Abstract
The Dlk1-Dio3 imprinted domain on mouse chromosome 12 contains three well-characterized paternally methylated differentially methylated regions (DMRs): IG-DMR, Gtl2-DMR, and Dlk1-DMR. These DMRs control the expression of many genes involved in embryonic development, inherited diseases, and human cancer in this domain. The first maternal methylation DMR discovered in this domain was the Meg8-DMR, the targets and biological function of which are still unknown. Here, using an enhancer-blocking assay, we first dissected the functional parts of the Meg8-DMR and showed that its insulator activity is dependent on the CCCTC-binding factor (CTCF) in MLTC-1. Results from RNA-seq showed that the deletion of the Meg8-DMR and its compartment CTCF binding sites, but not GGCG repeats, lead to the downregulation of numerous genes on chromosome 12, in particular the drastically reduced expression of Dlk1 and Rtl1 in the Dlk1-Dio3 domain, while differentially expressed genes are enriched in the MAPK pathway. In vitro assays revealed that the deletion of the Meg8-DMR and CTCF binding sites enhances cell migration and invasion by decreasing Dlk1 and activating the Notch1-Rhoc-MAPK/ERK pathway. These findings enhance research into gene regulation in the Dlk1-Dio3 domain by indicating that the Meg8-DMR functions as a long-range regulatory element which is dependent on CTCF binding sites and affects multiple genes in this domain.
1. Introduction
Genome imprinting is a classic epigenetic phenomenon in which some genes are expressed in a parental-origin-specific manner from just one of the two parental chromosomes [1]. To date, approximately 200 imprinted genes have been discovered in mammals [2]. In mice, by utilizing the “homologous recombination” technique, many studies have proved that imprinted genes play important roles not only in the growth of the embryo, placenta and neonate, but also in the regulation of behavior and neurological abnormalities [3]. The disorder of imprinted genes also leads to human diseases such as cancers and some syndromes including Prader–Willi and Angelman syndromes, Silver–Russell syndrome, Beckwith–Wiedemann syndrome, and Albright hereditary osteodystrophy and Uniparental disomy 14 [4].
The majority of imprinted genes are found in clusters, with each cluster being governed by a single imprinting control region (ICR) [5]. ICRs are typically coincident with germline differentially methylated regions (DMRs), which are locations where maternally and paternally derived alleles have distinct methylation statuses. Most of these ICRs acquire methylation in the female germline and are unmethylated in the male germline; only three regions, Igf2-H19, Dlk1-Dio3, and Rasgrf1, acquire methylation during spermatogenesis [6].
Being located on the distal of chromosome 12, the Dlk1-Dio3 imprinted domain contains three paternally expressed protein coding genes (Dlk1, Rtl1, and Dio3) and a series of maternally expressed non-coding genes (such as lncRNA Gtl2, Rian, Mirg, some miRNAs, and snoRNAs) [7,8,9]. In this domain, three paternally methylated DMRs have been well-researched. The most important is IG-DMR, located between Dlk1 and Gtl2, which is a germline DMR and has been proved to be a control element in this region in knockout mice. When it was deleted on a maternally inherited chromosome, maternal to paternal epigenotype transition was detected [10]. The other two DMRs are secondary somatic DMRs: Gtl2-DMR (Meg3-DMR), which flanks Gtl2′s 5′ end [11], and Dlk1-DMR, which is found in exon 5 of Dlk1 [12].
The allelic-specific binding of the CTCF has been identified in the Meg3-DMR and demonstrated to be important in the organization of sub-topologically associating domains (TADs) [13,14]. The CTCF insulator protein is enriched on the boundaries of TADs, which are 3D structures with abundant intra-domain connections, protecting genes and their regulatory components from one another [15]. Existing research recognizes the important role played by the CTCF in the regulation of gene expression in some imprinted domains [16]. It was discovered that a subset of gDMRs displayed the allelic recruitment of the CTCF [17]. In the typical Igf2-H19 domain, the CTCF binds to the unmethylated maternal allele of the H19-ICR, which functions as an insulator and controls the imprinted gene expression in this domain [18,19]. Furthermore, in one study, it was found that the insulator activity of the H19-ICR varies by cell type in mouse mutant models [20].
The first maternally methylated DMR in the Dlk1-Dio3 domain, named Meg8-DMR, was discovered in the second intron of Rian and acquired differential methylation prior to E7.5 [21]. It has been shown that the histone modifications H3K4me3 and H3K9me3 are both abundant in the Meg8-DMR, and that the CTCF binds to it non-allele-specifically in vivo [21]. However, the function of Meg8-DMR and the targets that it can regulate are unknown.
In this study, first, we demonstrated that the Meg8-DMR exhibits insulator activity that is dependent on CTCF binding sites. Then, we dissected the functions of Meg8-DMR and its components using the MLTC-1 cell line as a model. The superb down expression of genes on chromosome 12 was detected in the absence of the Meg8-DMR, as well as CTCF binding sites, especially the paternal genes Dlk1 and Rtl1 in the Dlk1-Dio3 domain. Furthermore, in MLTC-1, the deletion of the Meg8-DMR and CTCF binding sites increased cell migration and invasion and suppressed cell proliferation. When we overexpressed Dlk1 in Meg8-DMR-KO, cell invasion and migration were inhibited as the Notch1-Rhoc-MAPK/ERK signaling pathway was blocked. These findings imply that the Meg8-DMR is a long-range regulatory element that is dependent on CTCF binding sites and regulates several genes in the Dlk1-Dio3 domain.
2. Results
2.1. Meg8-DMR Acts as an Insulator Dependent on the CTCF Binding Sites
The Meg8-DMR, which is located in the second intron of Rian, contains conserved CTCF binding sites along with GGCG repeats (Figure 1A). Many known DMRs that bind to the CTCF, such as H19-ICR, have the enhancer-blocking properties of an insulator, which is one of the classic mechanisms which explain genomic imprinting [22]. To investigate whether the Meg8-DMR also has insulator activity, we used an enhancer-blocking assay, as described previously [23]. The MLTC-1 cell line is a suitable transfection host, and expression of the main genes in the Dlk1-Dio3 domain, such as Dlk1, Rtl1, Gtl2, Rian, and Mirg, can be detected. Furthermore, CTCF can bind to CTCF binding sites in the Meg8-DMR of MLTC-1 (Figure S1). As a result, we chose the MLTC-1 as a model for our assays. We generated constructs (PNI) in which fragments A–E (Figure 1A) could be inserted between the enhancer and promoter that was driving the neo gene (Figure 1B).
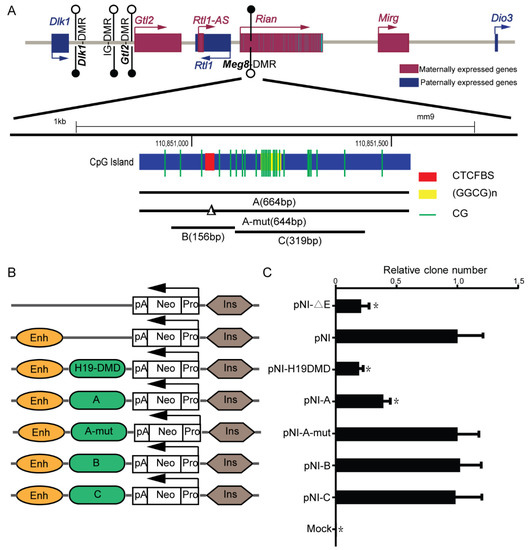
Figure 1.
