Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species
Abstract
1. Introduction
2. Results
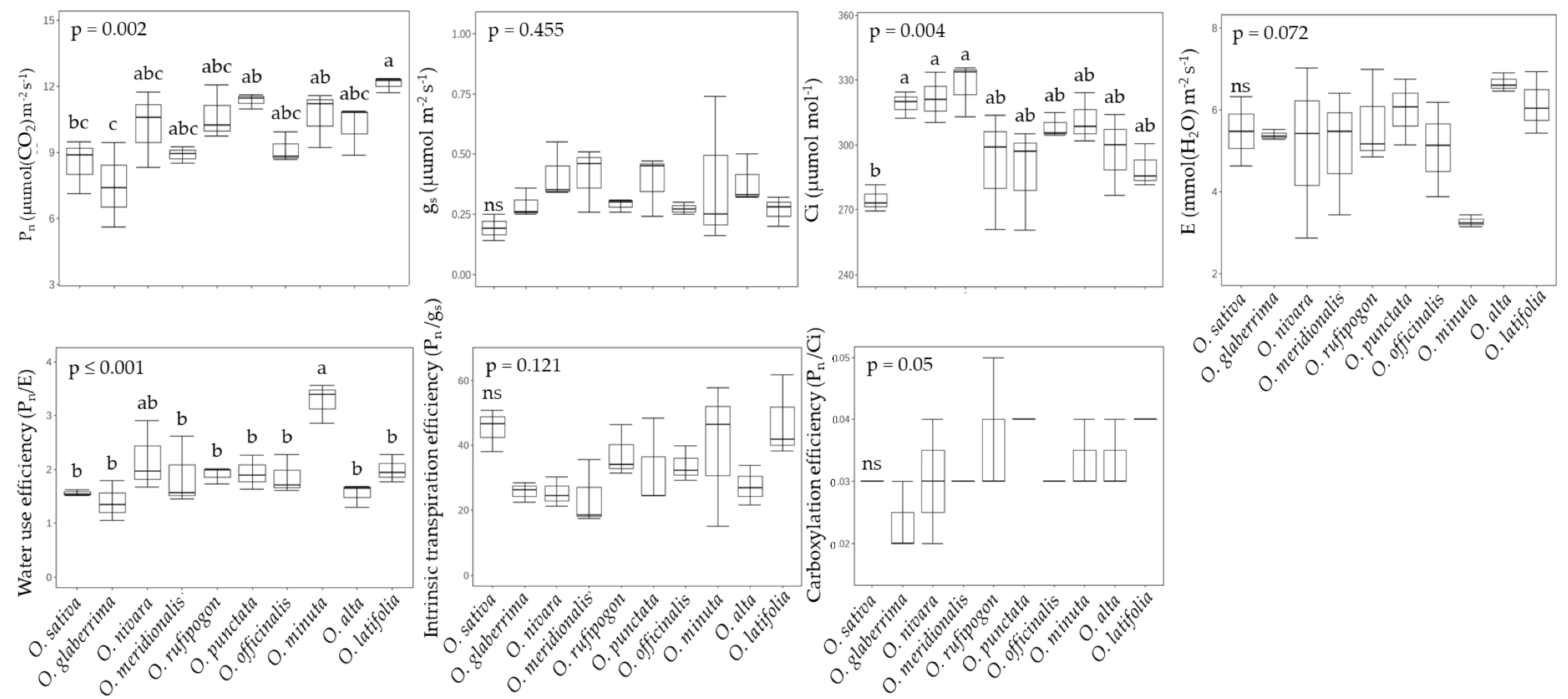
2.1. Growth and Photosynthetic Performance in Rice Species
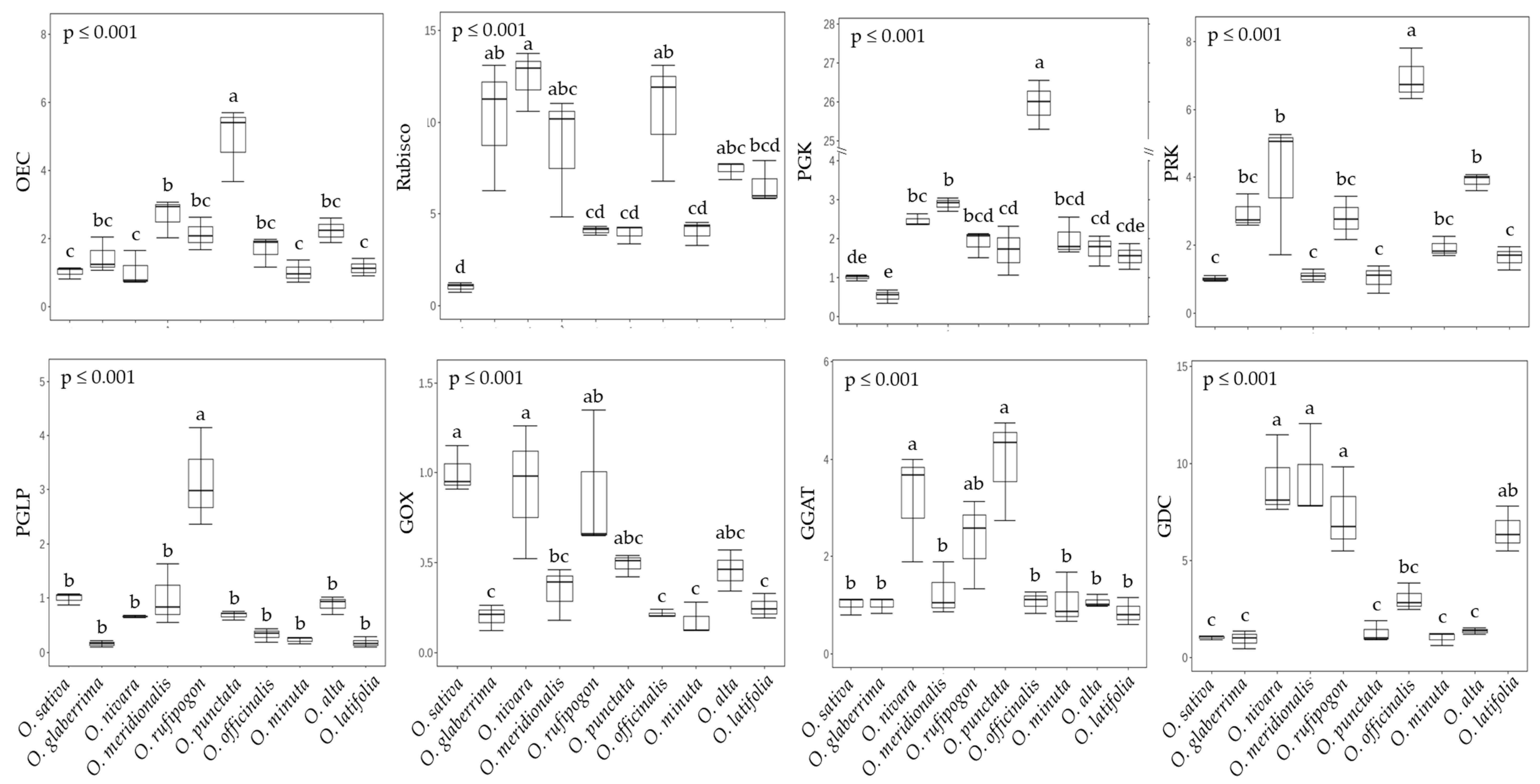
2.2. Photosynthesis and Photorespiration in Rice Species
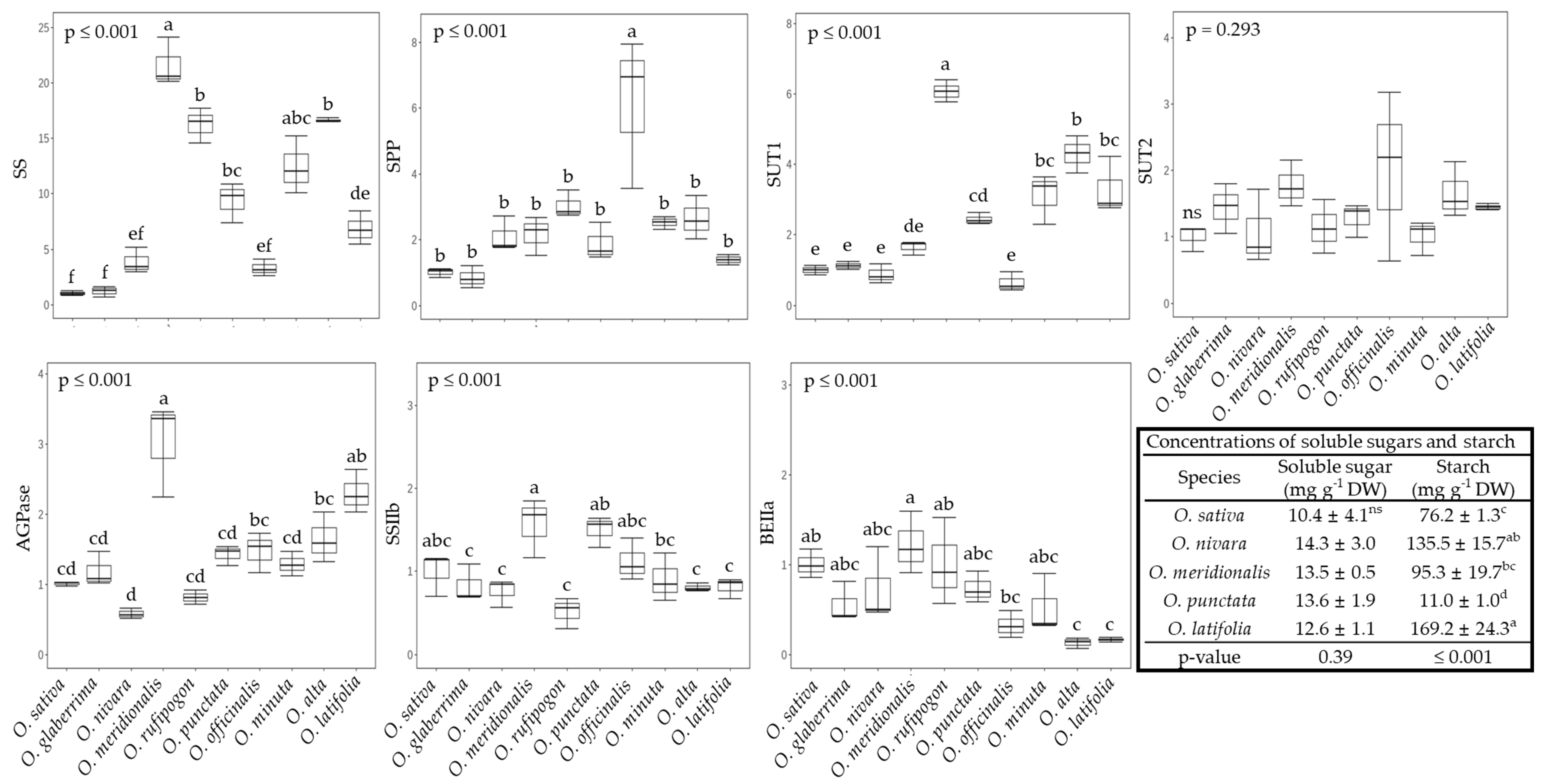
2.3. The Sucrose-to-Starch Transition and Sucrose Transport in Rice Species
2.4. Primary and Secondary Cell Wall Synthesis in Rice Species
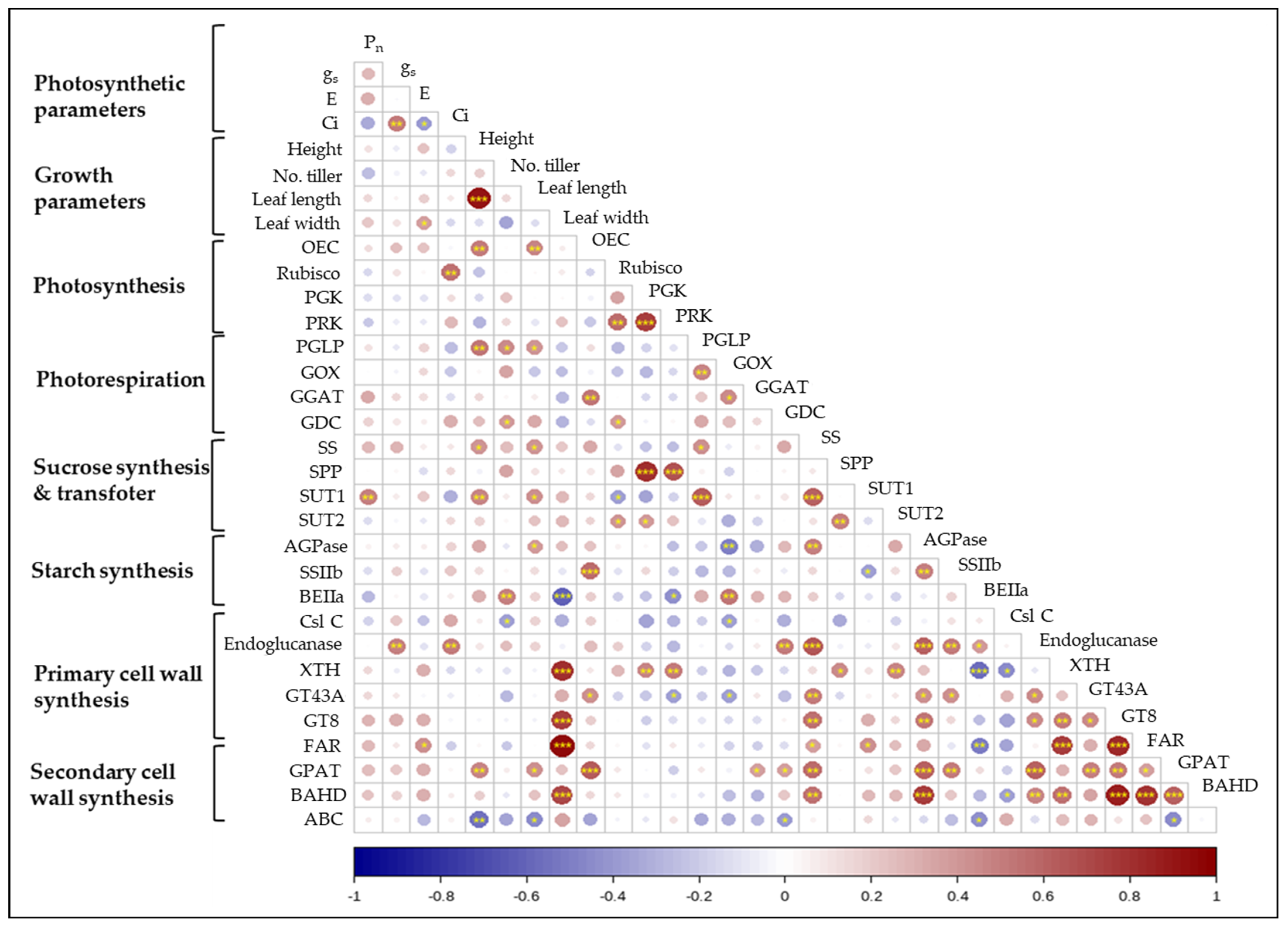
2.5. Correlation between Photosynthesis, Growth Parameters, and Gene Expression
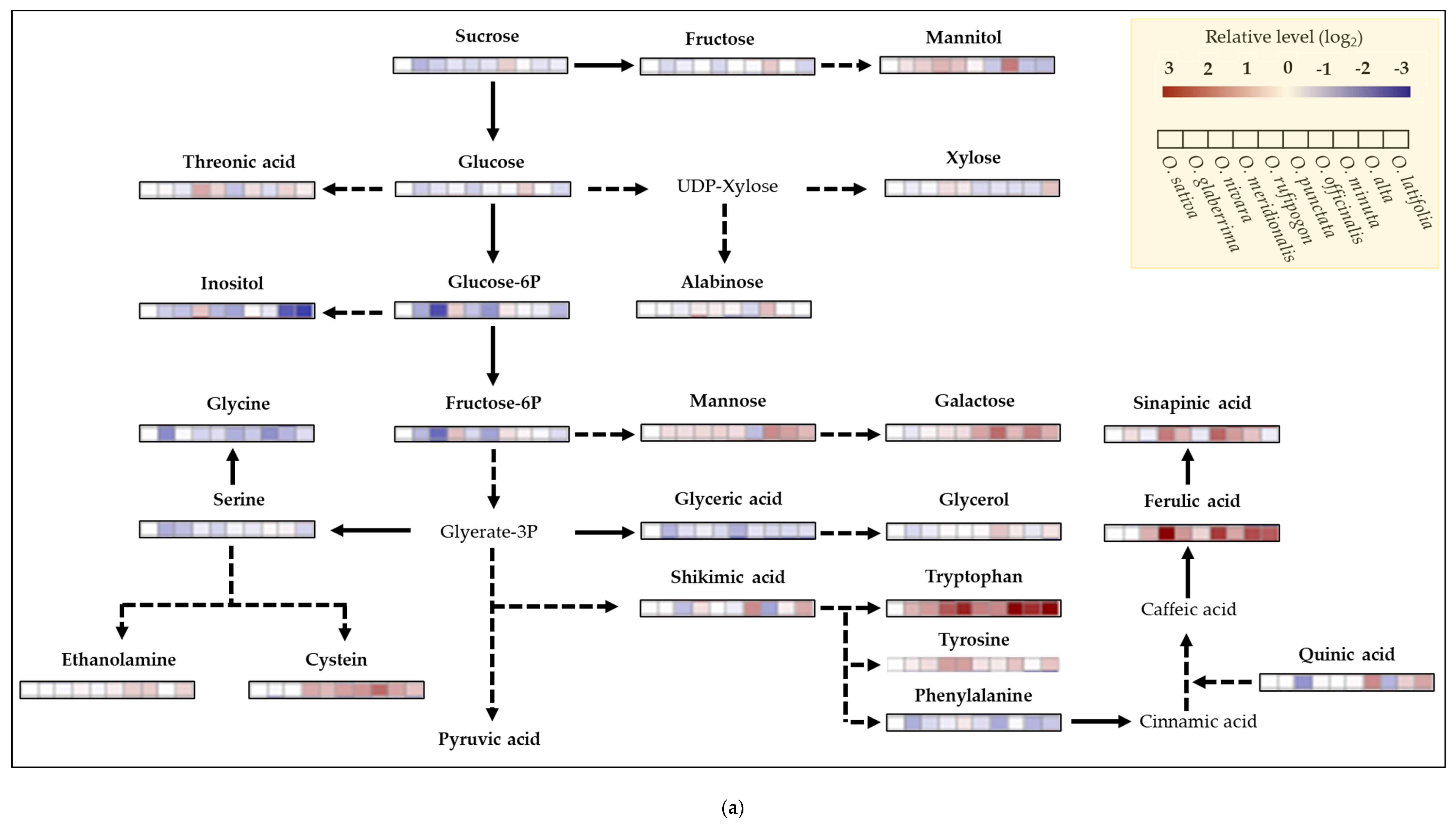
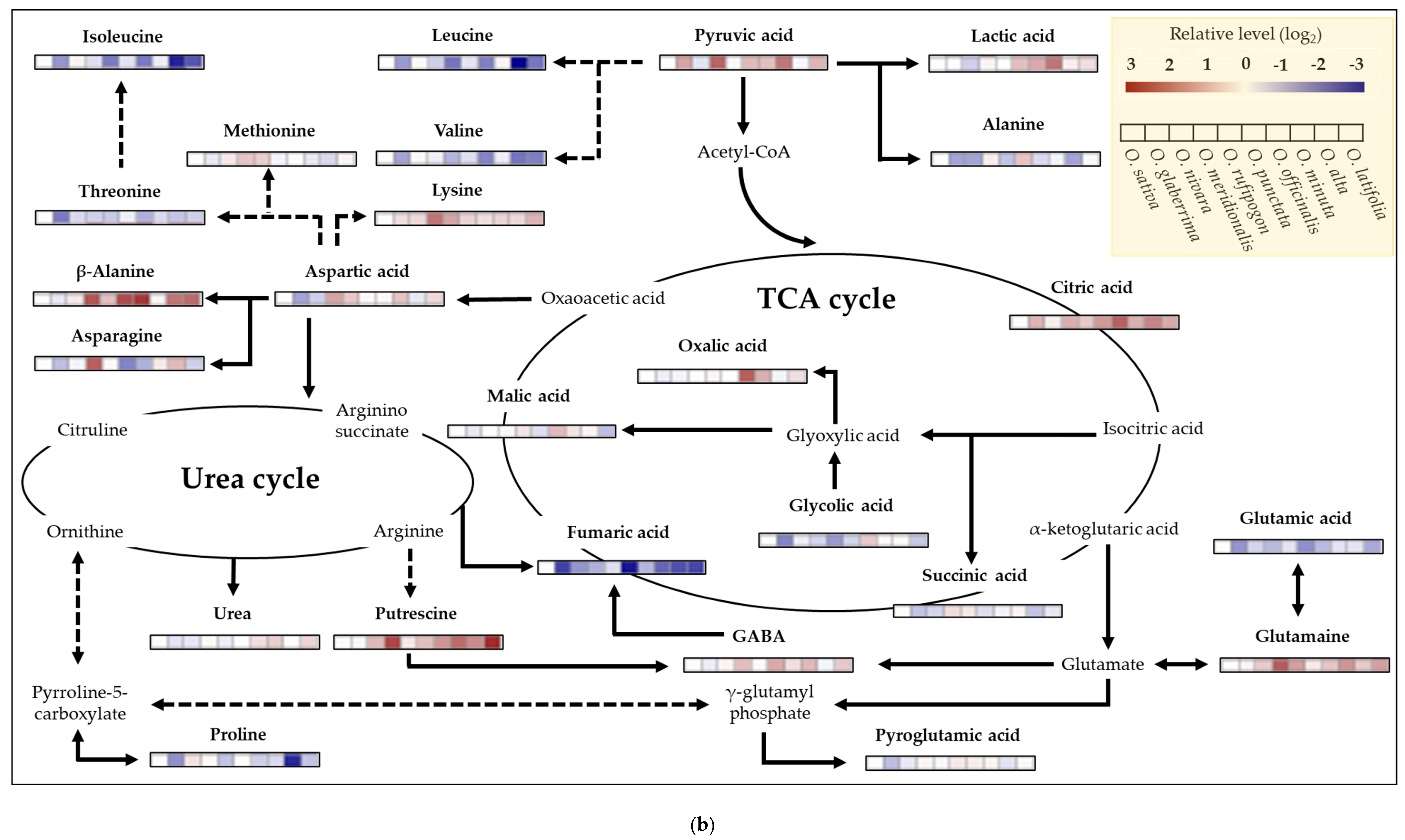
2.6. Metabolite Profiling of Rice Species
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. Phenotypic Characteristics and Photosynthesis
4.3. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
4.4. Extraction and Analysis of Polar Metabolites
4.5. Concentrations of Total Soluble Sugars and Starch
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raza, A.; Razzaq, A.; Mehmood, S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A.; Soni, J.K.; Pawar, N. Physiological response of C3, C4 and CAM plants in changeable climate. J. Pharm. Innov. 2017, 6, 70. [Google Scholar]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Ray, D.K.; West, P.C.; Clark, M.; Gerber, J.S.; Prishchepov, A.V.; Chatterjee, S. Climate change has likely already affected global food production. PLoS ONE 2019, 14, e0217148. [Google Scholar] [CrossRef]
- Wang, J.; Vanga, S.; Saxena, R.; Orsat, V.; Raghavan, V. Effect of Climate Change on the Yield of Cereal Crops: A Review. Climate 2018, 6, 41. [Google Scholar] [CrossRef]
