Evaluation of the Molecular Landscape in PD-L1 Positive Metastatic NSCLC: Data from Campania, Italy
Abstract
:1. Introduction
2. Results
2.1. Patient and Sample Characteristics
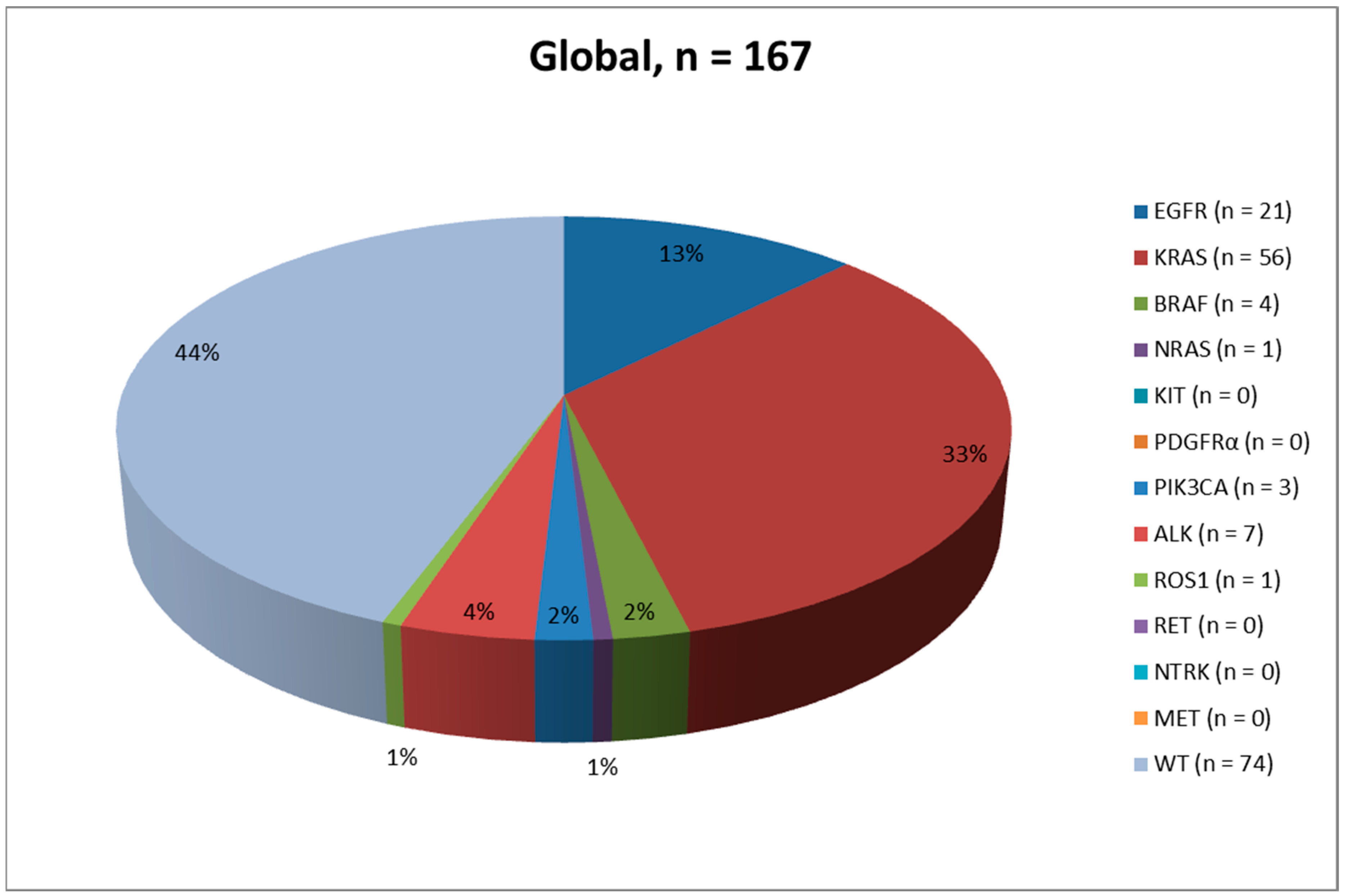
2.2. PD-L1 Status and Molecular Evaluation
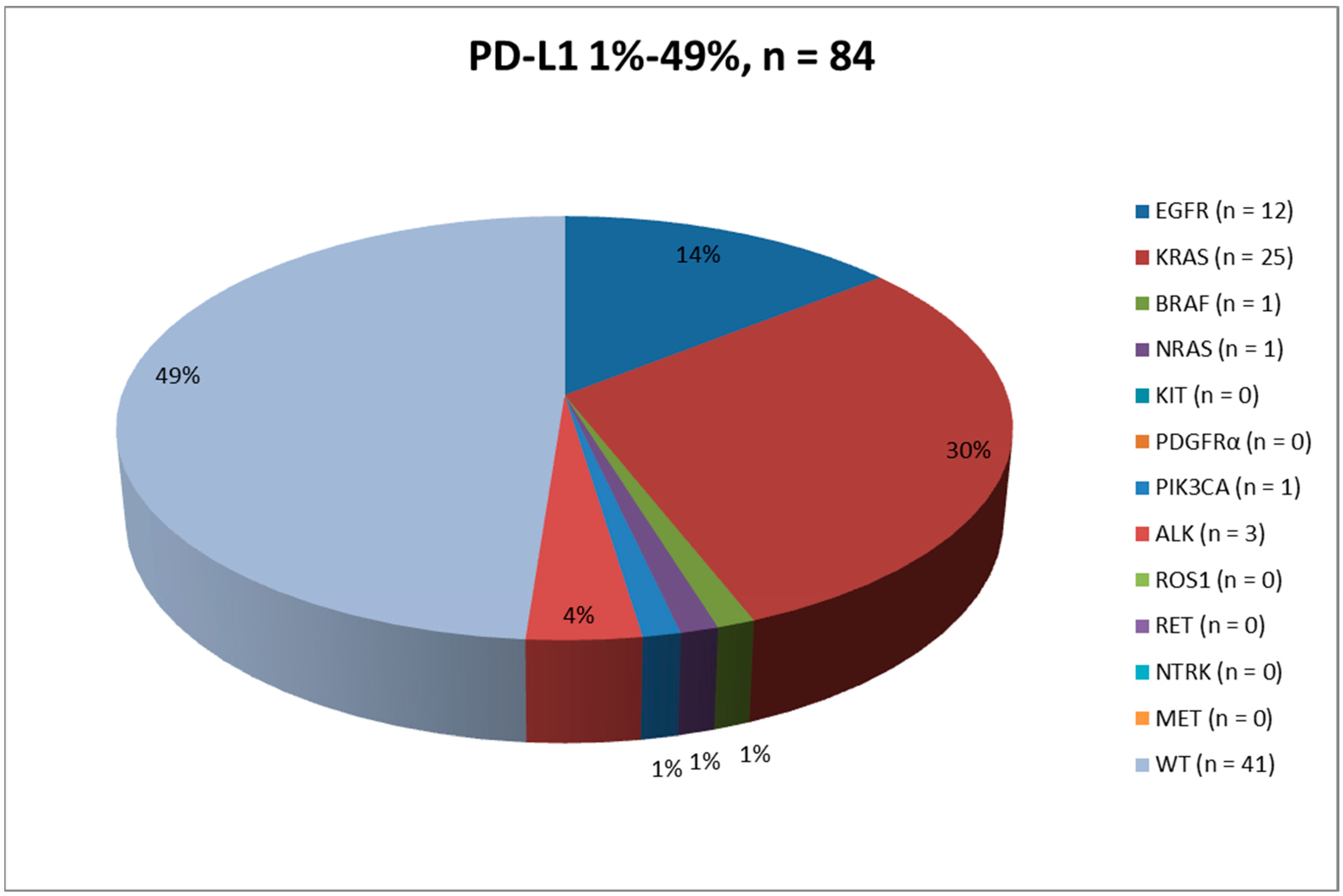
2.3. PD-L1 Expression: 1–49%
2.4. PD-L1 Expression: ≥50%
2.5. Clinical Management
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. IHC/ICC Analysis
4.3. Molecular Testing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Wu, Y.L.; Schuler, M.; Sebastian, M.; Popat, S.; Yamamoto, N.; Zhou, C.; Hu, C.-P.; O’Byrne, K.; Feng, J.; et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015, 16, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Smit, E.F.; Groen, H.J.M.; Mazieres, J.; Besse, B.; Helland, Å.; Giannone, V.; D’Amelio, A.M., Jr.; Zhang, P.; Mookerjee, B.; et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: An open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1307–1316. [Google Scholar] [CrossRef]
- Planchard, D.; Besse, B.; Groen, H.J.M.; Souquet, P.J.; Quoix, E.; Baik, C.S.; Barlesi, F.; Kim, T.M.; Mazieres, J.; Novello, S.; et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016, 17, 984–993. [Google Scholar] [CrossRef] [Green Version]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef]
- Hallin, J.; Engstrom, L.D.; Hargis, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.T.; Kim, D.-W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.-J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Shaw, A.T.; Kim, D.W.; Mehra, R.; Tan, D.S.; Felip, E.; Chow, L.Q.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.H.I.; Pérol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 829–838. [Google Scholar] [CrossRef]
- Camidge, D.R.; Kim, H.R.; Ahn, M.-J.; Yang, J.C.-H.; Han, J.-Y.; Lee, J.-S.; Hochmair, M.J.; Li, J.Y.-C.; Chang, G.-C.; Lee, K.H.; et al. Brigatinib versus Crizotinib in ALK-Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2027–2039. [Google Scholar] [CrossRef]
- Shaw, A.T.; Bauer, T.M.; de Marinis, F.; Felip, E.; Goto, Y.; Liu, G.; Mazieres, J.; Kim, D.-W.; Mok, T.; Polli, A.; et al. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N. Engl. J. Med. 2020, 383, 2018–2029. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Ou, S.H.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.T.; Solomon, B.J.; Chiari, R.; Riely, G.J.; Besse, B.; A Soo, R.; Kao, S.; Lin, C.-C.; Bauer, T.M.; Clancy, J.S.; et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: A multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol. 2019, 20, 1691–1701. [Google Scholar] [CrossRef]
- Drilon, A.; Siena, S.; Dziadziuszko, R.; Barlesi, F.; Krebs, M.G.; Shaw, A.T.; de Braud, F.; Rolfo, C.; Ahn, M.-J.; Wolf, J.; et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 261–270. [Google Scholar] [CrossRef]
- Passiglia, F.; Reale, M.L.; Cetoretta, V.; Novello, S. Immune-Checkpoint Inhibitors Combinations in Metastatic NSCLC: New Options on the Horizon? Immunotargets Ther. 2021, 10, 9–26. [Google Scholar] [CrossRef]