Meg8-DMR acts as an insulator dependent on the CTCF binding sites. (A) Schematic representation of the Dlk1-Dio3 imprinted domain. Maternally expressed noncoding RNAs are shown in red, whereas, protein coding paternally expressed RNAs are in blue. Black and white circles represent methylated and unmethylated. Meg8-DMR, which is methylated in the maternally inherited chromosome, but unmethylated in the paternally inherited chromosome, is located on the second intron of Rian and contains conserved CTCF-binding sites and GGCG repeats. (B) The constructs of the enhancer-blocking assay were prepared and transfected into MLTC-1 cells. Each construct contained a mouse β-globin 3′HS2 enhancer element and a human Aγ-globin promoter directing transcription of a neo reporter (Neo), and various test fragments were inserted between them. (C) The colony number for each test is expressed relative to the number observed with the control plasmid (PNI). Mock is the blank control group. Error bars, mean ± SEM. n = 3. p values were calculated using t-test. * p < 0.05.
The constructs were introduced into MLTC-1 cells. The clones were counted after four weeks of treatment with G418. When comparing the PNI with a construct without the enhancer, it was shown that the number of clones was reduced by 80%, similar to the previously characterized 1.6kb H19-DMD insulator [18] (Figure 1C). The presence of enhancer-blocking activity in the Meg8-DMR was confirmed by inserting segment A between the enhancer and the promoter, which reduced the number of clones by 60% (Figure 1C). Next, we removed the 20bp CTCF binding motif in fragment A and obtained a similar clone number to that of the PNI (Figure 1C), indicating that fragment A-mut lost enhancer-blocking activity. Based on these results, we concluded that the Meg8-DMR has an insulator function that is dependent on CTCF binding sites.
We also investigated segment B, which only included CTCF binding sites; however, the number of colonies was not considerably reduced (Figure 1C). This demonstrates that Meg8-DMR’s insulator activity is influenced by factors other than CTCF binding sites. Furthermore, the GGCG repeats were shown to be insufficient but necessary as fragment C also lacked the insulator function (Figure 1C).
2.2. Deletion of Meg8-DMR Reduced the Expression of Many Genes on Chromosome 12
To investigate the target(s) and function of the Meg8-DMR, we generated cell line models that knocked out the Meg8-DMR and its predicted functional elements using the CRISPR/Cas9 system. Three sgRNAs were synthesized and inserted in a pairwise combinatorial manner into a modified PX458 vector [24], targeting the full-length Meg8-DMR, CTCF-binding sites (CTCFBS) and GGCG repeats ((GGCG)n) (Figure 2A). Then, they were independently transferred into MLTC-1 cells. Next, using limiting dilution, we were able to obtain monoclonal cells. The genomic DNA isolated from the clones was then amplified using the outer and inner primers, resulting in the identification of three homozygous cell lines (Figure 2B). The peaks on the chromatogram from DNA sequencing revealed that these three clones were correctly deleted versions of the Meg8-DMR, CTCFBS, and (GGCG)n, and they were given the names Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO, respectively (Figure 2C).
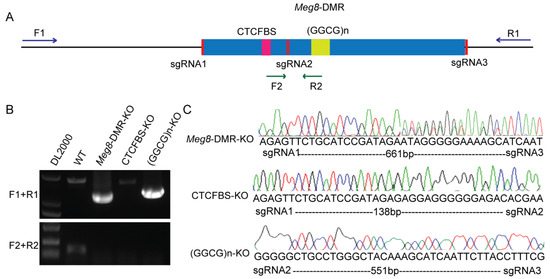
Figure 2.
Generation of Meg8-DMR knockout cell line models via CRISPR/Cas9. (A) A schematic of CRISPR/Cas9-mediated deletion of Meg8-DMR, CTCFBS, and (GGCG)n. (B) PCR identified homozygous clones, F1/R1primers were located outside of Meg8-DMR, and F2/R2 primers were located inside of Meg8-DMR. (C) The deletion clones of Meg8-DMR, CTCFBS and (GGCG)n were identified through sequencing results (Meg8-DMR-KO has double peaks close to the targets because the different copies of the chromosomes may be repaired in various ways).
To learn more about the involvement of the Meg8-DMR in MLTC-1 gene regulation, we used high-throughput sequencing to profile mRNA expression in Meg8-DMR-KO, CTCFBS-KO, (GGCG)n-KO, and WT of MLTC-1. In comparison to the WT, these three cell lines had 476, 485, and 392 differentially expressed genes (DEGs), respectively (Figure 3A). In the Meg8-DMR-KO and CTCFBS-KO cell lines, the majority of the down-regulated genes were found on chromosome 12; however, in the (GGCG)n-KO cell line, the genes were evenly distributed across all chromosomes (Figure 3B). Then, we looked at all the genes detected on chromosome 12, where the Meg8-DMR was discovered. In addition to the DEGs, there were a large number of other genes with decreased expression in the Meg8-DMR-KO and CTCFBS-KO cell lines, particularly those in and near the Dlk1-Dio3 imprinted domain (Figure 3C). Furthermore, RT-qPCR analysis revealed a significant reduction in the expression of the paternally inherited protein coding gene Dlk1, while maternally expressed lncRNAs (Gtl2, Rian, and Mirg) were up-regulated in Meg8-DMR-KO and CTCFBS-KO compared with WT (Figure 3D). Similarly, strand-specific RT-PCR analysis also showed a significant reduction in the expression of the paternally inherited protein coding gene Rtl1 in Meg8-DMR-KO and CTCFBS-KO. Additionally, the expression of these genes in (GGCG)n-KO was comparable to that in WT (Figure 3D).
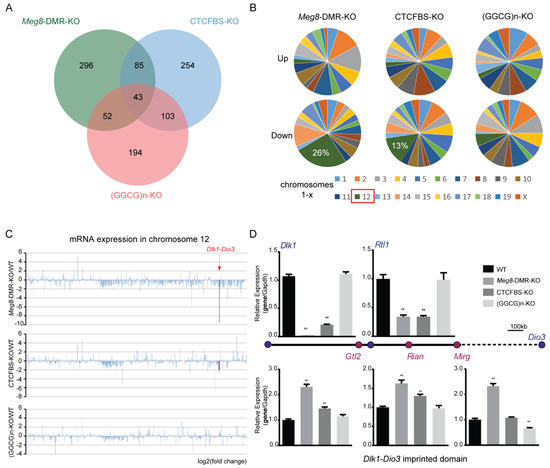
Figure 3.
RNA−seq analysis of the Meg8−DMR deletion cell lines. (A) Venn diagram of upregulated genes and downregulated genes in Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO. (B) Distribution of upregulated genes and downregulated genes in Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO on chromosomes. (C) RNA-seq demonstrates log-fold changes of mRNA expression on chromosome 12 in Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO compared with WT. Significantly reduced expression of Dlk1 and Rtl1 is observed in Meg8-DMR-KO and CTCFBS-KO. Dlk1 and Rtl1 were marked in red. (D) RT-qPCR shows a dramatic reduction in paternal Dlk1 expression and slightly increased expression of maternal Gtl2, Rian, and Mirg lncRNAs in Meg8-DMR-KO and CTCFBS-KO compared with WT. Strand-specific RT-PCR shows a dramatic reduction in paternal Rtl1 expression in Meg8-DMR-KO and CTCFBS-KO compared with WT. The relative expression level was normalized by Gapdh. Error bars, mean ± SEM. n ≥ 3. p values were calculated using t-test. ** p < 0.01.