- Ray, D.K.; Gerber, J.S.; Macdonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Lobell, D.B.; Asseng, S. Comparing estimates of climate change impacts from process-based and statistical crop models. Environ. Res. Lett. 2017, 12, 015001. [Google Scholar] [CrossRef]
- Zhu, X.-G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Timm, S.; Bauwe, H. The variety of photorespiratory phenotypes—Employing the current status for future research directions on photorespiration. Plant Biol. 2013, 15, 737–747. [Google Scholar] [CrossRef]
- Christin, P.A.; Osborne, C.P.; Chatelet, D.S.; Columbus, J.T.; Besnard, G.; Hodkinson, T.R.; Garrison, L.M.; Vorontsova, M.S.; Edwards, E.J. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl. Acad. Sci. USA 2013, 110, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Hibberd, J.M.; Sheehy, J.E.; Langdale, J.A. Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr. Opin. Plant Biol. 2008, 11, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Sage, T.L.; Sage, R.F. The Functional Anatomy of Rice Leaves: Implications for Refixation of Photorespiratory CO2 and Efforts to Engineer C4 Photosynthesis into Rice. Plant Cell Physiol. 2009, 50, 756–772. [Google Scholar] [CrossRef] [PubMed]
- Sitch, L.A. Incompatibility Barriers Operating in Crosses of Oryza sativa with Related Species and Genera; Springer: Boston, MA, USA, 1990. [Google Scholar]
- Vaughan, D.A. Genus Oryza L. Current Status of Taxonomy; The International Rice Research Institute: Makati, Philippines, 1989. [Google Scholar]
- Yeo, M.E.; Yeo, A.R.; Flowers, T.J. University of Sussex, Brighton BN1 9QG (United Kingdom)), Photosynthesis and photorespiration in the genus Oryza. J. Exp. Bot. 1994, 45, 553–560. [Google Scholar] [CrossRef]
