- Hoeltig, D.; Rohde, J.; Frase, R.; Nietfeld, F.; Waldmann, K.H.; Valentin-Weigand, P.; Meens, J. Multi-organ spreading of Actinobacillus pleuropneumoniae serovar 7 in weaned pigs during the first week after experimental infection. Vet. Res. 2018, 49, 97. [Google Scholar] [CrossRef]
- Obradovic, M.R.; Segura, M.; Segalés, J.; Gottschalk, M. Review of the speculative role of co-infections in Streptococcus suis-associated diseases in pigs. Vet. Res. 2021, 52, 49. [Google Scholar] [CrossRef]
- Vötsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef]
- Gottschalk, M. Streptococcosis. In Diseases of Swine; Locke, A.K., Zimmerman, J., Ramirez, A., Schwartz, K.J., Stevenson, G., Eds.; Wiley-Blackwell: Ames, IA, USA, 2012; pp. 841–855. ISBN 978-0-8138-2267-9. [Google Scholar]
- Stygar, A.H.; Niemi, J.K.; Oliviero, C.; Laurila, T.; Heinonen, M. Economic value of mitigating Actinobacillus pleuropneumoniae infections in pig fattening herds. Agric. Syst. 2016, 144, 113–121. [Google Scholar] [CrossRef]
- Neila-Ibáñez, C.; Casal, J.; Hennig-Pauka, I.; Stockhofe-Zurwieden, N.; Gottschalk, M.; Migura-García, L.; Pailler-García, L.; Napp, S. Stochastic Assessment of the Economic Impact of Streptococcus suis-Associated Disease in German, Dutch and Spanish Swine Farms. Front. Vet. Sci. 2021, 8, 958. [Google Scholar] [CrossRef]
- Ma, F.; Chang, X.; Wang, G.; Zhou, H.; Ma, Z.; Lin, H.; Fan, H. Streptococcus suis Serotype 2 Stimulates Neutrophil Extracellular Traps Formation via Activation of p38 MAPK and ERK1/2. Front. Immunol. 2018, 9, 2854. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.R.; Ma, A.C.; Tavener, S.A.; McDonald, B.; Goodarzi, Z.; Kelly, M.M.; Patel, K.D.; Chakrabarti, S.; McAvoy, E.; Sinclair, G.D.; et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007, 13, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Bouti, P.; Webbers, S.D.S.; Fagerholm, S.C.; Alon, R.; Moser, M.; Matlung, H.L.; Kuijpers, T.W. β2 Integrin Signaling Cascade in Neutrophils: More Than a Single Function. Front. Immunol. 2021, 11, 619925. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.C.; Doerschuk, C.M. The role of CD18 in the production and release of neutrophils from the bone marrow. Lab. Investig. 2010, 90, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk, M.; Stege, H.; Grabbe, S.; Bros, M. β2 Integrins—Multi-Functional Leukocyte Receptors in Health and Disease. Int. J. Mol. Sci. 2020, 21, 1402. [Google Scholar] [CrossRef]
- Aulik, N.A.; Hellenbrand, K.M.; Klos, H.; Czuprynski, C.J. Mannheimia haemolytica and its leukotoxin cause neutrophil extracellular trap formation by bovine neutrophils. Infect. Immun. 2010, 78, 4454–4466. [Google Scholar] [CrossRef]
- Kamp, E.M.; Stockhofe-Zurwieden, N.; van Leengoed, L.A.; Smits, M.A. Endobronchial inoculation with Apx toxins of Actinobacillus pleuropneumoniae leads to pleuropneumonia in pigs. Infect. Immun. 1997, 65, 4350–4354. [Google Scholar] [CrossRef]
- Li, S.C.; Cheng, Y.T.; Wang, C.Y.; Wu, J.Y.; Chen, Z.W.; Wang, J.P.; Lin, J.H.; Hsuan, S.L. Actinobacillus pleuropneumoniae exotoxin ApxI induces cell death via attenuation of FAK through LFA-1. Sci. Rep. 2021, 11, 1753. [Google Scholar] [CrossRef]
- Chiers, K.; De Waele, T.; Pasmans, F.; Ducatelle, R.; Haesebrouck, F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet. Res. 2010, 41, 65. [Google Scholar] [CrossRef]
- Bleuzé, M.; Gottschalk, M.; Segura, M. Neutrophils in Streptococcus suis Infection: From Host Defense to Pathology. Microorganisms 2021, 9, 2392. [Google Scholar] [CrossRef]
- Vötsch, D.; Willenborg, M.; Oelemann, W.M.R.; Brogden, G.; Valentin-Weigand, P. Membrane Binding, Cellular Cholesterol Content and Resealing Capacity Contribute to Epithelial Cell Damage Induced by Suilysin of Streptococcus suis. Pathogens 2019, 9, 33. [Google Scholar] [CrossRef]
