High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants
Abstract
1. Introduction
2. Results
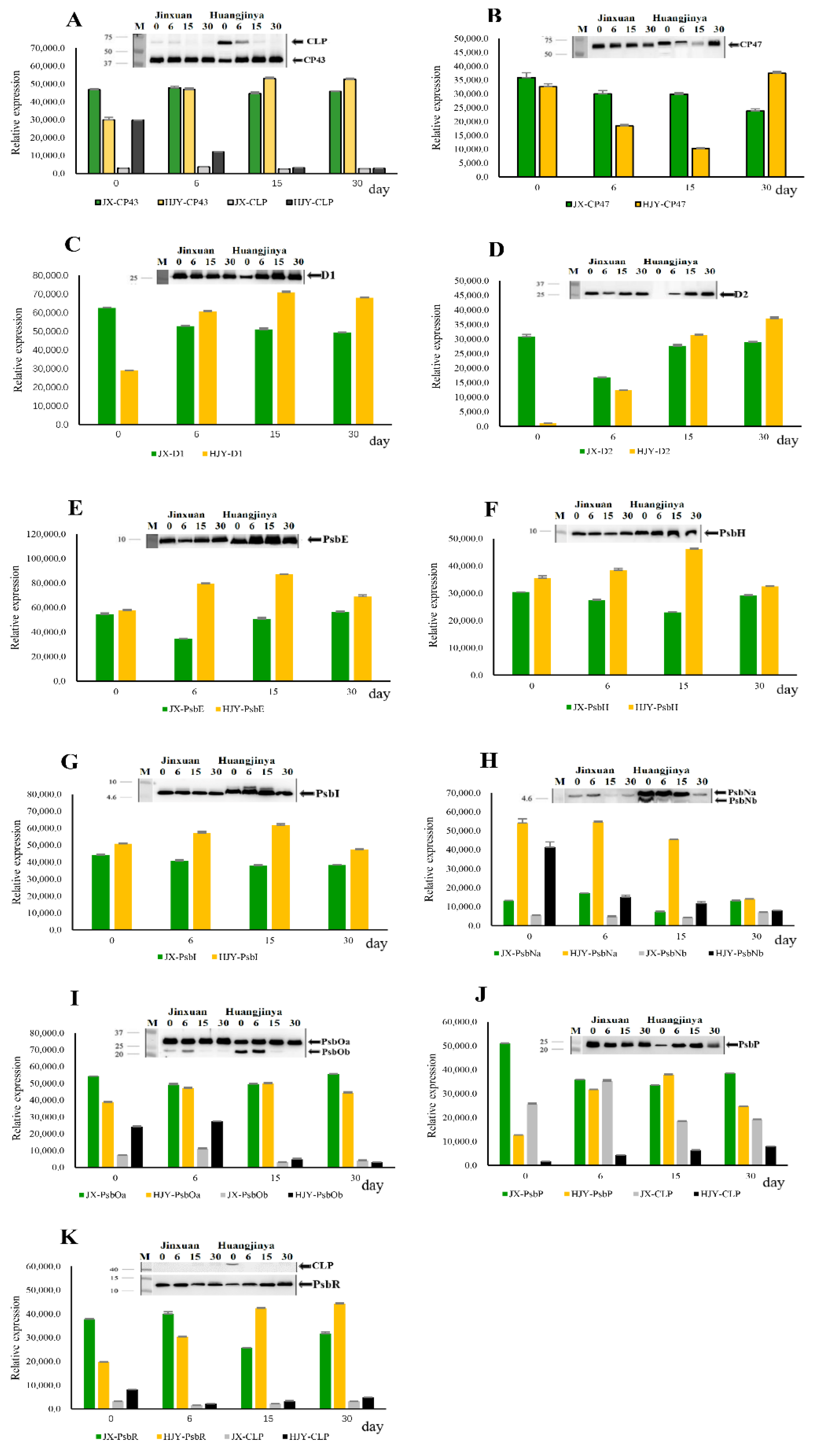
2.1. Effects of HL on PSII Protein Subunits
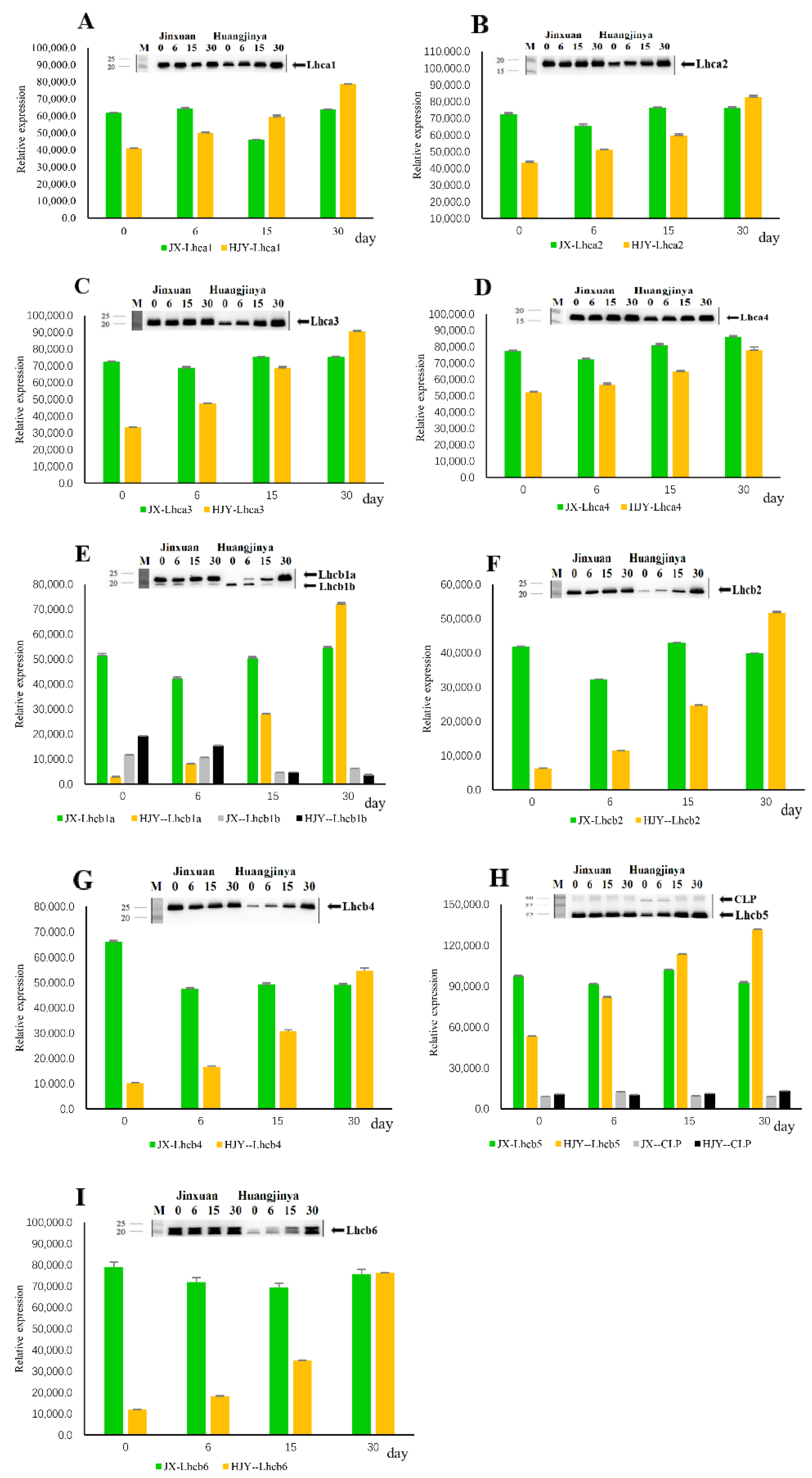