- Xiuqin, Z.; Ming, Z.; Juntao, X.; Wenzhu, Z.; Dongming, G.; Meiyun, W.; Jun, L.; Ning, Q. Stomata Characteristics of Leaves of High-photosynthetic Efficiency Progenies from a Cross between O.sativa and O.rufipogon and Their Parents. Acta Agron. Sin. 2003, 29, 216–221. [Google Scholar]
- Zhao, M.; Ding, Z.; Lafitte, R.; Sacks, E.; Dimayuga, G.; Holt, D. Photosynthetic characteristics in Oryza species. Photosynthetica 2010, 48, 234–240. [Google Scholar] [CrossRef]
- Haigler, C.H.; Ivanova-Datcheva, M.; Hogan, P.S.; Salnikov, V.V.; Hwang, S.; Martin, K.; Delmer, D.P. Carbon partitioning to cellulose synthesis. Plant Mol. Biol. 2001, 47, 29–51. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport and metabolism control carbon partitioning between stem and grain in rice. J. Exp. Bot. 2021, 72, 4355–4372. [Google Scholar] [CrossRef]
- Fernie, A.R.; Bachem, C.W.B.; Helariutta, Y.; Neuhaus, H.E.; Prat, S.; Ruan, Y.-L.; Stitt, M.; Sweetlove, L.J.; Tegeder, M.; Wahl, V.; et al. Synchronization of developmental, molecular and metabolic aspects of source–sink interactions. Nat. Plants 2020, 6, 55–66. [Google Scholar] [CrossRef]
- Flügel, F.; Timm, S.; Arrivault, S.; Florian, A.; Stitt, M.; Fernie, A.R.; Bauwe, H. The Photorespiratory Metabolite 2-Phosphoglycolate Regulates Photosynthesis and Starch Accumulation in Arabidopsis. Plant Cell 2017, 29, 2537–2551. [Google Scholar] [CrossRef]
- Vaughan, D.A.; Morishima, H.; Kadowaki, K. Diversity in the Oryza genus. Curr. Opin. Plant Biol. 2003, 6, 139–146. [Google Scholar] [CrossRef]