- Hirsch, F.R.; McElhinny, A.; Stanforth, D.; Ranger-Moore, J.; Jansson, M.; Kulangara, K.; Richardson, W.; Towne, P.; Hanks, D.; Vennapusa, B.; et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J. Thorac. Oncol. 2017, 12, 208–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, M.S.; Kerr, K.M.; Kockx, M.; Beasley, M.B.; Borczuk, A.C.; Botling, J.; Bubendorf, L.; Chirieac, L.; Chen, G.; Chou, T.Y.; et al. PD-L1 Immunohistochemistry Comparability Study in Real-Life Clinical Samples: Results of Blueprint Phase 2 Project. J. Thorac. Oncol. 2018, 13, 1302–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, T.S.K.; Wu, Y.L.; Kudaba, I.; Kowalski, D.M.; Cho, B.C.; Turna, H.Z.; Castro, G., Jr.; Srimuninnimit, V.; Laktionov, K.K.; Bondarenko, I.; et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial. Lancet 2019, 393, 1819–1830. [Google Scholar] [CrossRef]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Akinboro, O.; Larkins, E.; Pai-Scherf, L.H.; Mathieu, L.N.; Ren, Y.; Cheng, J.; Fiero, M.H.; Fu, W.; Bi, Y.; Kalavar, S.; et al. FDA Approval Summary: Pembrolizumab, Atezolizumab, and Cemiplimab-rwlc as single agents for first-line treatment of advanced/metastatic PD-L1 high NSCLC. Clin. Cancer Res. 2022, 28, 2221–2228. [Google Scholar] [CrossRef] [PubMed]
- Addeo, A.; Passaro, A.; Malapelle, U.; Banna, G.L.; Subbiah, V.; Friedlaender, A. Immunotherapy in non-small cell lung cancer harbouring driver mutations. Cancer Treat. Rev. 2021, 96, 102179. [Google Scholar] [CrossRef] [PubMed]
- Lisberg, A.; Cummings, A.; Goldman, J.W.; Bornazyan, K.; Reese, N.; Wang, T.; Coluzzi, P.; Ledezma, B.; Mendenhall, M.; Hunt, J.; et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients with Advanced NSCLC. J. Thorac. Oncol. 2018, 13, 1138–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigliar, E.; Iaccarino, A.; Campione, S.; Campanino, M.R.; Clery, E.; Pisapia, P.; De Luca, C.; Bellevicine, C.; Malapelle, U.; De Dominicis, G.; et al. PD-L1 expression in cell-blocks of non-small cell lung cancer: The impact of prolonged fixation. Diagn. Cytopathol. 2020, 48, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Vigliar, E.; Malapelle, U.; Iaccarino, A.; Acanfora, G.; Pisapia, P.; Clery, E.; De Luca, C.; Bellevicine, C.; Troncone, G. PD-L1 expression on routine samples of non-small cell lung cancer: Results and critical issues from a 1-year experience of a centralised laboratory. J. Clin. Pathol. 2019, 72, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; De Luca, C.; Smeraglio, R.; Pisapia, P.; Sgariglia, R.; Nacchio, M.; Russo, M.; Serra, N.; Rocco, D.; Battiloro, C.; et al. Performance analysis of SiRe next-generation sequencing panel in diagnostic setting: Focus on NSCLC routine samples. J. Clin. Pathol. 2019, 72, 38–45. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Gragnano, G.; Pisapia, P.; Vigliar, E.; Malapelle, U.; Bellevicine, C.; Troncone, G. EGFR mutation detection on lung cancer cytological specimens by the novel fully automated PCR-based Idylla EGFR Mutation Assay. J. Clin. Pathol. 2017, 70, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Gragnano, G.; Nacchio, M.; Sgariglia, R.; Conticelli, F.; Iaccarino, A.; De Luca, C.; Troncone, G.; Malapelle, U. Performance evaluation of a fully closed real-time PCR platform for the detection of KRAS p.G12C mutations in liquid biopsy of patients with non-small cell lung cancer. J. Clin. Pathol. 2021, 75, 350–353. [Google Scholar] [CrossRef]
- De Luca, C.; Pepe, F.; Iaccarino, A.; Pisapia, P.; Righi, L.; Listì, A.; Greco, L.; Gragnano, G.; Campione, S.; De Dominicis, G.; et al. RNA-Based Assay for Next-Generation Sequencing of Clinically Relevant Gene Fusions in Non-Small Cell Lung Cancer. Cancers 2021, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.; O’Sullivan, B.; Hughes, F.; Mullis, T.; Smith, M.; Trim, N.; Taniere, P. The Clinicopathological and Molecular Associations of PD-L1 Expression in Non-small Cell Lung Cancer: Analysis of a Series of 10,005 Cases Tested with the 22C3 Assay. Pathol. Oncol. Res. 2020, 26, 79–89. [Google Scholar] [CrossRef]
