A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response
Abstract
1. Introduction
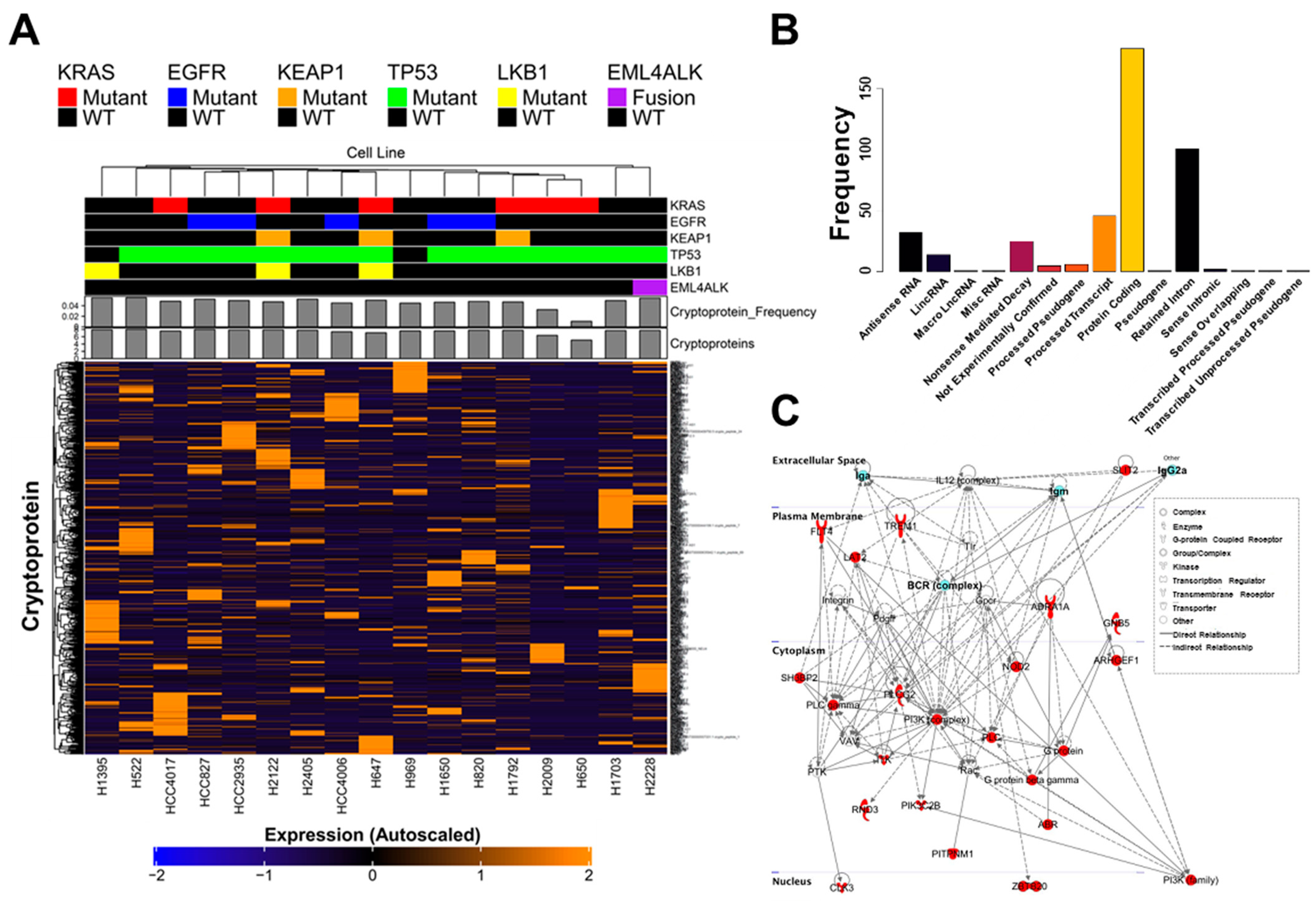
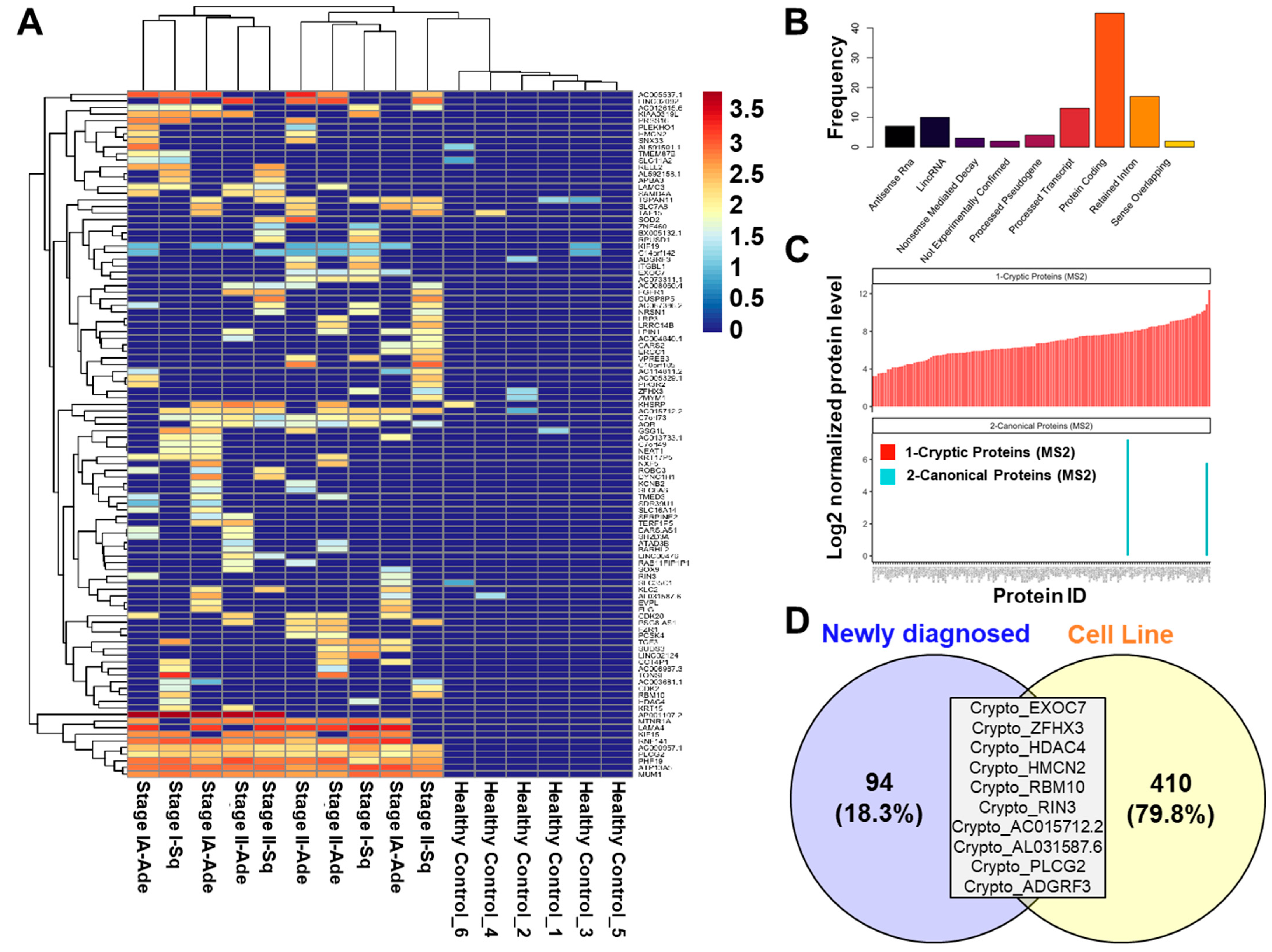
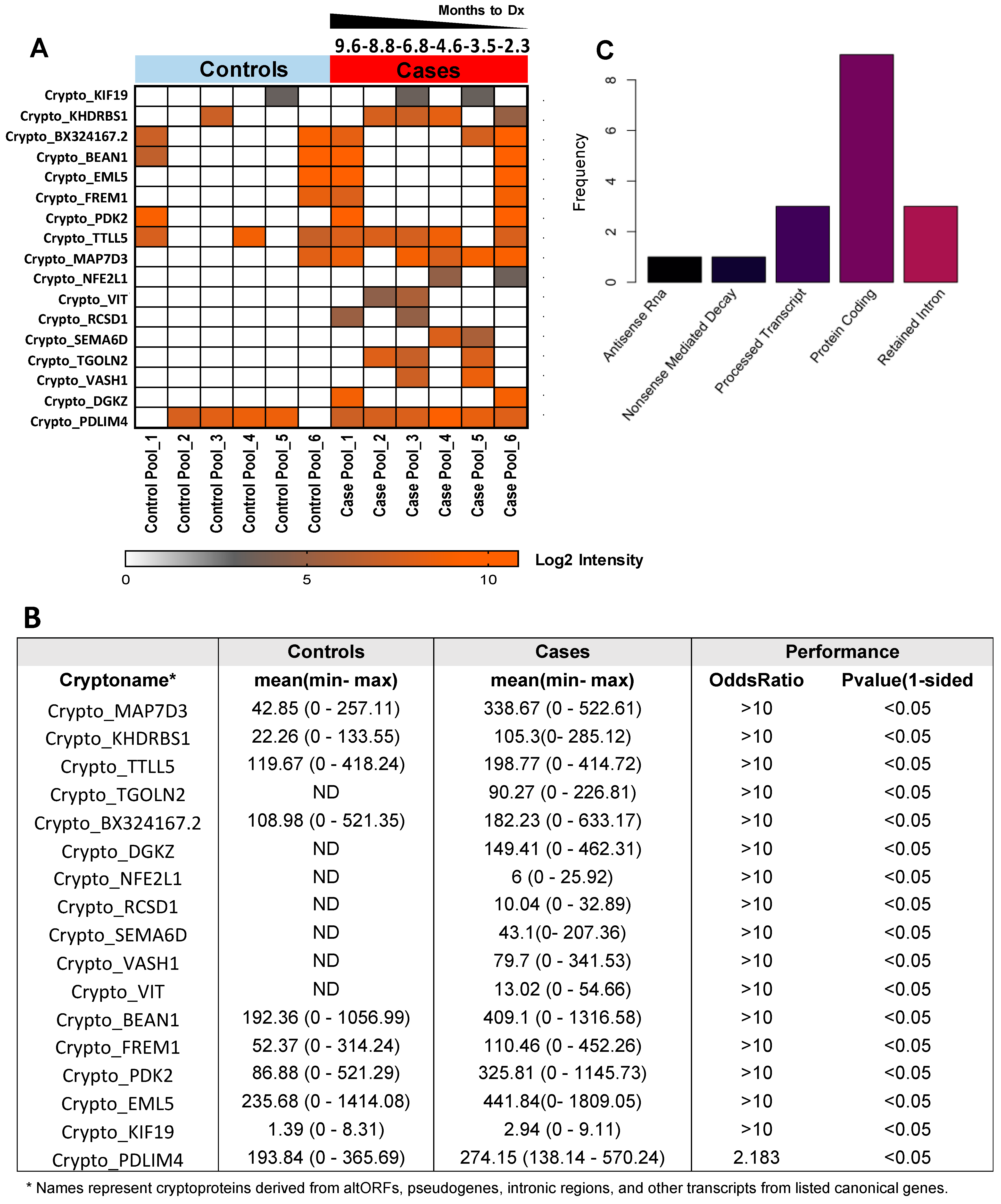
2. Results
3. Discussion
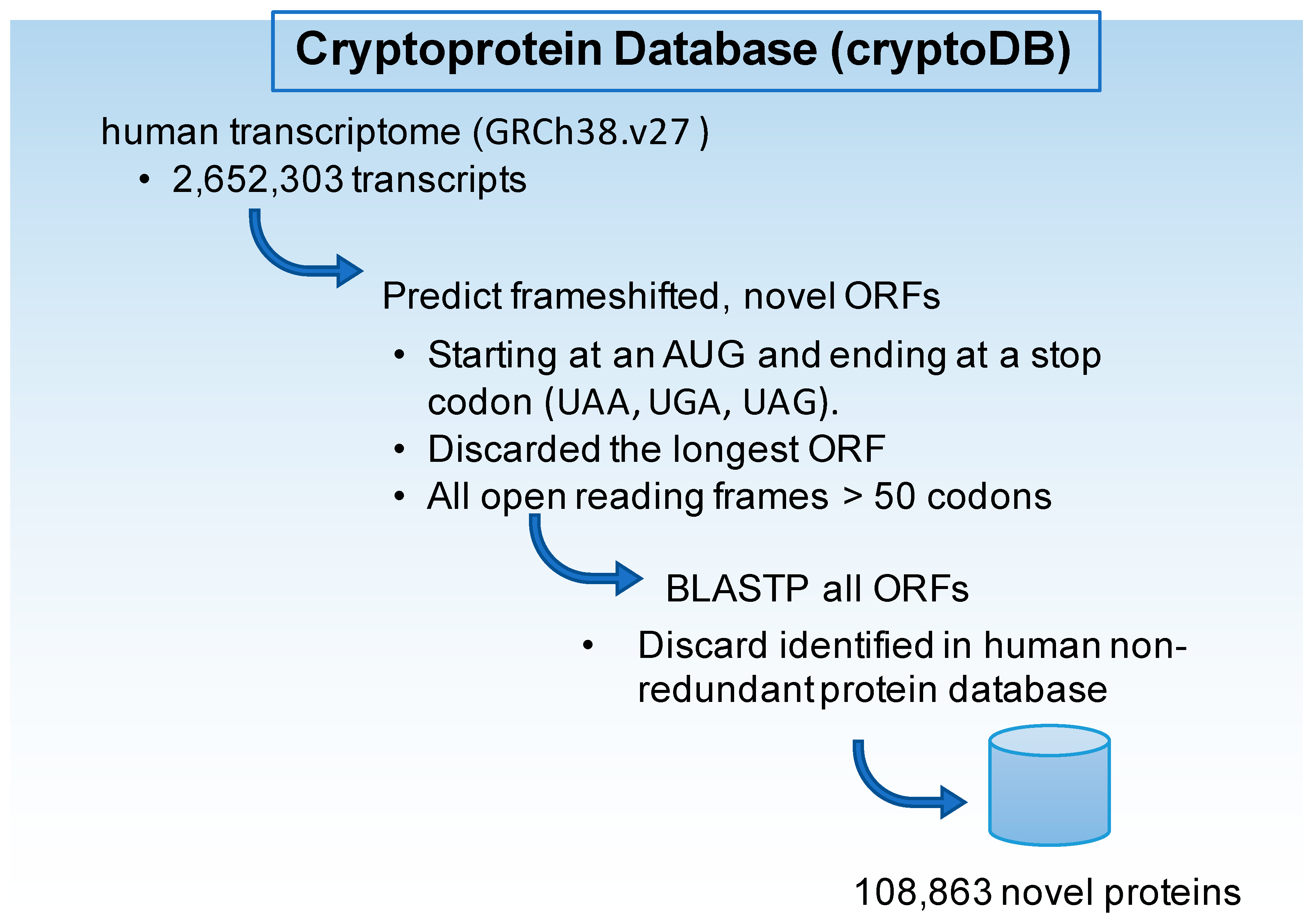
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsou, P.; Katayama, H.; Ostrin, E.J.; Hanash, S.M. The Emerging Role of B Cells in Tumor Immunity. Cancer Res. 2016, 76, 5597–5601. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.-N.; Boisson, A.; Duvillier, H. Tumor-infiltrating B cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, e129641. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S. Harnessing immunity for cancer marker discovery. Nat. Biotechnol. 2003, 21, 37–38. [Google Scholar] [CrossRef]
- Le Naour, F. Identification of tumor antigens by using proteomics. Methods Mol. Biol. 2007, 360, 327–334. [Google Scholar] [PubMed]
- Pereira-Faca, S.R.; Kuick, R.; Puravs, E.; Zhang, Q.; Krasnoselsky, A.L.; Phanstiel, D.; Qiu, J.; Misek, D.E.; Hinderer, R.; Tammemagi, M.; et al. Identification of 14-3-3 theta as an antigen that induces a humoral response in lung cancer. Cancer Res. 2007, 67, 12000–12006. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, H.; Goodell, V.; Disis, M.L. Humoral Immunity Directed against Tumor-Associated Antigens As Potential Biomarkers for the Early Diagnosis of Cancer. J. Proteome Res. 2008, 7, 1388–1394. [Google Scholar] [CrossRef]
- Babel, I.; Barderas, R.; Díaz-Uriarte, R.; Martínez-Torrecuadrada, J.L.; Sanchez-Carbayo, M.; Casal, J.I. Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol. Cell. Proteom. 2009, 8, 2382–2395. [Google Scholar] [CrossRef]