- Banaticla-Hilario, M.C.N.; Sosef, M.S.M.; McNally, K.L.; Hamilton, N.R.S.; Van Den Berg, R.G. Ecogeographic Variation in the Morphology of Two Asian Wild Rice Species, Oryza nivara and Oryza rufipogon. Int. J. Plant Sci. 2013, 174, 896–909. [Google Scholar] [CrossRef]
- Kasem, S.; Waters, D.L.E.; Rice, N.; Shapter, F.M.; Henry, R.J. Whole grain morphology of Australian rice species. Plant Genet. Resour. 2010, 8, 74–81. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Kiran, T.V.; Rao, Y.V.; Subrahmanyam, D.; Rani, N.S.; Bhadana, V.P.; Rao, P.R.; Voleti, S.R. Variation in leaf photosynthetic characteristics in wild rice species. Photosynthetica 2013, 51, 350–358. [Google Scholar] [CrossRef]
- Kondamudi, R.; Swamy, K.N.; Rao, Y.V.; Kiran, T.V.; Suman, K.; Rao, D.S.; Rao, P.R.; Subrahmanyam, D.; Sarla, N.; Ramana Kumari, B.; et al. Gas exchange, carbon balance and stomatal traits in wild and cultivated rice (Oryza sativa L.) genotypes. Acta Physiol. Plant. 2016, 38, 160. [Google Scholar] [CrossRef]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory Metabolism: Genes, Mutants, Energetics, and Redox Signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef]
- Zhou, Q.; Yu, Q.; Wang, Z.; Pan, Y.; Lv, W.; Zhu, L.; Chen, R.; He, G. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ. 2013, 36, 1476–1489. [Google Scholar] [CrossRef]
- Mohanan, K.V.; Mini, C.B. Relative Contribution of Rice Tillers of Different Status Towards Yield. Int. J. Plant Breed. Genet. 2007, 2, 9–12. [Google Scholar] [CrossRef]
- Hu, M.; Lv, S.; Wu, W.; Fu, Y.; Liu, F.; Wang, B.; Li, W.; Gu, P.; Cai, H.; Sun, C.; et al. The domestication of plant architecture in African rice. Plant J. 2018, 94, 661–669. [Google Scholar] [CrossRef]