To further understand the effect of the Meg8-DMR and its component deficiencies in MLTC-1, we compared the DEGs of Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO to WT using KEGG pathway enrichment analysis and gene ontology (GO) enrichment analysis. The MAPK pathway was enriched in all three cell lines, and it was more significant in Meg8-DMR-KO and CTCFBS-KO than in (GGCG)n-KO, according to the KEGG pathway enrichment analysis (Figure 4A–C). The GO enrichment analysis revealed that the cellular component was enriched in the cytoskeleton of Meg8-DMR-KO and CTCFBS-KO but not in ((GGCG)n-KO (Figure 4D).
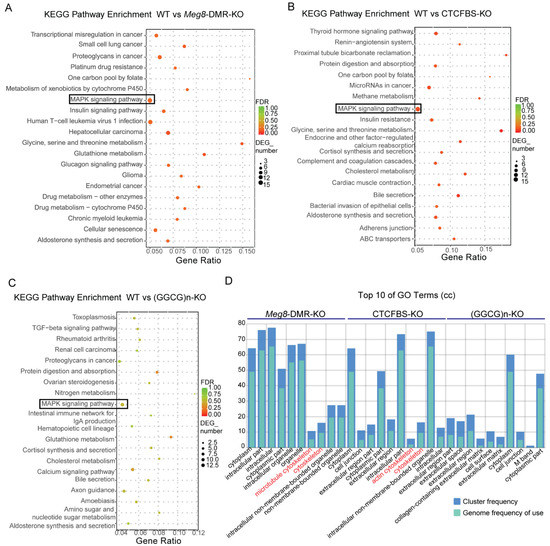
Figure 4.
DEGs Functional Analysis of the Meg8-DMR deletion cell lines. (A–C) Top 20 of KEGG pathway enrichment of genes differentially expressed in Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO. (D) Top 10 GO terms of cellular component (CC) of GO enrichment analysis of differentially expressed genes in Meg8-DMR-KO, CTCFBS-KO and (GGCG)n-KO.
2.3. Meg8-DMR Deletion Enhanced Migration and Invasion of MLTC-1
In order to understand the biological function of Meg8-DMR, we utilized various assays in vitro. According to the results of GO analysis, we initially detected the impact of the deletion of Meg8-DMR on the actin cytoskeleton in MLTC-1 cells by examining the Phalloidin-California Red Conjugate staining of F-actin. The results showed that the cytoskeleton distribution of Meg8-DMR-KO and CTCFBS-KO changed, with filopodia in Meg8-DMR-KO being noticeably more abundant, and filopodia in CTCFBS-KO also being more plentiful than those in WT and (GGCG)n-KO (Figure 5A). The mobility of cells was then examined based on this outcome. In comparison with WT, the migratory speed in Meg8-DMR-KO was much higher, whereas the speed in CTCFBS-KO was also higher but not significant, and the speed in (GGCG)n-KO was essentially unchanged (Figure 5B). Similarly, Transwell analysis revealed that Meg8-DMR-KO and CTCFBS-KO had greater abilities to migrate and invade than WT and (GGCG)n-KO, and that the change in Meg8-DMR-KO was more significant than that in CTCFBS-KO (Figure 5C).
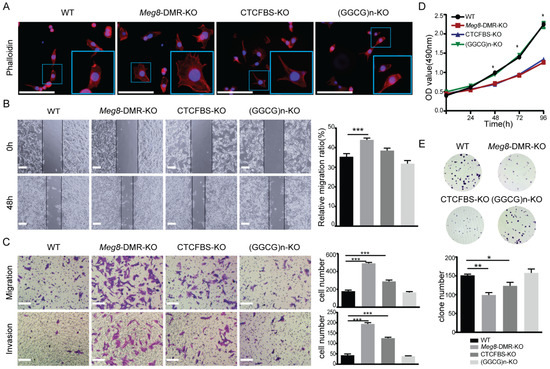
Figure 5.
Meg8-DMR deletion enhanced migration and invasion of MLTC-1. (A) Immunofluorescence staining of Phalloidin-California Red Conjugate in WT, Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO. (B) Confluent monolayers of four cell lines were wounded and after being incubated for an additional 48 h, the relative migration ratio was calculated. (C) Migration and invasion of four cell lines were measured via Transwell assay. (D) Cell proliferation of four cell lines was measured via MTT assay. (E) Colonies grown from cells of four cell lines were counted. Error bars, mean ± SEM. n ≥ 3. p values were calculated using t-test. * p < 0.05, ** p < 0.01, *** p < 0.001, scale bars = 200 μm.
Furthermore, we explored the effects of Meg8-DMR deletion on cell growth using the MTT assay. Interestingly, Meg8-DMR and CTCFBS deletion dramatically decreased cell growth at 72 and 96 h, despite the shift in cell mobility (Figure 5D). The clone creation assay was then utilized to confirm the effects of the Meg8-DMR on proliferation. Consistent with the MTT experiment, the data demonstrated that the efficiency of colony formation in Meg8-DMR-KO and CTCFBS-KO was greatly reduced (Figure 5E). Both assays indicated that deleting (GGCG)n had no effect on cell proliferation (Figure 5D,E).
2.4. Overexpression of Dlk1 in Meg8-DMR-KO Suppressed Cells’ Migration and Invasion by Blocking Notch1-Rhoc-MAPK/ERK
We attempted to rescue Dlk1 expression in the Meg8-DMR-KO clone to study the mechanisms by which Meg8-DMR loss can lead to alterations in MLTC-1 cells, as Dlk1 is one of the most significantly changed genes in the deletion of the Meg8-DMR. The Western blot results demonstrated that the overexpression of Dlk1 was achieved after transfecting pcDNA3.1-Dlk1 into Meg8-DMR-KO (Figure 6A). The exchange of Meg8-DMR-KO was then observed after Dlk1 was overexpressed. First, by examining the Phalloidin-California Red Conjugate staining of F-actin, we discovered that the overexpression of Dlk1 repaired the cytoskeleton distribution alteration caused by Meg8-DMR deletion (Figure 6B). Furthermore, when Dlk1 was overexpressed, the migratory speed of Meg8-DMR-KO was reduced in a wound-healing assay (Figure 6C). Similarly, in Transwell assays, we found that overexpressing Dlk1 inhibited Meg8-DMR-KO migration and invasion (Figure 6D). We then tested some proteins that may have contributed to this process in order to thoroughly examine its molecular underpinnings. According to many studies, Dlk1 is identified as a Notch family protein and the best studied noncanonical Notch ligand [25,26], so we first tested Notch1, one of the four Notch receptors, which has been verified to directly interact with Dlk1 in both yeast and mammalian two-hybrid systems [27,28]. Then, we looked at Rhoc, which is a protein that plays a key role in cell migration [29]. Taking the results of the KEGG analysis, we also tested the Erk1/2. Notch1, Rhoc, and p-Erk1/2 were increased when Meg8-DMR was deleted, and they decreased when Dlk1 was overexpressed in Meg8-DMR-KO (Figure 6A). In conclusion, the deletion of the Meg8-DMR decreased Dlk1 and increased MLTC-1 migration and invasion via Notch1-Rhoc-MAPK/ERK.
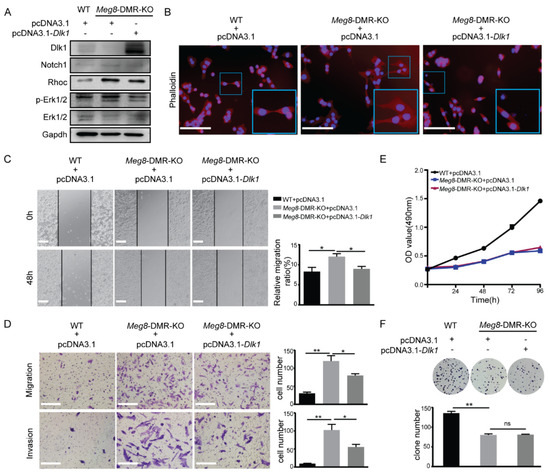
Figure 6.