- Massoner, P.; Lueking, A.; Goehler, H.; Höpfner, A.; Kowald, A.; Kugler, K.G.; Amersdorfer, P.; Horninger, W.; Bartsch, G.; Schulz-Knappe, P. Serum-autoantibodies for discovery of prostate cancer specific biomarkers. Prostate 2012, 72, 427–436. [Google Scholar] [CrossRef]
- Babel, I.; Barderas, R.; Diaz-Uriarte, R.; Moreno, V.; Suarez, A.; Fernandez-Aceñero, M.J.; Salazar, R.; Capellá, G.; Casal, J.I. Identification of MST1/STK4 and SULF1 proteins as autoantibody targets for the diagnosis of colorectal cancer by using phage microarrays. Mol. Cell. Proteom. 2011, 10, M110.001784. [Google Scholar] [CrossRef]
- Hong, S.-H.; Misek, D.E.; Wang, H.; Puravs, E.; Giordano, T.J.; Greenson, J.K.; Brenner, D.E.; Simeone, D.M.; Logsdon, C.D.; Hanash, S.M. An autoantibody-mediated immune response to calreticulin isoforms in pancreatic cancer. Cancer Res. 2004, 64, 5504–5510. [Google Scholar] [CrossRef][Green Version]
- Nam, M.J.; Kee, M.K.; Kuick, R.; Hanash, S.M. Identification of defensin α6 as a potential biomarker in colon adenocarcinoma. J. Biol. Chem. 2005, 280, 8260–8265. [Google Scholar] [CrossRef] [PubMed]
- Landry, C.R.; Zhong, X.; Nielly-Thibault, L.; Roucou, X. Found in translation: Functions and evolution of a recently discovered alternative proteome. Curr. Opin. Struct. Biol. 2015, 32, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.P.; Loughran, G.; Atkins, J.F. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. USA 2008, 105, 10079–10084. [Google Scholar] [CrossRef] [PubMed]
- Laumont, C.M.; Daouda, T.; Laverdure, J.-P.; Bonneil, É.; Caron-Lizotte, O.; Hardy, M.-P.; Granados, D.P.; Durette, C.; Lemieux, S.; Thibault, P. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun. 2016, 7, 10238. [Google Scholar] [CrossRef]