- Zhao, M.; Renee, J.; Erik, S.; Glenn, D.; Xiuqin, Z. Variation in Leaf Photosynthesis among Wild Species in Genus Oryza and among the Progeny of Selected Crosses of the Wild Species with a Rice Cultivar. Available online: http://www.regional.org.au/au/asa/2004/poster/2/7/1/1060_zhaom.htm (accessed on 2 July 2022).
- Haritha, G.; Vishnukiran, T.; Yugandhar, P.; Sarla, N.; Subrahmanyam, D. Introgressions from Oryza rufipogon Increase Photosynthetic Efficiency of KMR3 Rice Lines. Rice Sci. 2017, 24, 85–96. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Jathar, V.; Ranjan, A. High photosynthesis rate in two wild rice species is driven by leaf anatomy mediating high Rubisco activity and electron transport rate. J. Exp. Bot. 2021, 72, 7119–7135. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.L.; Scafaro, A.P.; Atwell, B.J. Photosynthetic traits of Australian wild rice (Oryza australiensis) confer tolerance to extreme daytime temperatures. Plant Mol. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, N.; Tohya, M.; Nakashima, T.; Ueno, O. Variations in structural, biochemical, and physiological traits of photosynthesis and resource use efficiency in Amaranthus species (NAD-ME-type C4). Plant Prod. Sci. 2017, 20, 300–312. [Google Scholar] [CrossRef]
- Sanchez, P.L.; Wing, R.A.; Brar, D.S. The Wild Relative of Rice: Genomes and Genomics. In Genetics and Genomics of Rice; Springer: New York, NY, USA, 2013; pp. 9–25. [Google Scholar]
- Shanmugam, S.; Boyett, V.A.; Khodakovskaya, M. Enhancement of drought tolerance in rice by silencing of the OsSYT-5 gene. PLoS ONE 2021, 16, e0258171. [Google Scholar] [CrossRef]
- Ogren, W.L. Photorespiration: Pathways, Regulation, and Modification. Annu. Rev. Plant Physiol. 1984, 35, 415–442. [Google Scholar] [CrossRef]