Overexpression of Dlk1 in Meg8−DMR−KO suppressed cells’ migration and invasion by blocking Notch1−Rhoc−MAPK/ERK. (A) Western blot shows the expression level of Dlk1 was reduced while Nocth1 Rhoc and p-ERK1/2 were activated in Meg8-DMR-KO. When the overexpression vector pcDNA3.1-Dlk1 was transfected into Meg8-DMR-KO, the expression of Nocth1, Rhoc, and p-ERK1/2 was reduced. (B) Immunofluorescence staining of Phalloidin-California Red Conjugate in WT, Meg8-DMR-KO, and Meg8-DMR-KO with Dlk1 overexpression. (C) Confluent monolayers of cells of WT, Meg8-DMR-KO and Meg8-DMR-KO with Dlk1 overexpression were wounded and after being incubated for an additional 48 h, the relative migration ratio was calculated. (D) Migration and invasion of WT, Meg8-DMR-KO, and Meg8-DMR-KO with Dlk1 overexpression were analyzed. (E) Cell proliferation of WT, Meg8-DMR-KO and Meg8-DMR-KO with Dlk1 overexpression was measured via MTT assay. (F) Colonies grown from cells of WT, Meg8-DMR-KO, and Meg8-DMR-KO with Dlk1 overexpression were counted. Error bars, mean ± SEM. n ≥ 3. p values were calculated using t-test. * p < 0.05, ** p < 0.01, ns (non-significant), scale bars = 200 μm.
We also used MTT and colony formation assays to determine whether Dlk1 overexpression had any influence on cell proliferation, and the results demonstrate that it had no effect in Meg8-DMR-KO (Figure 6E,F).
2.5. Absence of Meg8-DMR Has No Impact on the Methylation of Other DMRs in the Dlk1-Dio3 Domain
Including the IG-DMR, which serves as the imprinting control region, there are three known paternally methylated DMRs which can regulate the expression of imprinted genes in the Dlk1-Dio3 domain [30]. We employed bisulfite sequencing to examine DNA methylation in WT, Meg8-DMR-KO, CTCFBS-KO, and (GGCG)n-KO cell lines to determine whether the deletion of the Meg8-DMR can affect the methylation status of these three DMRs.
The results showed that the Dlk1-DMR and IG-DMR were hypermethylated in the WT MLTC-1 cell line, but the Gtl2-DMR was hypomethylated in one half and hypermethylated in the other half (Figure 7A). The methylation status of these three DMRs in the examined cell lines and WT were not significantly different (Figure 7A). This implies that the absence of the Meg8-DMR does not affect the methylation of Dlk1-DMR, IG-DMR, and Gtl2-DMR and in the present case, the deleted Meg8-DMR itself may have contributed to the regulation of gene expression.
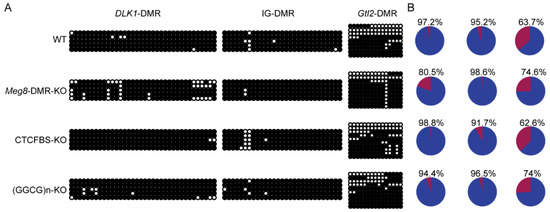
Figure 7.
Methylation of the Dlk1-Dio3 domain in the absence of Meg8-DMR. (A) Bisulfite sequencing analysis of Dlk1-DMR, IG-DMR, and Gtl2-DMR in each cell line. Each CpG dinucleotide is represented with a circle. Each row of circles represents an individual clone sequenced. Black and white circles represent methylated and unmethylated CpGs, respectively. (B) Percentage statistics of methylation status.
3. Discussion
Numerous studies have emphasized the role of imprinted genes in the control of embryonic growth, development, and the etiology of major disorders since their discovery in the 1980s [31]. Dlk1-Dio3, like Igf2-H19, is an imprinted domain in which both maternal and paternal uniparental disomies cause fetal mortality, and it also acts as a parthenogenesis blocker [32,33]. This suggests that the imprinted genes in this region are crucial for growth and development. The important functions of various genes in the Dlk1-Dio3 domain in mammalian development and disease incidence have been established using gene knockout models. As a result, a thorough analysis of epigenetic modifications in this region is necessary to understand the regulation of imprinting and gene expression.
In our investigation, we utilized genetic engineering and targeted epigenetic elements editing in MLTC-1 to dissect the regulatory mechanisms of the Meg8-DMR, the first discovered maternal DMR that has not been well characterized in the Dlk1-Dio3 domain. For the first time, our findings describe the genes that the Meg8-DMR targets, as well as the phenotypes in MLTC-1 when distinct portions of the Meg8-DMR are knocked out. More importantly, our research sheds light on the structure of the Meg8-DMR and identifies the critical role played by CTCF binding sites.
The insulator model, which relies on the transcription factor CTCF that bonds in DMRs to regulate imprinted gene expression in a parental-origin-specific manner in some imprinted clusters, is one of the cis-acting silencing mechanisms that are proposed to govern imprinting [34]. In a common scenario, H19-DMD, a gDMR in the H19-Igf2 domain, acts as an insulator, which is described as an element that sits between the enhancer and the promoter and prevents them from interacting, and subsequently serves as the ICR for this region [18,35,36]. This model has also been identified in other DMRs such as the Rasgrf1-DMR, KvDMR1, and Grb10-DMR [37,38,39]. Using the enhancer blocking experiment in MLTC-1, the Meg8-DMR, which contains conserved CTCF binding sites, was shown to function as an insulator as well as the H19-DMD. In the Meg8-DMR insulator model, the CTCF binding sites are the core element, but this element alone cannot block the enhancer. This means that GGCG repeats are required in this model. Tandem repeats are a common occurrence in imprinting centers [40] and some of them serve as a high frequency of binding sites for transcription factors and participate in the regulation of imprinted gene expression. The tandem repeat in the H19-IGF2 intergenic DMR concentrates the methylation-restricted binding of the key transcription factors ZFP57 and CTCF [41,42]. In the recombination of tandem repeats, recurring imprinting errors can be seen [43]. Additionally, the tandem repeats in the IG-DMR have also been demonstrated to be useful through deletion model construction [44]. However, some repeats are nonfunctional, such as the long-interspersed elements (LINE-1) in the DlK1-Dio3 domain, the deletion of which does not disrupt imprinting and does not result in aberrant phenotypes in mutant mice [45].
Through CRISPR knockout experiments, we revealed that the Meg8-DMR is a functional element in the regulation of gene expression on chromosome 12, especially in the Dlk1-Dio3 domain. An identical gene expression pattern can be obtained by removing the entire Meg8-DMR or simply the CTCF binding sites. This is in line with the findings from the insulator activity assay, and CTCF binding sites were shown to be important in the Meg8-DMR. The cell morphology and behavior assays showed a similar pattern of results. The ablation of the Meg8-DMR altered the distribution of F-actin and increased MLTC-1 cell invasion and migration while decreasing proliferation considerably. When the CTCF-binding sites were knocked out, the phenotypes followed a similar pattern, but not to the same extent. Meanwhile, some data suggest GGCG repeats could be a possible supporter of the Meg8-DMR, because the deletion of GGCG repeats produces fewer alterations in gene expression and essentially no changes in cell morphology and behavior when compared to the Meg8-DMR and CTCF binding sites.