- Karatrasoglou, E.A.; Chatziandreou, I.; Sakellariou, S.; Stamopoulos, K.; Kavantzas, N.; Lazaris, A.C.; Korkolopoulou, P.; Saetta, A.A. Association between PD-L1 expression and driver gene mutations in non-small cell lung cancer patients: Correlation with clinical data. Virchows Arch. 2020, 477, 207–217. [Google Scholar] [CrossRef]
- Sumimoto, H.; Takano, A.; Teramoto, K.; Daigo, Y. RAS–Mitogen-Activated Protein Kinase Signal Is Required for Enhanced PD-L1 Expression in Human Lung Cancers. PLoS ONE 2016, 11, e0166626. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Fang, W.; Lin, Z.; Peng, P.; Wang, J.; Zhan, J.; Hong, S.; Huang, J.; Liu, L.; Sheng, J.; et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol. Immunother. 2017, 66, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MMiura, Y.; Sunaga, N. Role of Immunotherapy for Oncogene-Driven Non-Small Cell Lung Cancer. Cancers 2018, 10, 245. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.-Y.; Zhong, W.-Z.; Zhang, X.-C.; Su, J.; Xie, Z.; Liu, S.-Y.; Tu, H.-Y.; Chen, H.-J.; Sun, Y.-L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.K.; Man, J.; Lord, S.; Cooper, W.; Links, M.; Gebski, V.; Herbst, R.S.; Gralla, R.J.; Mok, T.; Yang, J.C. Clinical and Molecular Characteristics Associated with Survival among Patients Treated with Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Bodor, J.N.; Bauman, J.R.; Handorf, E.A.; Ross, E.A.; Clapper, M.L.; Treat, J. Real-world progression-free survival (rwPFS) and the impact of PD-L1 and smoking in driver-mutated non-small cell lung cancer (NSCLC) treated with immunotherapy. J. Cancer Res. Clin. Oncol. 2022; Epub ahead of print. [Google Scholar] [CrossRef]
- Falk, A.T.; Yazbeck, N.; Guibert, N.; Chamorey, E.; Paquet, A.; Ribeyre, L.; Bence, C.; Zahaf, K.; Leroy, S.; Marquette, C.-H.; et al. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer 2018, 121, 70–75. [Google Scholar] [CrossRef]
- Herbst, R.S.; Lopes, G.; Kowalski, D.M.; Kasahara, K.; Wu, Y.L.; De Castro, G., Jr.; Cho, B.C.; Turna, H.Z.; Cristescu, R.; Aurora-Garg, D.; et al. LBA4 Association of KRAS mutational status with response to pembrolizumab monotherapy given as first-line therapy for PD-L1-positive advanced non-squamous NSCLC in Keynote-042. Ann. Oncol. 2019, 30, xi63–xi64. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.B.; Mhanna, L.; Cortot, A.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Peters, S.; Gettinger, S.; Johnson, M.L.; Jänne, P.A.; Garassino, M.C.; Christoph, D.; Toh, C.K.; Rizvi, N.A.; Chaft, J.E.; Costa, E.C.; et al. Phase II Trial of Atezolizumab as First-Line or Subsequent Therapy for Patients with Programmed Death-Ligand 1–Selected Advanced Non–Small-Cell Lung Cancer (BIRCH). J. Clin. Oncol. 2017, 35, 2781–2789. [Google Scholar] [CrossRef]
- Garassino, M.C.; Cho, B.-C.; Kim, J.-H.; Mazières, J.; Vansteenkiste, J.; Lena, H.; Jaime, J.C.; Gray, J.E.; Powderly, J.; Chouaid, C.; et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): An open-label, single-arm, phase 2 study. Lancet Oncol. 2018, 19, 521–536. [Google Scholar] [CrossRef]
- Frisone, D.; Friedlaender, A.; Malapelle, U.; Banna, G.; Addeo, A. A BRAF new world. Crit. Rev. Oncol. Hematol. 2020, 152, 103008. [Google Scholar] [CrossRef]