- Kozaki, A.; Takeba, G. Photorespiration protects C3 plants from photooxidation. Nature 1996, 384, 557–560. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Z.; Zhang, Q.; Han, X.; Gu, X.; Lu, T. The molecular cloning and clarification of a photorespiratory mutant, oscdm1, using enhancer trapping. Front. Genet. 2015, 6, 226. [Google Scholar] [CrossRef][Green Version]
- Levey, M.; Timm, S.; Mettler-Altmann, T.; Luca Borghi, G.; Koczor, M.; Arrivault, S.; PM Weber, A.; Bauwe, H.; Gowik, U.; Westhoff, P. Efficient 2-phosphoglycolate degradation is required to maintain carbon assimilation and allocation in the C4 plant Flaveria bidentis. J. Exp. Bot. 2019, 70, 575–587. [Google Scholar] [CrossRef]
- Kubis, A.; Bar-Even, A. Synthetic biology approaches for improving photosynthesis. J. Exp. Bot. 2019, 70, 1425–1433. [Google Scholar] [CrossRef]
- Schulze, S.; Westhoff, P.; Gowik, U. Glycine decarboxylase in C3, C4 and C3–C4 intermediate species. Curr. Opin. Plant Biol. 2016, 31, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Vanloocke, A.; Bernacchi, C.J.; Ort, D.R. The Costs of Photorespiration to Food Production Now and in the Future. Annu. Rev. Plant Biol. 2016, 67, 107–129. [Google Scholar] [CrossRef] [PubMed]
- Busch, F.A. Photorespiration in the context of Rubisco biochemistry, CO2 diffusion and metabolism. Plant J. 2020, 101, 919–939. [Google Scholar] [CrossRef] [PubMed]
- Lindén, P.; Keech, O.; Stenlund, H.; Gardeström, P.; Moritz, T. Reduced mitochondrial malate dehydrogenase activity has a strong effect on photorespiratory metabolism as revealed by13C labelling. J. Exp. Bot. 2016, 67, 3123–3135. [Google Scholar] [CrossRef]