When we rescued Dlk1 expression in the Meg8-DMR deletion cell line, the distribution of F-actin was recovered, and the migration and invasion of cells were suppressed compared to the Meg8-DMR-KO cells without Dlk1 overexpression. Additionally, Notch1 was reduced company with Rhoc and p-ERK1/2. These results are consistent with the findings of a number of previous studies on other cells. Despite the fact that many contradictory studies from different models suggest that Dlk1 can stimulate, inhibit, or have no effect on Notch signaling, it is widely accepted that Dlk1 acts as a Notch1 inhibitor in some cases [46,47,48,49] via cis-inhibition by competing with conventional ligands [50]. In cervical cancer, Notch1 has been reported to lead to changes in migration and invasion by regulating Rhoc [51] and it has also been found to make similar phenotypic contributions with Rhoc in some other cancers such as non-small-cell lung cancer [52] and ovarian cancer [53]. Rhoc, a member of the RHO GTPase family, has been proven to enhance cancer cells migration, invasion, and metastasis through the regulation of cytoskeletal organization [54]. In highly metastatic inflammatory breast cancer, Rhoc can induce motility and invasion through the activation of the MAPK/ERK pathway [55]. These results reveal that the deletion of the Meg8-DMR enhanced the migration and invasion of MLTC-1 through lowering Dlk1 and then activating the Notch1-Rhoc-MAPK/ERK pathway.
However, the cell growth and colony formation of MLTC-1 were not restored when Dlk1 was overexpressed. This effect of deletion of the Meg8-DMR on MLTC-1 could be influenced by other genes. Previous research has shown that many genes in Dlk1-Dio3 have a relationship with cell proliferation. MEG3, a human ortholog of Gtl2, was considered to be a LncRNA tumor suppresser and its re-expression inhibits tumor cell proliferation and colony formation [56]. The suppression of RTL1 was found to inhibit the growth of melanoma cells [57]. It has also been reported that increased MEG8, a human ortholog of Rian, expression inhibited the proliferation of trophoblast and vascular smooth muscle cell lines [58,59], but MIRG inhibition markedly reduced the activity of bone marrow macrophages in terms of cell proliferation [60]. For all of these genes that changed after Meg8-DMR deletion and which may have effects on the proliferation ability of cells, further research is required to investigate the mechanism by which Meg8-DMR deletion affects the cell growth of MLTC-1. Besides, a study examined the role of MEG8 and MEG3 in migration and invasion, with both MEG8 and MEG3 expression causing epithelial–mesenchymal transition-related cell morphological changes and increased cell motility [61]. Additionally, in BEAS-2B cells, MEG8 overexpression facilitated cell proliferation, invasion, and migration [62]. The ability of the HCC cells to proliferate, migrate, and invade was significantly suppressed by MEG8 inhibition [63]. Considering these findings, the relationship between these genes and the cell phenotypes in Meg8-DMR mutant cells requires further investigation.
In addition, there are many microRNAs in the Dlk1-Dio3 domain whose targets can be found on all the chromosomes. We used the ENCORI (https://starbase.sysu.edu.cn/) to analyze the differentially expressed genes which may be the targets of microRNAs. The results are listed in Table S5. In the Meg8-DMR-KO cell line 347 of 476 DEGs could be the targets of 80 microRNAs in the Dlk1-Dio3 domain, and 278 of these genes were outside chromosome 12. For each gene may be the target of multiple microRNAs and each microRNA can also target multiple genes, the correlation between DEGs and microRNAs in the Dlk1-Dio3 is complex and worth exploring for future research.
According to several studies, the IG-DMR and Gtl2-DMR regulate imprinted genes in the Dlk1-Dio3 domain in a coordinated and long-range manner. When the IG-DMR was deleted on the maternally inherited chromosome, all the genes in the cluster lost imprinting, the expression of maternally expressed noncoding genes was silenced, and the paternally expressed protein coding genes had the opposite expression [10]. The same gene expression pattern silencing of maternally expressed genes and activation of PEGs was also reported in the mat deletion of the Gtl2-DMR in mice [64,65]. In Meg8-DMR-KO and CTCFBS-KO, the methylation status of these two DMRs was maintained, indicating the direct regulatory function of the Meg8-DMR by itself, rather than through the other two DMRs. Although the majority of known differentially methylated regions have regulatory functions in non-methylated alleles [1,66], the Meg8-DMR which is methylated (data not shown) in MLTC-1 is still functional. This may because the CTCF binds to this binding site in a biallelic manner by not actually containing CpG dinucleotide [21]. In our study, the necessity of CTCF binding to the Meg8-DMR was shown through the insulator activity assay and the deletion of CTCF binding sites, which resulted in the change in gene expression and cell phenotype. More research is needed to determine how the Meg8-DMR regulates gene expression in the Dlk1-Dio3 domain through binding with the CTCF.
4. Materials and Methods
4.1. Cell Culture
The MLTC-1 (ATCC CRL-2065) cells were cultured in RPIM1640 (GIBCO, Carlsbad, CA, USA) containing 10% fetal bovine serum with penicillin/streptomycin at 37 °C with 5% CO2 in a humidified incubator.
4.2. The Enhancer Blocking Assay
The enhancer blocking assay was performed as described previously [23]. The human Aγ-globin promoter and a mouse β-globin 3′HS2 enhancer element were used to activate a gene for neomycin resistance in each reporter. 2.5 × 105 cells of MLTC-1 were seeded in each well of 12-well plates; after 12 h, an equal number of moles of each construct was transfected into the MLTC-1 cells in each well using Lipofectamine™ 3000 (#L3000015, Invitrogen, Carlsbad, CA, USA), and cells of one well were not transfected with any constructs as a blank control. Then, 48 h after transfection, 5 × 105 cells of each well were seeded in new dishes and cultured in the medium containing 150 μg/mL G418. After 3–4 weeks, when the cells in the blank control were all dead, these cells were fixed with 4% PFA for 30 min and then stained with 0.1% crystal violet for 30 min. At last, the number of colonies was counted and normalized to that obtained with pNI. An equal number of moles of each construct were transfected into the MLTC-1 cells in order to normalize the copy number among the various mutants. After 4 weeks, the number of colonies that were resistant to G418 was counted. Three independent replications were performed. Primer sequences of the fragments that were inserted in each reporter are listed in Table S1.
4.3. CRISPR/Cas9-Mediated Deletion of Meg8-DMR, CTCF Binding Sites and GGCG Repeats
The single-guide RNAs for deletion were designed by using http://crispr.mit.edu/ (accessed on 3 November 2014), and http://crispr.dbcls.jp/ (accessed on 8 September 2016). SgRNAs were annealed with NEBuffer2 for 2 min at 90 °C, 1 h at 37 °C, and finally 1 h at room temperature. BbsI and BsaI (NEB, Ispawich, MA, USA) were used to insert annealed double-stranded DNA into the CRISPR/Cas9 PX458 vector, which was modified [24]. Then, the purified recombinant plasmids were transfected into MLTC-1 cells in 24-well plates. After being screened using puromycin, the cells were seeded into 96-well plates with limiting dilution. Two weeks later, the DNA of the clones was isolated and then identified via PCR using external and internal primers of Meg8-DMR. Primer sequences are listed in Table S1.
4.4. DNA Extraction and Methylation Analysis
Each cell line’s genomic DNA was obtained using a conventional procedure that included phenol/chloroform extraction and ethanol precipitation. The isolated DNA was modified with bisulfite according to the manufacturer’s instructions using the EZ DNA Methylation-Gold Kit (Zymo Research, cat# D5005, Irvine, CA, USA). ZymoTaq™ DNA Polymerase (Zymo Research, cat#E2001, Irvine, CA, USA) was used to amplify bisulfite-treated materials using a nested PCR. Table S1 lists the primer sequences. The PCR products were cloned into the pMD19T vector. The subclones were then identified and chosen for sequencing.