- Dudnik, E.; Peled, N.; Nechushtan, H.; Wollner, M.; Onn, A.; Agbarya, A.; Moskovitz, M.; Keren, S.; Popovits-Hadari, N.; Urban, D.; et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J. Thorac. Oncol. 2018, 13, 1128–1137. [Google Scholar] [CrossRef] [Green Version]
- Tan, I.; Stinchcombe, T.E.; Ready, N.E.; Crawford, J.; Datto, M.B.; Nagy, R.J.; Lanman, R.B.; Gu, L.; Clarke, J.M. Therapeutic outcomes in non-small cell lung cancer with BRAF mutations: A single institution, retrospective cohort study. Transl. Lung Cancer Res. 2019, 8, 258–267. [Google Scholar] [CrossRef]
- Gainor, J.F.; Shaw, A.T.; Sequist, L.V.; Fu, X.; Azzoli, C.G.; Piotrowska, Z.; Huynh, T.G.; Zhao, L.; Fulton, L.; Schultz, K.R.; et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non–Small Cell Lung Cancer: A Retrospective Analysis. Clin. Cancer Res. 2016, 22, 4585–4593. [Google Scholar] [CrossRef] [Green Version]
- Bylicki, O.; Guisier, F.; Monnet, I.; Doubre, H.; Gervais, R.; Janicot, H.; Perol, M.; Fournel, P.; Lamy, R.; Auliac, J.B.; et al. Efficacy and safety of programmed cell-death-protein-1 and its ligand inhibitors in pretreated patients with epidermal growth-factor receptor-mutated or anaplastic lymphoma kinase-translocated lung adenocarcinoma. Medicine 2020, 99, e18726. [Google Scholar] [CrossRef]
- West, H.; McCleod, M.; Hussein, M.; Morabito, A.; Rittmeyer, A.; Conter, H.J.; Kopp, H.-G.; Daniel, D.; McCune, S.; Mekhail, T.; et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 924–937. [Google Scholar] [CrossRef]
- Socinski, M.A.; Jotte, R.M.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodríguez-Abreu, D.; Moro-Sibilot, D.; Thomas, C.A.; Barlesi, F.; et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N. Engl. J. Med. 2018, 378, 2288–2301. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Fujioka, N.; Patel, M. Immune Checkpoint Inhibitors in ROS1-Rearranged Non–Small Cell Lung Cancer: A Report of Two Cases. J. Thorac. Oncol. 2019, 14, e165–e167. [Google Scholar] [CrossRef]
- Hegde, A.; Andreev-Drakhlin, A.Y.; Roszik, J.; Huang, L.; Liu, S.; Hess, K.; Cabanillas, M.; Hu, M.I.; Busaidy, N.L.; Sherman, S.I.; et al. Responsiveness to immune checkpoint inhibitors versus other systemic therapies in RET-aberrant malignancies. ESMO Open 2020, 5, e000799. [Google Scholar] [CrossRef]
- Guisier, F.; Dubos-Arvis, C.; Viñas, F.; Doubre, H.; Ricordel, C.; Ropert, S.; Janicot, H.; Bernardi, M.; Fournel, P.; Lamy, R.; et al. Efficacy and Safety of Anti-PD-1 Immunotherapy in Patients with Advanced NSCLC With BRAF, HER2, or MET Mutations or RET Translocation: GFPC 01-2018. J. Thorac. Oncol. 2020, 15, 628–636. [Google Scholar] [CrossRef]
- Sabari, J.; Leonardi, G.; Shu, C.; Umeton, R.; Montecalvo, J.; Ni, A.; Chen, R.; Dienstag, J.; Mrad, C.; Bergagnini, I.; et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann. Oncol. 2018, 29, 2085–2091. [Google Scholar] [CrossRef]
- Pisapia, P.; Lozano, M.D.; Vigliar, E.; Bellevicine, C.; Pepe, F.; Malapelle, U.; Troncone, G. ALK and ROS1 testing on lung cancer cytologic samples: Perspectives. Cancer Cytopathol. 2017, 125, 817–830. [Google Scholar] [CrossRef] [Green Version]
- Conde, E.; Hernandez, S.; Prieto, M.; Martinez, R.; Lopez-Rios, F. Profile of Ventana ALK (D5F3) Companion Diagnostic Assay for Non-Small-Cell Lung Carcinomas. Expert Rev. Mol. Diagn. 2016, 16, 707–713. [Google Scholar] [CrossRef]
- Kerr, K.M.; López-Ríos, F. Precision medicine in NSCLC and pathology: How does ALK fit in the pathway? Ann. Oncol. 2016, 27 (Suppl. 3), iii16–iii24. [Google Scholar] [CrossRef]