- Tang, Y.; Li, X.; Lu, W.; Wei, X.; Zhang, Q.; Lv, C.; Song, N. Enhanced photorespiration in transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase gene contributes to alleviating low nitrogen stress. Plant Physiol. Biochem. 2018, 130, 577–588. [Google Scholar] [CrossRef]
- Timm, S.; Mielewczik, M.; Florian, A.; Frankenbach, S.; Dreissen, A.; Hocken, N.; Fernie, A.R.; Walter, A.; Bauwe, H. High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS ONE 2012, 7, e42809. [Google Scholar] [CrossRef]
- Zhang, J.; Li, D.; Xu, X.; Ziska, L.H.; Zhu, J.; Liu, G.; Zhu, C. The potential role of sucrose transport gene expression in the photosynthetic and yield response of rice cultivars to future CO2 concentration. Physiol. Plant. 2020, 168, 218–226. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, T.; Huang, L.; Wu, H.; Hu, F.; Zhang, F.; Zhu, L.; Fu, B. Comparative Metabolite Profiling and Hormone Analysis of Perennial and Annual Rice. J. Plant Biol. 2012, 55, 73–80. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Chung, J.-W.; Edwards, G.; Ryu, H.; Kim, T. Photosynthesis, Metabolite Composition and Anatomical Structure of Oryza sativa and Two Wild Relatives, O. grandiglumis and O. alta. Rice Sci. 2017, 24, 218–227. [Google Scholar] [CrossRef]
- Szareski, V.J.; Carvalho, I.R.; da Rosa, T.C.; Dellagostin, S.M.; de Pelegrin, A.J.; Barbosa, M.H.; dos Santos, O.P.; Muraro, D.S.; de Souza, V.Q.; Pedó, T.; et al. Oryza Wild Species: An Alternative for Rice Breeding under Abiotic Stress Conditions. Am. J. Plant Sci. 2018, 09, 1093–1104. [Google Scholar] [CrossRef]