4.5. RNA Extraction and RNA-Seq Analysis
Total RNAs were prepared from each cell line by RNAiso Plus (TaKaRa, Dalian, China) according to the manufacturer’s protocol and then given to Personal Bio Company for transcriptome sequencing. The library was then subjected to Paired-End sequencing on the Illumina Hiseq sequencing technology after it was produced. Raw data were filtered using Cutadapt, and quality control was performed using FastQC. Paired-end Fastq files were mapped to ENSEMBL Mouse assembly GRCm38 (mm10) using Tophat2. DESeq was used to analyze the differentially expressed genes (DEGs), and absolute fold change >2 or <0.5 and p-value < 0.05 were used to identify DEGs. All the DEGs of the three cell lines are listed in Tables S2–S4.
4.6. qRT-PCR and Strand-Specific RT-PCR
The cDNA was synthesized via reverse transcription using the PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa). Then, the qRT-PCR was performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa) on an ABI ViiA 7 system. The results were analyzed using the relative quantitative method, and the mRNA expression of genes was normalized with Gapdh. The primers for qRT-PCR are shown in Table S1.
A strand-specific reverse transcriptase reaction was performed using the method a previous study reported [67] for the detection of paternally expressed Rtl1 using the PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa). The PCR results were quantified with Image J 1.53e software (Wayne Rasband and contributors National Institutes of Health, USA, http://imagej.nih.gov/ij), and the results were normalized to Gapdh expression.
4.7. Immunofluorescence
The cells of each cell line were cultured on 48-well chamber slides and fixed with 4% PFA for 30 min. After being washed three times with PBS, fixed cells were incubated in 0.1% Triton X-100 in PBS for 5 min. Then, the cells were stained with Phalloidin-California Red Conjugate (#B8325, APExBIO) at 37 °C for 1 h. Finally, the slides with cells were incubated with DAPI (Invitrogen).
4.8. Wound Healing, Cell Migration and Invasion
For the wound-healing assay, each cell line was cultured into a confluent monolayer and then scratched with a 10 μL pipette tip. Medium, which was refreshed into serum-free, was added to culture cells to close the wounds for 48 h.
5 × 104 cells of each cell line were placed on the top sides of Transwell chambers (Corning, Kennebunk, ME, USA), after incubation for 24 h, while the Transwell chambers were coated with Matrigel (Corning, Bedford, MA, USA) for 48 h, and the lower surface cells were fixed with 4% paraformaldehyde (PFA) at room temperature for 30 min and stained with 0.1% crystal violet for 30 min. Four independent fields were counted under the microscope.
4.9. MTT and Colony Formation Assays
3 × 103 cells of each cell line were seeded in each well of 96-well plates. After every 24 h at the indicated time, 20 μL of MTT were added to 200 μL of culture medium per well. After 4 h of incubation at 37 °C, the medium was removed and 100 μL of dimethyl sulfoxide was added. The absorbance was measured at a wavelength of 490 nm in a plate reader (Tecan i-control infinite 200).
3 × 102 cells of each cell line were seeded in each well of 6-well plates, and after being cultured for 3 weeks, these cells were fixed with 4% PFA for 30 min and then stained with 0.1% crystal violet for 30 min, and then the number of colonies was counted.
4.10. Western Blotting Analysis
The total protein content was extracted from cells of each cell line by using RIPA lysis buffer and then determined by using the BCA method. The whole protein of each sample was separated using 10% SDS-PAGE and transferred onto PVDF membranes for blotting. The antibodies used in this study were Gapdh (#60004-1-Ig Proteintech, Wuhan, China), Dlk1 (#ab210471, Abcam, Cambridge, UK), Notch1 (#4ab094535, 4A BIOTECH, Beijing, China), Rhoc (#4ab090482, 4A BIOTECH, Beijing, China), Erk1/2 (#BF8004, Affinity Biosciences, Changzhou, China), and p-Erk1/2 (#AF1015, Affinity Biosciences, Changzhou, China). The full length of Dlk1 cDNA was cloned to pcDNA3.1 to overexpress Dlk1.
5. Conclusions
In this study, we demonstrated that the Meg8-DMR has insulator activity depending on CTCF-binding sites in MLTC-1. The deletion of the Meg8-DMR altered the expression of genes in the Dlk1-Dio3 imprint domain, which suppressed cell growth and promoted invasion and migration, with CTCF binding sites playing a key role. We preliminarily identified the targets and biology function of the Meg8-DMR in MLTC-1; however, more research is needed to understand how it regulates gene expression.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23158828/s1. Reference [24] is cited in the supplementary materials.
Author Contributions
Q.W. conceived the project and designed the experiment. X.H. performed the experiments and wrote the paper. H.H. designed the experiments and edited the paper. L.S. established stable transfected cell lines. S.C. and H.Y. carried out visualization and edited the paper. X.Z. performed the methylation analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (Nos.31771601).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
RNA-seq sequencing data have been deposited in GEO under accession number
Acknowledgments
We would like to thank Tie-bo Zeng for technical support with the enhancer-blocking assay.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, A.C.; Bourc’his, D. The discovery and importance of genomic imprinting. eLife 2018, 7, e42368. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Bartolomei, M.S. Genomic Imprinting in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G. Genomic imprinting disorders in humans: A mini-review. J. Assist. Reprod. Genet. 2009, 26, 477–486. [Google Scholar] [CrossRef]
- Lin, S.; Ferguson-Smith, A.C.; Schultz, R.M.; Bartolomei, M.S. Nonallelic Transcriptional Roles of CTCF and Cohesins at Imprinted Loci. Mol. Cell. Biol. 2011, 31, 3094–3104. [Google Scholar] [CrossRef]
- Cleaton, M.A.; Edwards, C.A.; Ferguson-Smith, A.C. Phenotypic Outcomes of Imprinted Gene Models in Mice: Elucidation of Pre- and Postnatal Functions of Imprinted Genes. Annu. Rev. Genom. Hum. Genet. 2014, 15, 93–126. [Google Scholar] [CrossRef]
- da Rocha, S.T.; Edwards, C.A.; Ito, M.; Ogata, T.; Ferguson-Smith, A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008, 24, 306–316. [Google Scholar] [CrossRef]
- Hagan, J.P.; O’Neill, B.L.; Stewart, C.L.; Kozlov, S.V.; Croce, C.M. At Least Ten Genes Define the Imprinted Dlk1-Dio3 Cluster on Mouse Chromosome 12qF1. PLoS ONE 2009, 4, e4352. [Google Scholar] [CrossRef]