- Rossi, G.; Ragazzi, M.; Tamagnini, I.; Mengoli, M.C.; Vincenzi, G.; Barbieri, F.; Piccioli, S.; Bisagni, A.; Vavala, T.; Righi, L.; et al. Does Immunohistochemistry Represent a Robust Alternative Technique in Determining Drugable Predictive Gene Alterations in Non-Small Cell Lung Cancer? Curr. Drug Targets 2017, 18, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Selinger, C.I.; Li, B.T.; Pavlakis, N.; Links, M.; Gill, A.J.; Lee, A.; Clarke, S.; Tran, T.N.; Lum, T.; Yip, P.Y.; et al. Screening for ROS1 gene rearrangements in non-small-cell lung cancers using immunohistochemistry with FISH confirmation is an effective method to identify this rare target. Histopathology 2017, 70, 402–411. [Google Scholar] [CrossRef] [Green Version]
- Malapelle, U.; Mayo de-Las-Casas, C.; Rocco, D.; Garzon, M.; Pisapia, P.; Jordana-Ariza, N.; Russo, M.; Sgariglia, R.; De Luca, C.; Pepe, F.; et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br. J. Cancer 2017, 116, 802–810. [Google Scholar] [CrossRef] [Green Version]
- Van Haele, M.; Vander Borght, S.; Ceulemans, A.; Wieërs, M.; Metsu, S.; Sagaert, X.; Weynand, B. Rapid clinical mutational testing of KRAS, BRAF and EGFR: A prospective comparative analysis of the Idylla technique with high-throughput next-generation sequencing. J. Clin. Pathol. 2020, 73, 35–41. [Google Scholar] [CrossRef]


| Global | 1–49% | ≥50% | |
|---|---|---|---|
| Total (%) | 167 (100.0) | 84 (100.0) | 83 (100.0) |
| Sex (%) | M: 103 (61.7) F: 64 (38.3) | M: 53 (63.1) F: 31 (36.9) | M: 50 (60.2) F: 33 (39.8) |
| Median Age (range) | 67.3 y (43–93 y) | 66.9 y (43–92 y) | 67.8 y (44–93 y) |
| Sample type (n; %) - subtype (n; %) | Histological (110, 65.9) - Biopsy (86, 78.2) - Resection (24, 21.8) Cytological (57, 34.1) - Cell block (52, 91.2) - Smear (5, 8.8) | Histological (53, 63.1) - Biopsy (40, 75.5) - Resection (13, 24.5) Cytological (31, 36.9) - Cell block (29, 93.5) - Smear (2, 6.5) | Histological (57, 68.7) - Biopsy (46, 80.7) - Resection (11, 19.3) Cytological (26, 31.3) - Cell block (23, 88.5) - Smear (3, 11.5) |
| Diagnosis (n, %) | ADC (62, 37.1) NSCLC favor ADC (58, 34.7) NSCLC NOS (32, 19.2) SqCC (8, 4.8) NSCLC favor SqCC (4, 2.4) ADC + SqCC (3, 1.8) | ADC (41, 48.8) NSCLC favor ADC (24, 28.6) NSCLC NOS (11, 13.1) SqCC (6, 7.1) NSCLC favor SqCC (1, 1.2) ADC + SqCC (1, 1.2) | NSCLC favor ADC (34, 41.0) ADC (21, 25.3) NSCLC NOS (21, 25.3) NSCLC favor SqCC (3, 3.6) SqCC (2, 2.4) ADC + SqCC (2, 2.4) |
| PD-L1 (n, %) | 1–49 (84, 50.3) ≥50 (83, 49.7) | - | - |
| Clone (n, %) | SP263 (134, 80.2) 22C3 (33, 19.8) | SP263 (67, 79.8) 22C3 (17, 20.2) | SP263 (67, 80.7) 22C3 (16, 19.3) |
| DNA based-biomarker molecular platform (n, %) | NGS (164, 98.2) RT-qPCR (3, 1.8) | NGS (83, 98.8) RT-qPCR (1, 1.2) | NGS (81, 97.6) RT-qPCR (2, 2.4) |
| Molecular results (n, %) | WT (74, 44.3) Mutated (93, 55.7) | WT (41, 48.8) Mutated (43, 51.2) | WT (33, 39.8) Mutated (50, 60.2) |
| DNA-based biomarkers (n, %) | EGFR (167, 100.0) - WT (146, 87.4) - mutated (21, 12.6) - p.L858R (9, 42.8) - p.E746_A750del (6, 28.4) - p.E709_T710insD (1, 4.8) - p.G719A + p.T790M (1, 4.8) - p.I744_K745insKIPVAI (1, 4.8) - p.E746_S752del (1, 4.8) - p.S768_D760dup (1, 4.8) - p.S768I (1, 4.8) KRAS (167, 100.0) - WT (111, 66.5) - mutated (56, 33.5) - p.G12C (27, 48.1) - p.G12V (13, 23.2) - p.G12A (5, 8.9) - p.G12D (3, 5.4) - p.Q61H (3, 5.4) - p.G13C (2, 3.6) - p.G12R (1, 1.8) - p.G13D (1, 1.8) - p.G13R (1, 1.8) BRAF (167, 100.0) - WT (163, 97.6) - mutated (4, 2.4) - p.V600E (2, 50.0) - p.G466V (1, 25.0) - p.G469A (1, 25.0) NRAS (164, 98.2) - WT (163, 99.4) - mutated (1, 0.6) - p.G12D (1, 100.0) KIT (164, 98.2) - WT (164, 100.0) PDGFRα (164, 98.2) - WT (164, 100.0) PIK3CA (164, 98.2) - WT (161, 98.2) - mutated (3, 1.8) - p.E545K (2, 66.7) - p.E542K (1, 33.3) | EGFR (84, 100.0) - WT (72, 85.7) - mutated (12, 14.3) - p.E746_A750del (4, 33.4) - p.L858R (4, 33.4) - p.E709_T710indD (1, 8.3) - p.G719 + p.T790M (1, 8.3) - p.S768_D760dup (1, 8.3) - p.S768I (1, 8.3) KRAS (84, 100.0) - WT (59, 70.2) - mutated (25, 29.8) - p.G12C (10, 40.0) - p.G12V (8, 32.0) - p.G13C (2, 8.0) - p.G12A (1, 4.0) - p.G12D (1, 4.0) - p.G12R (1, 4.0) - p.G13R (1, 4.0) - p.Q61H (1, 4.0) BRAF (84, 100.0) - WT (83, 98.8) - mutated (1, 1.2) - p.G469A (1, 100.0) NRAS (83, 98.8) - WT (82, 98.8) - mutated (1, 1.2) - p.G12D (1, 100.0) KIT (83, 98.8) - WT (83, 100.0) PDGFRα (83, 98.8) - WT (83, 100.0) PIK3CA (83, 98.8) - WT (82, 98.8) - mutated (1, 1.2) - p.E542K (1, 100.0) | EGFR (83, 100.0) - WT (74, 89.2) - mutated (9, 10.8) - p.L858R (5, 55.6) - p.E746_A750del (2, 22.2) - p.I744_K745insKIPVAI (1, 11.1) - p.E746_S752del (1, 11.1) KRAS (83, 100.0) - WT (52, 62.7) - mutated (31, 37.3) - p.G12C (17, 54.8) - p.G12V (5, 16.1) - p.G12A (4, 12.9) - p.G12D (2, 6.5) - p.Q61H (2, 6.5) - p.G13D (1, 3.2) BRAF (83, 100.0) - WT (80, 96.4) - mutated (3, 3.6) - p.V600E (2, 66.7) - p.G466V (1, 33.3) NRAS (81, 97.6) - WT (81, 100.0) KIT (81, 97.6) - WT (81, 100.0) PDGFRα (81, 97.6) - WT (81, 100.0) PIK3CA (81, 97.6) - WT (79, 97.5) - mutated (2, 2.5) - p.E545K (2, 100.0) |
| RNA-based biomarker assays (n, %) | IHC/ICC (152, 91.0) NGS (15, 9.0) | IHC/ICC (78, 92.9) NGS (6, 7.1) | IHC/ICC (74, 89.2) NGS (9, 10.8) |
| RNA-based biomarkers (n, %) | ALK (167, 100.0) - Negative/WT (160, 95.8) - Positive/rearranged (7, 4.2) ROS1 (167, 100.0) - Negative/WT (166, 99.4) - Positive/rearranged (1, 0.6) RET (15, 9.0) - WT (15, 100.0) NTRK (15, 9.0) - WT (15, 100.0) MET (15, 9.0) - WT (15, 100.0) | ALK (84, 100.0) - Negative/WT (81, 96.4) - Positive/rearranged (3, 3.6) ROS1 (84, 100.0) - Negative/WT (84, 100.0) RET (6, 7.1) - WT (6, 100.0) NTRK (6, 7.1) - WT (6, 100.0) MET (6, 7.1) - WT (6, 100.0) | ALK (83, 100.0) - Negative/WT (79, 95.2) - Positive/rearranged (4, 4.8) ROS1 (83, 100.0) - Negative/WT (82, 98.8) - Positive/rearranged (1, 1.2) RET (9, 10.8) - WT (9, 100.0) NTRK (9, 10.8) - WT (9, 100.0) MET (9, 10.8) - WT (9, 100.0) |
| Sex | Age | Sample Type | Sample Subtype | Site | Diagnosis | PD-L1 | Clone | Alteration | First Oncological Observation Date | Performance Status | First Line Treatment | First-Line Treatment Starting Date | First-Line Treatment End Date |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | 70 | Histological | Biopsy | Brain | NSCLC favor ADC | ≥50% | SP263 | KRAS exon 2 p.G12C | March 2019 | 1 | Pembrolizumab | May 2019 | Ongoing |
| F | 66 | Histological | Resection | Brain | ADC | ≥50% | SP263 | WT | February 2020 | 1 | Pembrolizumab | March 2021 | December 2021 |
| M | 77 | Histological | Biopsy | Lymphnode | NSCLC favor ADC | 1–49% | SP263 | KRAS exon 2 p.G12V | May 2020 | 1 | Carboplatino + Pemetrexed + Pembrolizumab | June 2020 | April 2021 |
| M | 75 | Histological | Biopsy | Lymphnode | ADC | ≥50% | SP263 | WT | April 2020 | 1 | Durvalumab | February 2021 | Ongoing |
| F | 75 | Histological | Biopsy | Lung | ADC | 1–49% | SP263 | KRAS exon 2 p.G12C | April 2020 | 0 | Carboplatino-Pemetrexed | July 2020 | Ongoing |
| M | 57 | Histological | Biopsy | Lung | ADC-SqCC | ≥50% | SP263 | WT | June 2020 | 0 | Pembrolizumab | July 2020 | Ongoing |