- Wu, D.; Li, Y.; Cao, Y.; Hu, R.; Wu, X.; Zhang, W.; Tao, W.; Xu, G.; Wang, X.; Zhang, Y. Increased glutamine synthetase by overexpression of TaGS1 improves grain yield and nitrogen use efficiency in rice. Plant Physiol. Biochem. 2021, 169, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, K.; Thatikunta, R.; Naik, D.S. Enzymatic Study of Glutamine Synthetase Activity for Validating Nitrogen Use Efficiency in Rice Genotypes. Int. J. Pure Appl. Biosci. 2017, 5, 1416–1423. [Google Scholar] [CrossRef]
- Kim, Y.B.; Park, S.-Y.; Park, C.H.; Park, W.T.; Kim, S.-J.; Ha, S.-H.; Arasu, M.V.; Al-Dhabi, N.A.; Kim, J.K.; Park, S.U. Metabolomics of differently colored Gladiolus cultivars. Appl. Biol. Chem. 2016, 59, 597–607. [Google Scholar] [CrossRef]
- Roe, J.H. The Determination of Sugar in Blood and Spinal Fluid with Anthrone Reagent. J. Biol. Chem. 1955, 212, 335–343. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, J.-Y.; Kim, M.-S.; Cho, Y.-G.; Fernie, A.R.; Sung, J. Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species. Int. J. Mol. Sci. 2022, 23, 8901. https://doi.org/10.3390/ijms23168901
Joo J-Y, Kim M-S, Cho Y-G, Fernie AR, Sung J. Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species. International Journal of Molecular Sciences. 2022; 23(16):8901. https://doi.org/10.3390/ijms23168901
Chicago/Turabian StyleJoo, Jae-Yeon, Me-Sun Kim, Yong-Gu Cho, Alisdair R. Fernie, and Jwakyung Sung. 2022. "Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species" International Journal of Molecular Sciences 23, no. 16: 8901. https://doi.org/10.3390/ijms23168901
APA StyleJoo, J.-Y., Kim, M.-S., Cho, Y.-G., Fernie, A. R., & Sung, J. (2022). Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species. International Journal of Molecular Sciences, 23(16), 8901. https://doi.org/10.3390/ijms23168901