- Seitz, M.; Baums, C.G.; Neis, C.; Benga, L.; Fulde, M.; Rohde, M.; Goethe, R.; Valentin-Weigand, P. Subcytolytic effects of suilysin on interaction of Streptococcus suis with epithelial cells. Vet. Microbiol. 2013, 167, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Benga, L.; Fulde, M.; Neis, C.; Goethe, R.; Valentin-Weigand, P. Polysaccharide capsule and suilysin contribute to extracellular survival of Streptococcus suis co-cultivated with primary porcine phagocytes. Vet. Microbiol. 2008, 132, 211–219. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Pian, Y.; Ren, Z.; Bi, L.; Yuan, Y.; Zheng, Y.; Jiang, Y.; Wang, F. Increased production of suilysin contributes to invasive infection of the Streptococcus suis strain 05ZYH33. Mol. Med. Rep. 2014, 10, 2819–2826. [Google Scholar] [CrossRef]
- Dąbrowska, D.; Jabłońska, E.; Garley, M.; Ratajczak-Wrona, W.; Iwaniuk, A. New Aspects of the Biology of Neutrophil Extracellular Traps. Scand. J. Immunol. 2016, 84, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.E.; Zimmermann, T.J.; Goosmann, C.; Alexander, T.; Hedberg, C.; Ziegler, S.; Zychlinsky, A.; Waldmann, H. Tetrahydroisoquinolines: New Inhibitors of Neutrophil Extracellular Trap (NET) Formation. ChemBioChem 2017, 18, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Ostafin, M.; Pruchniak, M.P.; Ciepiela, O.; Reznick, A.Z.; Demkow, U. Different procedures of diphenyleneiodonium chloride addition affect neutrophil extracellular trap formation. Anal. Biochem. 2016, 509, 60–66. [Google Scholar] [CrossRef]
- Stoiber, W.; Obermayer, A.; Steinbacher, P.; Krautgartner, W. The Role of Reactive Oxygen Species (ROS) in the Formation of Extracellular Traps (ETs) in Humans. Biomolecules 2015, 5, 702–723. [Google Scholar] [CrossRef]
- Matsuura, M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front. Immunol. 2013, 4, 109. [Google Scholar] [CrossRef]
- De Buhr, N.; von Köckritz-Blickwede, M. Detection, Visualization, and Quantification of Neutrophil Extracellular Traps (NETs) and NET Markers. Methods Mol. Biol. 2020, 2087, 425–442. [Google Scholar]
- Von Köckritz-Blickwede, M.; Chow, O.; Ghochani, M.; Nizet, V. Visualization and functional evaluation of phagocyte extracellular traps. Methods Microbiol. 2010, 37, 139–160. [Google Scholar]
- Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps: Is immunity the second function of chromatin? J. Cell Biol. 2012, 198, 773–783. [Google Scholar] [CrossRef]
- Onouchi, T.; Shiogama, K.; Mizutani, Y.; Takaki, T.; Tsutsumi, Y. Visualization of neutrophil extracellular traps and fibrin meshwork in human fibrinopurulent inflammatory lesions: III. Correlative light and electron microscopic study. Acta Histochem. Cytochem. 2016, 49, 141–147. [Google Scholar] [CrossRef]
- Golding, C.G.; Lamboo, L.L.; Beniac, D.R.; Booth, T.F. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci. Rep. 2016, 6, 26516. [Google Scholar] [CrossRef]
- Kamoshida, G.; Kikuchi-Ueda, T.; Nishida, S.; Tansho-Nagakawa, S.; Kikuchi, H.; Ubagai, T.; Ono, Y. Spontaneous formation of neutrophil extracellular traps in serum-free culture conditions. FEBS Open Bio 2017, 7, 877–886. [Google Scholar] [CrossRef]
- Nauseef, W.M.; Kubes, P. Pondering neutrophil extracellular traps with healthy skepticism. Cell. Microbiol. 2016, 18, 1349–1357. [Google Scholar] [CrossRef]
- Fingerhut, L.; Ohnesorge, B.; von Borstel, M.; Schumski, A.; Strutzberg-Minder, K.; Mörgelin, M.; Deeg, C.A.; Haagsman, H.P.; Beineke, A.; von Köckritz-Blickwede, M.; et al. Neutrophil Extracellular Traps in the Pathogenesis of Equine Recurrent Uveitis (ERU). Cells 2019, 8, 1528. [Google Scholar] [CrossRef]
- Jansen, R.; Briaire, J.; Smith, H.E.; Dom, P.; Haesebrouck, F.; Kamp, E.M.; Gielkens, A.L.; Smits, M.A. Knockout mutants of Actinobacillus pleuropneumoniae serotype 1 that are devoid of RTX toxins do not activate or kill porcine neutrophils. Infect. Immun. 1995, 63, 27–37. [Google Scholar] [CrossRef]
- Bossé, J.T.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef]
- Chabot-Roy, G.; Willson, P.; Segura, M.; Lacouture, S.; Gottschalk, M. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb. Pathog. 2006, 41, 21–32. [Google Scholar] [CrossRef]
- De Jonge, N.; Peckys, D.B. Live Cell Electron Microscopy Is Probably Impossible. ACS Nano 2016, 10, 9061–9063. [Google Scholar] [CrossRef]