2.2. Effects of HL on Subunits of Light-Harvesting Complexes
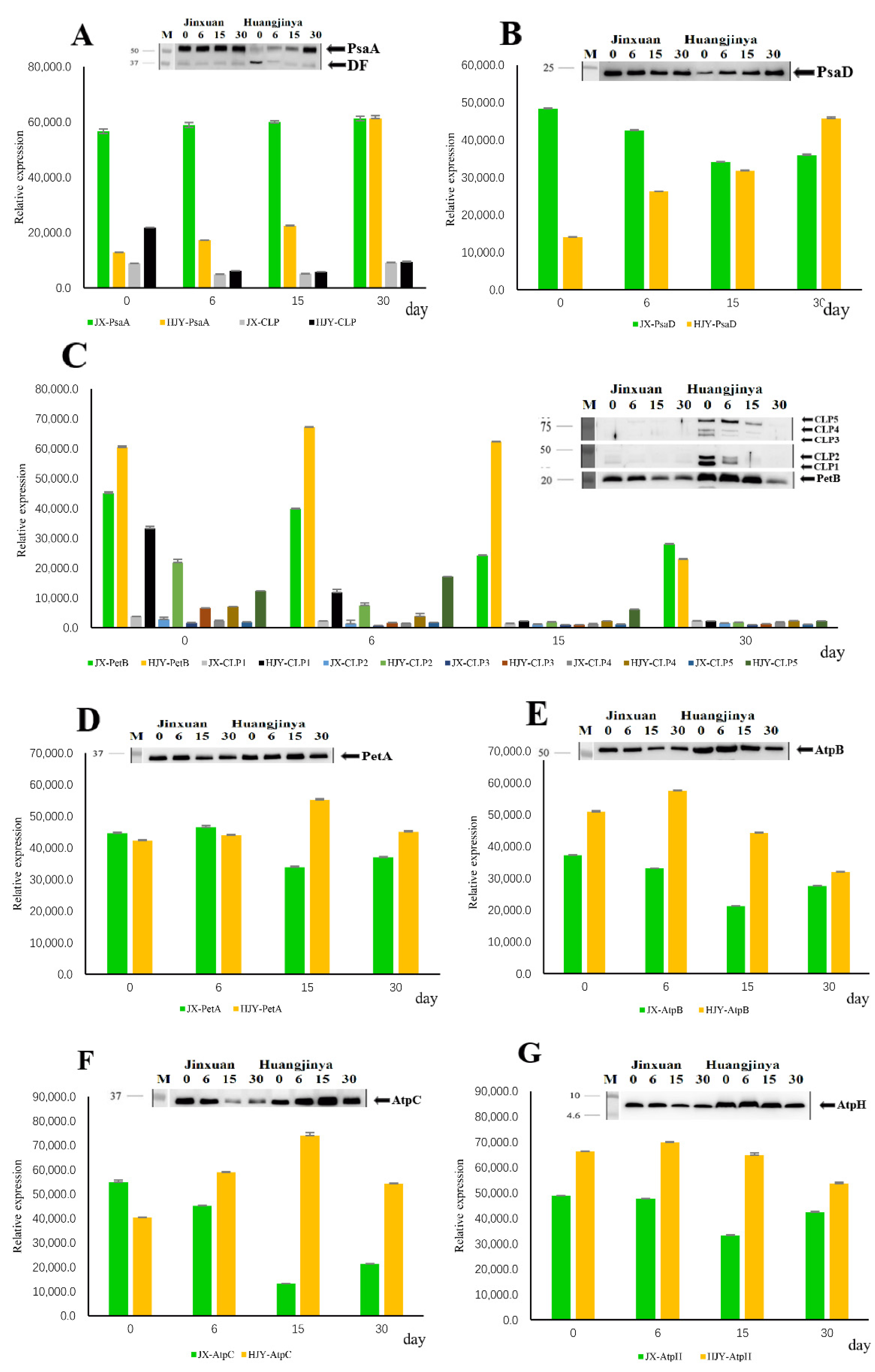