- Chong, C.; Müller, M.; Pak, H.; Harnett, D.; Huber, F.; Grun, D.; Leleu, M.; Auger, A.; Arnaud, M.; Stevenson, B.J. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 2020, 11, 1293. [Google Scholar] [CrossRef]
- Bartok, O.; Pataskar, A.; Nagel, R.; Laos, M.; Goldfarb, E.; Hayoun, D.; Levy, R.; Körner, P.-R.; Kreuger, I.Z.; Champagne, J. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature 2020, 590, 332–337. [Google Scholar] [CrossRef]
- Yan, Y.; Sun, N.; Wang, H.; Kobayashi, M.; Ladd, J.J.; Long, J.P.; Lo, K.C.; Patel, J.; Sullivan, E.; Albert, T. Whole Genome–Derived Tiled Peptide Arrays Detect Prediagnostic Autoantibody Signatures in Non–Small-Cell Lung Cancer. Cancer Res. 2019, 79, 1549–1557. [Google Scholar] [CrossRef]
- Hanash, S.M.; Ostrin, E.J.; Fahrmann, J.F. Blood based biomarkers beyond genomics for lung cancer screening. Transl. Lung Cancer Res. 2018, 7, 327. [Google Scholar] [CrossRef]
- Qiu, J.; Choi, G.; Li, L.; Wang, H.; Pitteri, S.J.; Pereira-Faca, S.R.; Krasnoselsky, A.L.; Randolph, T.W.; Omenn, G.S.; Edelstein, C. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J. Clin. Oncol. 2008, 26, 5060. [Google Scholar] [CrossRef]
- Pandolfi, P.P. Aberrant mRNA translation in cancer pathogenesis: An old concept revisited comes finally of age. Oncogene 2004, 23, 3134–3137. [Google Scholar] [CrossRef][Green Version]
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11, a032896. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.C.; Peters, H.L.; Taguchi, A.; Katayama, H.; Wang, H.; Momin, A.; Jolly, M.K.; Celiktas, M.; Rodriguez-Canales, J.; Liu, H. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc. Natl. Acad. Sci. USA 2016, 113, E1555–E1564. [Google Scholar] [CrossRef] [PubMed]