- Seitz, H.; Royo, H.; Bortolin, M.L.; Lin, S.P.; Ferguson-Smith, A.C.; Cavaillé, J. A Large Imprinted microRNA Gene Cluster at the Mouse Dlk1-Gtl2 Domain. Genome Res. 2004, 14, 1741–1748. [Google Scholar] [CrossRef]
- Lin, S.P.; Youngson, N.; Takada, S.; Seitz, H.; Reik, W.; Paulsen, M.; Cavaille, J.; Ferguson-Smith, A.C. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003, 35, 97–102. [Google Scholar] [CrossRef]
- Nowak, K.; Stein, G.; Powell, E.; He, L.M.; Naik, S.; Morris, J.; Marlow, S.; Davis, T.L. Establishment of paternal allele-specific DNA methylation at the imprinted mouseGtl2locus. Epigenetics 2011, 6, 1012–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gagne, A.; Hochman, A.; Qureshi, M.; Tong, C.; Arbon, J.; McDaniel, K.; Davis, T.L. Analysis of DNA methylation acquisition at the imprinted Dlk1 locus reveals asymmetry at CpG dyads. Epigenetics Chromatin 2014, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.L.; Wu, Y.Q.; Kwabi-Addo, B.; Coveler, K.J.; Reid Sutton, V.; Shaffer, L.G. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosom. Res. 2005, 13, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Llères, D.; Moindrot, B.; Pathak, R.; Piras, V.; Matelot, M.; Pignard, B.; Marchand, A.; Poncelet, M.; Perrin, A.; Tellier, V.; et al. CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains. Genome Biol. 2019, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Feil, R. Differential 3D chromatin organization and gene activity in genomic imprinting. Curr. Opin. Genet. Dev. 2020, 61, 17–24. [Google Scholar] [CrossRef]
- Prickett, A.R.; Barkas, N.; McCole, R.B.; Hughes, S.; Amante, S.M.; Schulz, R.; Oakey, R.J. Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res. 2013, 23, 1624–1635. [Google Scholar] [CrossRef]
- Bell, A.C.; Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000, 405, 482–485. [Google Scholar] [CrossRef]
- Schoenherr, C.J.; Levorse, J.M.; Tilghman, S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003, 33, 66–69. [Google Scholar] [CrossRef]
- Ideraabdullah, F.Y.; Thorvaldsen, J.L.; Myers, J.A.; Bartolomei, M.S. Tissue-specific insulator function at H19/Igf2 revealed by deletions at the imprinting control region. Hum. Mol. Genet. 2014, 23, 6246–6259. [Google Scholar] [CrossRef]
- Zeng, T.B.; He, H.J.; Han, Z.B.; Zhang, F.W.; Huang, Z.J.; Liu, Q.; Cui, W.; Wu, Q. DNA methylation dynamics of a maternally methylated DMR in the mouseDlk1-Dio3domain. FEBS Lett. 2014, 588, 4665–4671. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A.; Ferguson-Smith, A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007, 19, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; West, A.G.; Felsenfeld, G. The Protein CTCF Is Required for the Enhancer Blocking Activity of Vertebrate Insulators. Cell 1999, 98, 387–396. [Google Scholar] [CrossRef]
- Cui, S.; Wu, Q.; Liu, M.; Su, M.; Liu, S.; Shao, L.; Han, X.; He, H. EphA2 super-enhancer promotes tumor progression by recruiting FOSL2 and TCF7L2 to activate the target gene EphA2. Cell Death Dis. 2021, 12, 264. [Google Scholar] [CrossRef] [PubMed]
- Pittaway, J.F.H.; Lipsos, C.; Mariniello, K.; Guasti, L. The role of delta-like non-canonical Notch ligand 1 (DLK1) in cancer. Endocr. Relat. Cancer 2021, 28, R271–R287. [Google Scholar] [CrossRef]
- Perramón, M.; Jiménez, W. Pituitary Tumor-Transforming Gene 1/Delta like Non-Canonical Notch Ligand 1 Signaling in Chronic Liver Diseases. Int. J. Mol. Sci. 2022, 23, 6897. [Google Scholar] [CrossRef]
- Baladrón, V.; Ruiz-Hidalgo, M.J.; Nueda, M.L.; Díaz-Guerra, M.J.; García-Ramírez, J.J.; Bonvini, E.; Gubina, E.; Laborda, J. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp. Cell Res. 2005, 303, 343–359. [Google Scholar] [CrossRef]
- Traustadóttir, G.; Jensen, C.H.; Thomassen, M.; Beck, H.C.; Mortensen, S.B.; Laborda, J.; Baladrón, V.; Sheikh, S.P.; Andersen, D.C. Evidence of non-canonical NOTCH signaling: Delta-like 1 homolog (DLK1) directly interacts with the NOTCH1 receptor in mammals. Cell. Signal. 2016, 28, 246–254. [Google Scholar] [CrossRef]
- Lou, Y.; Jiang, Y.; Liang, Z.; Liu, B.; Li, T.; Zhang, D. Role of RhoC in cancer cell migration. Cancer Cell Int. 2021, 21, 527. [Google Scholar] [CrossRef]
- Takada, S.; Paulsen, M.; Tevendale, M.; Tsai, C.E.; Kelsey, G.; Cattanach, B.M.; Ferguson-Smith, A.C. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: Implications for imprinting control from comparison with Igf2-H19. Hum. Mol. Genet. 2002, 11, 77–86. [Google Scholar] [CrossRef]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; the Erice Imprinting Group. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Georgiades, P.; Watkins, M.; Surani, M.A.; Ferguson-Smith, A.C. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 2000, 127, 4719–4728. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Kumagai, T.; Kawahara, M.; Ogawa, H.; Hiura, H.; Obata, Y.; Takano, R.; Kono, T. Regulated expression of two sets of paternally imprinted genes is necessary for mouse parthenogenetic development to term. Reproduction 2006, 131, 481–488. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef]
- Hark, A.T.; Schoenherr, C.J.; Katz, D.J.; Ingram, R.S.; Levorse, J.M.; Tilghman, S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000, 405, 486–489. [Google Scholar] [CrossRef]
- Thorvaldsen, J.L.; Duran, K.L.; Bartolomei, M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998, 12, 3693–3702. [Google Scholar] [CrossRef]
- Yoon, B.; Herman, H.; Hu, B.; Park, Y.J.; Lindroth, A.; Bell, A.; West, A.G.; Chang, Y.; Stablewski, A.; Piel, J.C.; et al. Rasgrf1 Imprinting Is Regulated by a CTCF-Dependent Methylation-Sensitive Enhancer Blocker. Mol. Cell. Biol. 2005, 25, 11184–11190. [Google Scholar] [CrossRef]
- Fitzpatrick, G.V.; Pugacheva, E.M.; Shin, J.-Y.; Abdullaev, Z.; Yang, Y.; Khatod, K.; Lobanenkov, V.V.; Higgins, M.J. Allele-Specific Binding of CTCF to the Multipartite Imprinting Control Region KvDMR1. Mol. Cell. Biol. 2007, 27, 2636–2647. [Google Scholar] [CrossRef]
- Shiura, H.; Nakamura, K.; Hikichi, T.; Hino, T.; Oda, K.; Suzuki-Migishima, R.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal Chromosome 11 leading to severe pre- and postnatal growth retardation. Hum. Mol. Genet. 2009, 18, 1424–1438. [Google Scholar] [CrossRef]
- Hutter, B.; Helms, V.; Paulsen, M. Tandem repeats in the CpG islands of imprinted genes. Genomics 2006, 88, 323–332. [Google Scholar] [CrossRef]