| M | 77 | Histological | Resection | Lung | ADC | ≥50% | SP263 | KRAS exon 2 p.G12C | February 2020 | 0 | Pembrolizumab | September 2021 | March 2022 |
| F | 69 | Histological | Biopsy | Lung | NSCLC favor ADC | 1–49% | SP263 | BRAF exon 11 p.G469A | June 2020 | 2 | Carboplatino + Pemetrexed + Pembrolizumab | September 2020 | February 2021 |
| F | 69 | Histological | Biopsy | Lung | NSCLC favor ADC | ≥50% | SP263 | EML4(6)-ALK(20) | September 2020 | 2 | Brigatinib | February 2021 | June 2021 |
| F | 68 | Histological | Resection | Lymphnode | ADC | 1–49% | SP263 | WT | April 2019 | 0 | Carboplatino + Pemetrexed | April 2019 | Ongoing with only pemetrexed |
| F | 69 | Cytological | Cell block | Soft tissue | ADC | 1–49% | SP263 | EGFR exon 20 p.S768_D760dup | December 2019 | 1 | Carboplatino + Pemetrexed + Pembrolizumab | February 2020 | September 2020 |
| M | 57 | Histological | Biopsy | Lung | NSCLC-NOS | ≥50% | SP263 | KRAS exon 2 p.G12C | February 2020 | 2 | Pembrolizumab | March 2020 | March 2020 |
| F | 55 | Cytological | Cell block | Lung | NSCLC-NOS | ≥50% | SP263 | WT | December 2020 | 1 | Pembrolizumab | January 2021 | Ongoing |
| M | 62 | Histological | Biopsy | Pleura | ADC | 1–49% | SP263 | ALK positive | April 2021 | 1 | Alectinib | April 2021 | Ongoing |
| M | 72 | Cytological | Cell block | Lung | NSCLC favor ADC | 1–49% | 22C3 | WT | June 2018 | 2 | Carboplatin + Pemetrexed | July 2018 | September 2018 |
| M | 78 | Cytological | Smear | Lung | ADC | ≥50% | SP263 | WT | May 2019 | 2 | Carboplatin | June 2019 | September 2019 |
| M | 72 | Cytological | Cell block | Lung | NSCLC favor ADC | 1–49% | SP263 | KRAS exon 2 p.G12C | March 2019 | 1 | Carboplatin + Pemetrexed | March 2019 | January 2020 |
| F | 73 | Histological | Biopsy | Lung | NSCLC favor ADC | ≥50% | SP263 | KRAS exon 2 p.G12V | June 2019 | 0 | Pembrolizumab | June 2019 | January 2021 |
| M | 79 | Histological | Biopsy | Lung | ADC | 1–49% | SP263 | WT | September 2019 | 0 | Carboplatin + Pemetrexed | October 2019 | January 2020 |
| M | 49 | Cytological | Cell block | Lung | NSCLC favor ADC | ≥50% | SP263 | WT | December 2019 | 0 | Pembrolizumab | December 2019 | Ongoing |
| M | 60 | Histological | Biopsy | Lung | NSCLC favor ADC | ≥50% | SP263 | KRAS exon 3 p.Q61H | December 2019 | 0 | Pembrolizumab | January 2020 | April 2020 |
| F | 62 | Histological | Biopsy | Brain | NSCLC-NOS | 1–49% | SP263 | EGFR exon 19 p.E746_A750del | December 2020 | 0 | Osimertinib | January 2021 | Ongoing |
| F | 58 | Histological | Resection | Brain | ADC | 1–49% | SP263 | WT | March 2021 | 0 | Carboplatin + Pemetrexed + Pembrolizumab | April 2021 | Ongoing |
| M | 61 | Cytological | Cell block | Lung | ADC | 1–49% | 22C3 | WT | July 2018 | 0 | Cisplatino + Pemetrexed | July 2018 | September 2018 |
| F | 56 | Histological | Biopsy | Lung | NSCLC favor ADC | 1–49% | 22C3 | KRAS exon 2 p.G13C | December 2018 | 2 | Carboplatin + Pemetrexed | January 2019 | August 2019 |
| M | 71 | Histological | Biopsy | Lung | NSCLC favor ADC | 1–49% | SP263 | KRAS exon 2 p.G12V | May 2019 | 1 | Cisplatin + Pemetrexed | May 2019 | July 2019 |
| M | 48 | Histological | Biopsy | Pleura | NSCLC favor ADC | 1–49% | SP263 | WT | June 2019 | 1 | Cisplatin + Pemetrexed | June 2019 | October 2019 |
| F | 67 | Cytological | Cell block | Lymphnode | NSCLC favor ADC | 1–49% | SP263 | WT | December 2019 | 2 | Carboplatin + Pemetrexed | January 2020 | January 2020 |