- Schliekelman, M.J.; Taguchi, A.; Zhu, J.; Dai, X.; Rodriguez, J.; Celiktas, M.; Zhang, Q.; Chin, A.; Wong, C.-H.; Wang, H. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015, 75, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Lacas, B.; Le Teuff, G.; Hainaut, P.; Jänne, P.A.; Pignon, J.-P.; Le Chevalier, T.; Seymour, L.; Douillard, J.-Y.; Graziano, S. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non–small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2017, 35, 2018. [Google Scholar] [CrossRef]
- Ameli Mojarad, M.; Ameli Mojarad, M.; Pourmahdian, A. Long Non-coding RNA snaR Promotes Proliferation in EGFR Wild Type Non-Small Cell Lung Cancer Cells. Int. J. Mol. Cell. Med. 2021, 10, 258–264. [Google Scholar]
- Passaro, A.; Stenzinger, A.; Peters, S. Tumor Mutational Burden as a Pan-cancer Biomarker for Immunotherapy: The Limits and Potential for Convergence. Cancer Cell 2020, 38, 624–625. [Google Scholar] [CrossRef]
- Brierley, I.; Jenner, A.J.; Inglis, S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992, 227, 463–479. [Google Scholar] [CrossRef]
- Dinman, J.D. Slippery ribosomes prefer shapeshifting mRNAs. Proc. Natl. Acad. Sci. USA 2019, 116, 19225–19227. [Google Scholar] [CrossRef]
- Lambert, J.-M.; Ashi, M.O.; Srour, N.; Delpy, L.; Saulière, J. Mechanisms and Regulation of Nonsense-Mediated mRNA Decay and Nonsense-Associated Altered Splicing in Lymphocytes. Int. J. Mol. Sci. 2020, 21, 1335. [Google Scholar] [CrossRef]
- Rozov, A.; Demeshkina, N.; Westhof, E.; Yusupov, M.; Yusupova, G. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015, 6, 7251. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Marsh, T.; Irajizad, E.; Patel, N.; Murage, E.; Vykoukal, J.; Dennison, J.B.; Do, K.-A.; Ostrin, E.; Spitz, M.R.; et al. Blood-Based Biomarker Panel for Personalized Lung Cancer Risk Assessment. J. Clin. Oncol. 