- Beygo, J.; Citro, V.; Sparago, A.; De Crescenzo, A.; Cerrato, F.; Heitmann, M.; Rademacher, K.; Guala, A.; Enklaar, T.; Anichini, C.; et al. The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum. Mol. Genet. 2013, 22, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Sparago, A.; Cerrato, F.; Riccio, A. Is ZFP57 binding to H19/IGF2:IG-DMR affected in Silver-Russell syndrome? Clin. Epigenetics 2018, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- De Crescenzo, A.; Coppola, F.; Falco, P.; Bernardo, I.; Ausanio, G.; Cerrato, F.; Falco, L.; Riccio, A. A novel microdeletion in the IGF2/H19 imprinting centre region defines a recurrent mutation mechanism in familial Beckwith–Wiedemann syndrome. Eur. J. Med Genet. 2011, 54, e451–e454. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Hara, S.; Kato, T.; Tamano, M.; Muramatsu, A.; Asahara, H.; Takada, S. A tandem repeat array in IG-DMR is essential for imprinting of paternal allele at the Dlk1–Dio3 domain during embryonic development. Hum. Mol. Genet. 2018, 27, 3283–3292. [Google Scholar] [CrossRef]
- Soares, M.L.; Edwards, C.A.; Dearden, F.L.; Ferrón, S.R.; Curran, S.; Corish, J.A.; Rancourt, R.C.; Allen, S.E.; Charalambous, M.; Ferguson-Smith, M.A.; et al. Targeted deletion of a 170-kb cluster of LINE-1 repeats and implications for regional control. Genome Res. 2018, 28, 345–356. [Google Scholar] [CrossRef]
- Surmacz, B.; Noisa, P.; Risner-Janiczek, J.R.; Hui, K.; Ungless, M.; Cui, W.; Li, M. DLK1 Promotes Neurogenesis of Human and Mouse Pluripotent Stem Cell-Derived Neural Progenitors Via Modulating Notch and BMP Signalling. Stem Cell Rev. Rep. 2012, 8, 459–471. [Google Scholar] [CrossRef]
- Rodríguez, P.; Higueras, M.A.; González-Rajal, A.; Alfranca, A.; Fierro-Fernández, M.; García-Fernández, R.A.; Ruiz-Hidalgo, M.J.; Monsalve, M.; Rodríguez-Pascual, F.; Redondo, J.M.; et al. The non-canonical NOTCH ligand DLK1 exhibits a novel vascular role as a strong inhibitor of angiogenesis. Cardiovasc. Res. 2012, 93, 232–241. [Google Scholar] [CrossRef]
- Müller, D.; Cherukuri, P.; Henningfeld, K.; Poh, C.H.; Wittler, L.; Grote, P.; Schlüter, O.; Schmidt, J.; Laborda, J.; Bauer, S.R.; et al. Dlk1 Promotes a Fast Motor Neuron Biophysical Signature Required for Peak Force Execution. Science 2014, 343, 1264–1266. [Google Scholar] [CrossRef]
- García-Gallastegui, P.; Ibarretxe, G.; Garcia-Ramírez, J.J.; Baladrón, V.; Aurrekoetxea, M.; Nueda, M.L.; Naranjo, A.I.; Santaolalla, F.; Rey, A.S.D.; Laborda, J.; et al. DLK1 regulates branching morphogenesis and parasympathetic innervation of salivary glands through inhibition of NOTCH signalling. Biol. Cell 2014, 106, 237–253. [Google Scholar] [CrossRef]
- Traustadóttir, G.; Lagoni, L.V.; Ankerstjerne, L.B.S.; Bisgaard, H.C.; Jensen, C.H.; Andersen, D.C. The imprinted gene Delta like non-canonical Notch ligand 1 (Dlk1) is conserved in mammals, and serves a growth modulatory role during tissue development and regeneration through Notch dependent and independent mechanisms. Cytokine Growth Factor Rev. 2019, 46, 17–27. [Google Scholar] [CrossRef]
- Srivastava, S.; Ramdass, B.; Nagarajan, S.; Rehman, M.; Mukherjee, G.; Krishna, S. Notch1 regulates the functional contribution of RhoC to cervical carcinoma progression. Br. J. Cancer 2010, 102, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ren, H.; Jiang, J.; Wang, Q.; Wudu, M.; Zhang, Q.; Su, H.; Wang, C.; Jiang, L.; Qiu, X. KIAA 0247 inhibits growth, migration, invasion of non-small-cell lung cancer through regulating the Notch pathway. Cancer Sci. 2018, 109, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.F.; Zhao, Y.; Lu, H.; Yang, X.F.; Xiu, Y.L.; Zhao, S.; Liu, J.M.; Zhu, Z.T.; Sun, H.Z.; Liu, Y.P.; et al. The role of RhoC in epithelial-to-mesenchymal transition of ovarian carcinoma cells. BMC Cancer 2014, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Pranatharthi, A.; Ross, C.; Srivastava, S. RhoC: A fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res. 2019, 38, 328. [Google Scholar] [CrossRef]
- van Golen, K.L.; Bao, L.W.; Pan, Q.; Miller, F.R.; Wu, Z.F.; Merajver, S.D. Mitogen activated protein kinase pathway is involved in RhoC GTPase induced motility, invasion and angiogenesis in inflammatory breast cancer. Clin. Exp. Metastasis 2002, 19, 301–311. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef]
- Fan, G.; Ye, D.; Zhu, S.; Xi, J.; Guo, X.; Qiao, J.; Wu, Y.; Jia, W.; Wang, G.; Fan, G.; et al. RTL1 promotes melanoma proliferation by regulating Wnt/β-catenin signalling. Oncotarget 2017, 8, 106026–106037. [Google Scholar] [CrossRef]
- Sheng, F.; Sun, N.; Ji, Y.; Ma, Y.; Ding, H.; Zhang, Q.; Yang, F.; Li, W. Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion. J. Cell. Biochem. 2019, 120, 17378–17390. [Google Scholar] [CrossRef]
- Zhang, B.; Dong, Y.; Zhao, Z. LncRNA MEG8 regulates vascular smooth muscle cell proliferation, migration and apoptosis by targeting PPARα. Biochem. Biophys. Res. Commun. 2019, 510, 171–176. [Google Scholar] [CrossRef]
- Ling, L.; Hu, H.L.; Liu, K.Y.; Ram, Y.I.; Gao, J.L.; Cao, Y.M. Long noncoding RNA MIRG induces osteoclastogenesis and bone resorption in osteoporosis through negative regulation of miR-1897. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10195–10203. [Google Scholar] [CrossRef]
- Terashima, M.; Ishimura, A.; Wanna-Udom, S.; Suzuki, T. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J. Biol. Chem. 2018, 293, 18016–18030. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Shang, P.; Song, X. LncRNA MEG8 promotes tumor progression of non-small cell lung cancer via regulating miR-107/CDK6 axis. Anti-Cancer Drugs 2020, 31, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Yan, W.; Li, Q.Y.; Zhu, A.K.; Tan, B.Q.; Dong, R.; Zou, X.Z.; Liu, T. LncRNA MEG8 plays an oncogenic role in hepatocellular carcinoma progression through miR-367-3p/14-3-3ζ/TGFβR1 axis. Neoplasma 2021, 68, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cheunsuchon, P.; Nakayama, Y.; Lawlor, M.W.; Zhong, Y.; Rice, K.A.; Zhang, L.; Zhang, X.; Gordon, F.E.; Lidov, H.G.; et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 2010, 137, 2643–2652. [Google Scholar] [CrossRef]
- Zhu, W.; Botticelli, E.M.; Kery, R.E.; Mao, Y.; Wang, X.; Yang, A.; Wang, X.; Zhou, J.; Zhang, X.; Soberman, R.J.; et al. Meg3-DMR, not the Meg3 gene, regulates imprinting of the Dlk1-Dio3 locus. Dev. Biol. 2019, 455, 10–18. [Google Scholar] [CrossRef]
- Barlow, D.P. Genomic Imprinting: A Mammalian Epigenetic Discovery Model. Annu. Rev. Genet. 2011, 45, 379–403. [Google Scholar] [CrossRef]
- Riordan, J.D.; Keng, V.W.; Tschida, B.R.; Scheetz, T.E.; Bell, J.B.; Podetz-Pedersen, K.M.; Moser, C.D.; Copeland, N.G.; Jenkins, N.A.; Roberts, L.R.; et al. Identification of Rtl1, a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis. PLoS Genet. 2013, 9, e1003441. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).