| M | 70 | Cytological | Cell block | Lymphnode | NSCLC favor ADC | 1–49% | SP263 | KRAS exon 2 p.G12V | June 2021 | 1 | Carboplatin + Gemcitabina | July 2021 | September 2021 |
| F | 74 | Cytological | Cell block | Lung | ADC | 1–49% | SP263 | KRAS exon 2 p.G12C | September 2020 | 1 | Pemetrexed + Pembrolizumab | October 2020 | October 2020 |
| M | 70 | Histological | Biopsy | Lung | NSCLC favor ADC | ≥50% | SP263 | WT | June 2021 | 3 | Supportive care | - | - |
| M | 77 | Cytological | Cell block | Lung | NSCLC favor SqCC | 1–49% | SP263 | WT | April 2021 | 2 | Atezolizumab | May 2021 | August 2021 |
| F | 71 | Cytological | Cell block | Lymphnode | NSCLC favor ADC | ≥50% | SP263 | KRAS exon 2 p.G12A | February 2021 | 1 | Pembrolizumab | March 2021 | Ongoing |
| F | 76 | Histological | Biopsy | Lung | SqCC | 1–49% | 22C3 | WT | August 2018 | 1 | Nivolumab | January 2019 | January 2019 |
| M | 72 | Cytological | Cell block | Lymphnode | NSCLC favor ADC | 1–49% | 22C3 | WT | October 2017 | 1 | Cisplatin + Pemetrexed | October 2017 | November 2017 |
| M | 46 | Histological | Biopsy | Brain | NSCLC favor ADC | 1–49% | SP263 | WT | May 2019 | 0 | Cisplatin + Pemetrexed | June 2019 | January 2020 |
| M | 73 | Histological | Resection | Lung | ADC | 1–49% | SP263 | WT | June 2020 | 0 | Carboplatin + Pemetrexed | August 2020 | October 2020 |
| M | 59 | Cytological | Cell block | Lung | NSCLC favor ADC | ≥50% | SP263 | WT | July 2020 | 0 | Pembrolizumab | August 2020 | Ongoing |
| F | 64 | Histological | Biopsy | Lung | ADC | 1–49% | SP263 | WT | September 2020 | 1 | Carboplatin + Pemetrexed + Pembrolizumab | October 2020 | November 2020 |
| M | 75 | Histological | Biopsy | Liver | ADC | 1–49% | SP263 | EGFR exon 19 p.E746_A750del | January 2021 | 1 | Osimertinib | January 2021 | Ongoing |
| M | 68 | Histological | Biopsy | Lung | ADC | ≥50% | SP263 | KRAS exon 2 p.G12C | December 2020 | 1 | Pembrolizumab | December 2020 | Ongoing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisapia, P.; Iaccarino, A.; De Luca, C.; Acanfora, G.; Bellevicine, C.; Bianco, R.; Daniele, B.; Ciampi, L.; De Felice, M.; Fabozzi, T.; et al. Evaluation of the Molecular Landscape in PD-L1 Positive Metastatic NSCLC: Data from Campania, Italy. Int. J. Mol. Sci. 2022, 23, 8541. https://doi.org/10.3390/ijms23158541
Pisapia P, Iaccarino A, De Luca C, Acanfora G, Bellevicine C, Bianco R, Daniele B, Ciampi L, De Felice M, Fabozzi T, et al. Evaluation of the Molecular Landscape in PD-L1 Positive Metastatic NSCLC: Data from Campania, Italy. International Journal of Molecular Sciences. 2022; 23(15):8541. https://doi.org/10.3390/ijms23158541
Chicago/Turabian StylePisapia, Pasquale, Antonino Iaccarino, Caterina De Luca, Gennaro Acanfora, Claudio Bellevicine, Roberto Bianco, Bruno Daniele, Luisa Ciampi, Marco De Felice, Teresa Fabozzi, and et al. 2022. "Evaluation of the Molecular Landscape in PD-L1 Positive Metastatic NSCLC: Data from Campania, Italy" International Journal of Molecular Sciences 23, no. 15: 8541. https://doi.org/10.3390/ijms23158541
APA StylePisapia, P., Iaccarino, A., De Luca, C., Acanfora, G., Bellevicine, C., Bianco, R., Daniele, B., Ciampi, L., De Felice, M., Fabozzi, T., Formisano, L., Giordano, P., Gridelli, C., Ianniello, G. P., Libroia, A., Maione, P., Nacchio, M., Pagni, F., Palmieri, G., ... Malapelle, U. (2022). Evaluation of the Molecular Landscape in PD-L1 Positive Metastatic NSCLC: Data from Campania, Italy. International Journal of Molecular Sciences, 23(15), 8541. https://doi.org/10.3390/ijms23158541