2020, 40, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Ostrin, E.J.; Bantis, L.E.; Wilson, D.O.; Patel, N.; Wang, R.; Kundnani, D.; Adams-Haduch, J.; Dennison, J.B.; Fahrmann, J.F.; Chiu, H.T.; et al. Contribution of a Blood-Based Protein Biomarker Panel to the Classification of Indeterminate Pulmonary Nodules. J. Thorac. Oncol. 2021, 16, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Fahrmann, J.F.; Kim, K.; DeFelice, B.C.; Taylor, S.L.; Gandara, D.R.; Yoneda, K.Y.; Cooke, D.T.; Fiehn, O.; Kelly, K.; Miyamoto, S. Investigation of metabolomic blood biomarkers for detection of adenocarcinoma lung cancer. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Aieta, M.; Tartarone, A.; Pezzuto, A.; Facchinetti, A.; Santini, D.; Ulivi, P.; Ludovini, V.; Possidente, L.; Fiduccia, P.; et al. A fully automated assay to detect the expression of pan-cytokeratins and of EML4-ALK fusion protein in circulating tumour cells (CTCs) predicts outcome of non-small cell lung cancer (NSCLC) patients. Transl. Lung Cancer Res. 2021, 10, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hanash, S. Intact-protein analysis system for discovery of serum-based disease biomarkers. In Serum/Plasma Proteomics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 69–85. [Google Scholar]
- Kobayashi, M.; Katayama, H.; Irajizad, E.; Vykoukal, J.V.; Fahrmann, J.F.; Kundnani, D.L.; Yu, C.-Y.; Cai, Y.; Hsiao, F.C.; Yang, W.-L. Proteome Profiling Uncovers an Autoimmune Response Signature That Reflects Ovarian Cancer Pathogenesis. Cancers 2020, 12, 485. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Tanaka, I.; Irajizad, E.; Mao, X.; Dennison, J.B.; Murage, E.; Casabar, J.; Mayo, J.; Peng, Q.; Celiktas, M.; et al. Mutational Activation of the NRF2 Pathway Upregulates Kynureninase Resulting in Tumor Immunosuppression and Poor Outcome in Lung Adenocarcinoma. Cancers 2022, 14, 2543. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, I.; Dayde, D.; Tai, M.C.; Mori, H.; Solis, L.M.; Tripathi, S.C.; Fahrmann, J.F.; Unver, N.; Parhy, G.; Jain, R.; et al. SRGN-Triggered Aggressive and Immunosuppressive Phenotype in a Subset of TTF-1-Negative Lung Adenocarcinomas. J. Natl. Cancer Inst. 2022, 114, 290–301. [Google Scholar] [CrossRef]
- Taguchi, A.; Politi, K.; Pitteri, S.J.; Lockwood, W.W.; Faca, V.M.; Kelly-Spratt, K.; Wong, C.H.; Zhang, Q.; Chin, A.; Park, K.S.; et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell 2011, 20, 289–299. [Google Scholar] [CrossRef]
- Goodman, G.E.; Thornquist, M.D.; Balmes, J.; Cullen, M.R.; Meyskens, F.L., Jr.; Omenn, G.S.; Valanis, B.; Williams, J.H., Jr. The Beta-Carotene and Retinol Efficacy Trial: Incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping β-carotene and retinol supplements. J. Natl. Cancer Inst. 2004, 96, 1743–1750. [Google Scholar] [CrossRef]
- Capello, M.; Vykoukal, J.V.; Katayama, H.; Bantis, L.E.; Wang, H.; Kundnani, D.L.; Aguilar-Bonavides, C.; Aguilar, M.; Tripathi, S.C.; Dhillon, D.S. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat. Commun. 2019, 10, 254. [Google Scholar] [CrossRef]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.M.; Sickmann, A.; Martens, L.; Zahedi, R.P. Peptide identification quality control. Proteomics 2011, 11, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.; Claassen, M.; Schrimpf, S.P.; Jovanovic, M.; Schmidt, A.; Buhmann, J.M.; Hengartner, M.O.; Aebersold, R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteom. 2009, 8, 2405–2417. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Joh, Y.S.; Kim, H.; Paek, E.; Lee, S.-W.; Hwang, K.-B. Evaluating the effect of database inflation in proteogenomic search on sensitive and reliable peptide identification. BMC Genom. 2016, 17, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteom. 2010, 73, 2092–2123. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irajizad, E.; Fahrmann, J.F.; Long, J.P.; Vykoukal, J.; Kobayashi, M.; Capello, M.; Yu, C.-Y.; Cai, Y.; Hsiao, F.C.; Patel, N.; et al. A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response. Int. J. Mol. Sci. 2022, 23, 8933. https://doi.org/10.3390/ijms23168933
Irajizad E, Fahrmann JF, Long JP, Vykoukal J, Kobayashi M, Capello M, Yu C-Y, Cai Y, Hsiao FC, Patel N, et al. A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response. International Journal of Molecular Sciences. 2022; 23(16):8933. https://doi.org/10.3390/ijms23168933
Chicago/Turabian StyleIrajizad, Ehsan, Johannes F. Fahrmann, James P. Long, Jody Vykoukal, Makoto Kobayashi, Michela Capello, Chuan-Yih Yu, Yining Cai, Fu Chung Hsiao, Nikul Patel, and et al. 2022. "A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response" International Journal of Molecular Sciences 23, no. 16: 8933. https://doi.org/10.3390/ijms23168933
APA StyleIrajizad, E., Fahrmann, J. F., Long, J. P., Vykoukal, J., Kobayashi, M., Capello, M., Yu, C.-Y., Cai, Y., Hsiao, F. C., Patel, N., Park, S., Peng, Q., Dennison, J. B., Kato, T., Tai, M. C., Taguchi, A., Kadara, H., Wistuba, I. I., Katayama, H., ... Ostrin, E. J. (2022). A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response. International Journal of Molecular Sciences, 23(16), 8933. https://doi.org/10.3390/ijms23168933