- Thiam, H.R.; Wong, S.L.; Qiu, R.; Kittisopikul, M.; Vahabikashi, A.; Goldman, A.E.; Goldman, R.D.; Wagner, D.D.; Waterman, C.M. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture. Proc. Natl. Acad. Sci. USA 2020, 117, 7326–7337. [Google Scholar] [CrossRef]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef]
- Manfredi, A.A.; Ramirez, G.A.; Rovere-Querini, P.; Maugeri, N. The Neutrophil’s Choice: Phagocytose vs Make Neutrophil Extracellular Traps. Front. Immunol. 2018, 9, 288. [Google Scholar] [CrossRef]
- Manfredi, A.A.; Covino, C.; Rovere-Querini, P.; Maugeri, N. Instructive influences of phagocytic clearance of dying cells on neutrophil extracellular trap generation. Clin. Exp. Immunol. 2015, 179, 24–29. [Google Scholar] [CrossRef]
- Hennig-Pauka, I.; Baltes, N.; Jacobsen, I.; Stratmann-Selke, J.; Gerlach, G.-F.; Selbitz, H.-J.; Waldmann, K.-H. Study of the virulence of Actinobacillus pleuropneumoniae in finishing pigs as a basis for vaccination development. Berl. Munch. Tierarztl. Wochenschr. 2008, 121, 189–197. [Google Scholar]
- Smith, H.E.; Damman, M.; Van Der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756. [Google Scholar] [CrossRef]
- Meurer, M.; Öhlmann, S.; Bonilla, M.C.; Valentin-Weigand, P.; Beineke, A.; Hennig-Pauka, I.; Schwerk, C.; Schroten, H.; Baums, C.G.; von Köckritz-Blickwede, M.; et al. Role of Bacterial and Host DNases on Host-Pathogen Interaction during Streptococcus suis Meningitis. Int. J. Mol. Sci. 2020, 21, 5289. [Google Scholar] [CrossRef]
- Baien, S.H.; Langer, M.N.; Heppelmann, M.; von Köckritz-Blickwede, M.; de Buhr, N. Comparison Between K3EDTA and Lithium Heparin as Anticoagulant to Isolate Bovine Granulocytes from Blood. Front. Immunol. 2018, 9, 1570. [Google Scholar] [CrossRef]
- Bonilla, M.C.; Fingerhut, L.; Alfonso-Castro, A.; Mergani, A.; Schwennen, C.; von Köckritz-Blickwede, M.; de Buhr, N. How Long Does a Neutrophil Live?—The Effect of 24 h Whole Blood Storage on Neutrophil Functions in Pigs. Biomedicines 2020, 8, 278. [Google Scholar] [CrossRef]
- Stirling, J.W.; Graff, P.S. Antigen unmasking for immunoelectron microscopy: Labeling is improved by treating with sodium ethoxide or sodium metaperiodate, then heating on retrieval medium. J. Histochem. Cytochem. 1995, 43, 115–123. [Google Scholar] [CrossRef]
- Roth, J. Post-embedding cytochemistry with gold-labelled reagents: A review. J. Microsc. 1986, 143, 125–137. [Google Scholar] [CrossRef]
- Beineke, A.; Bennecke, K.; Neis, C.; Schröder, C.; Waldmann, K.-H.; Baumgärtner, W.; Valentin-Weigand, P.; Baums, C.G. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet. Microbiol. 2008, 128, 423–430. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonilla, M.C.; Quiros, O.N.; Wendt, M.; Hennig-Pauka, I.; Mörgelin, M.; von Köckritz-Blickwede, M.; de Buhr, N. New Insights into Neutrophil Extracellular Trap (NETs) Formation from Porcine Neutrophils in Response to Bacterial Infections. Int. J. Mol. Sci. 2022, 23, 8953. https://doi.org/10.3390/ijms23168953
Bonilla MC, Quiros ON, Wendt M, Hennig-Pauka I, Mörgelin M, von Köckritz-Blickwede M, de Buhr N. New Insights into Neutrophil Extracellular Trap (NETs) Formation from Porcine Neutrophils in Response to Bacterial Infections. International Journal of Molecular Sciences. 2022; 23(16):8953. https://doi.org/10.3390/ijms23168953
Chicago/Turabian StyleBonilla, Marta C., Oriana N. Quiros, Michael Wendt, Isabel Hennig-Pauka, Matthias Mörgelin, Maren von Köckritz-Blickwede, and Nicole de Buhr. 2022. "New Insights into Neutrophil Extracellular Trap (NETs) Formation from Porcine Neutrophils in Response to Bacterial Infections" International Journal of Molecular Sciences 23, no. 16: 8953. https://doi.org/10.3390/ijms23168953
APA StyleBonilla, M. C., Quiros, O. N., Wendt, M., Hennig-Pauka, I., Mörgelin, M., von Köckritz-Blickwede, M., & de Buhr, N. (2022). New Insights into Neutrophil Extracellular Trap (NETs) Formation from Porcine Neutrophils in Response to Bacterial Infections. International Journal of Molecular Sciences, 23(16), 8953. https://doi.org/10.3390/ijms23168953