2.3. Effects of HL on Partial Subunits of Electron Transport Complexes
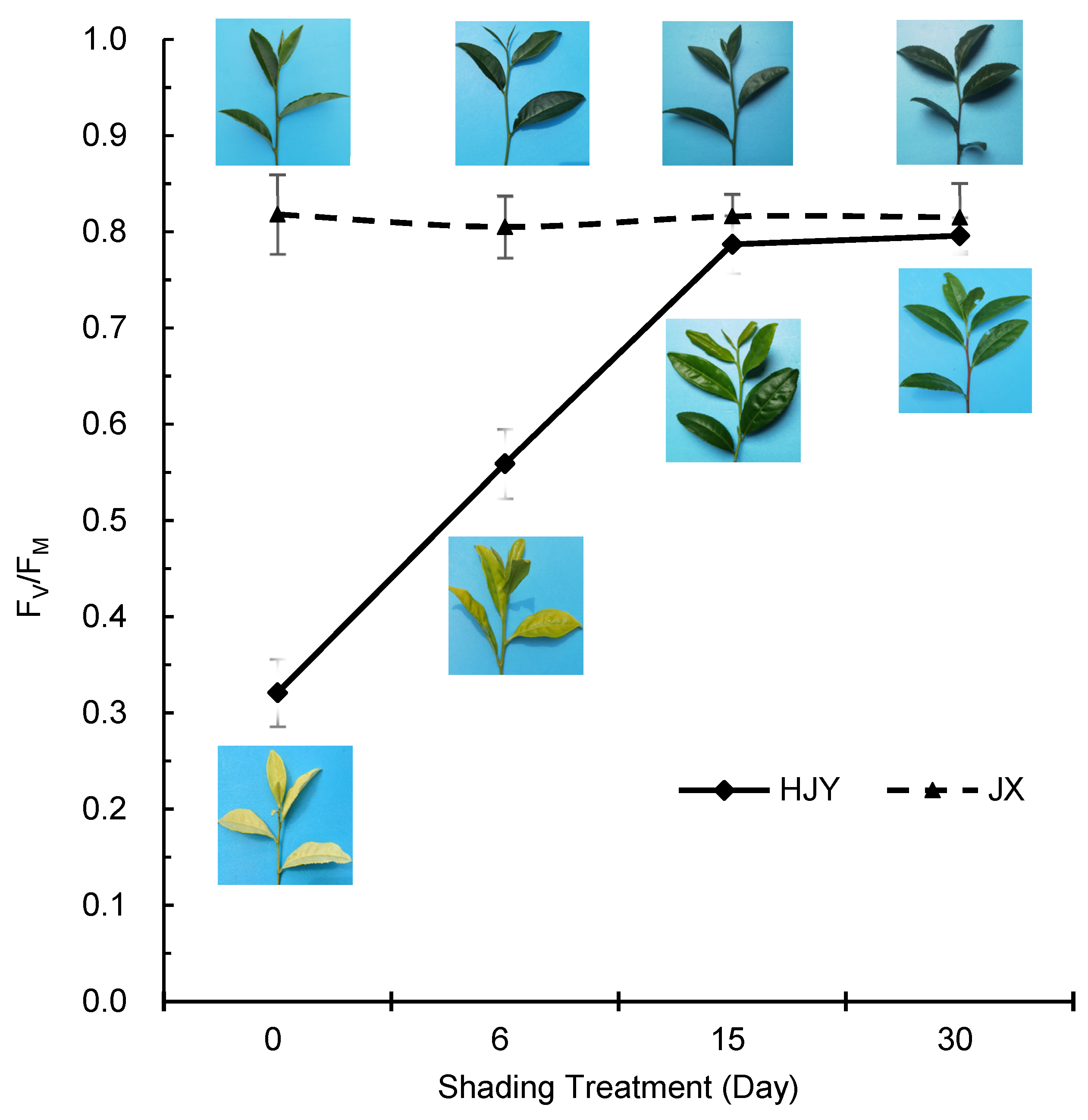
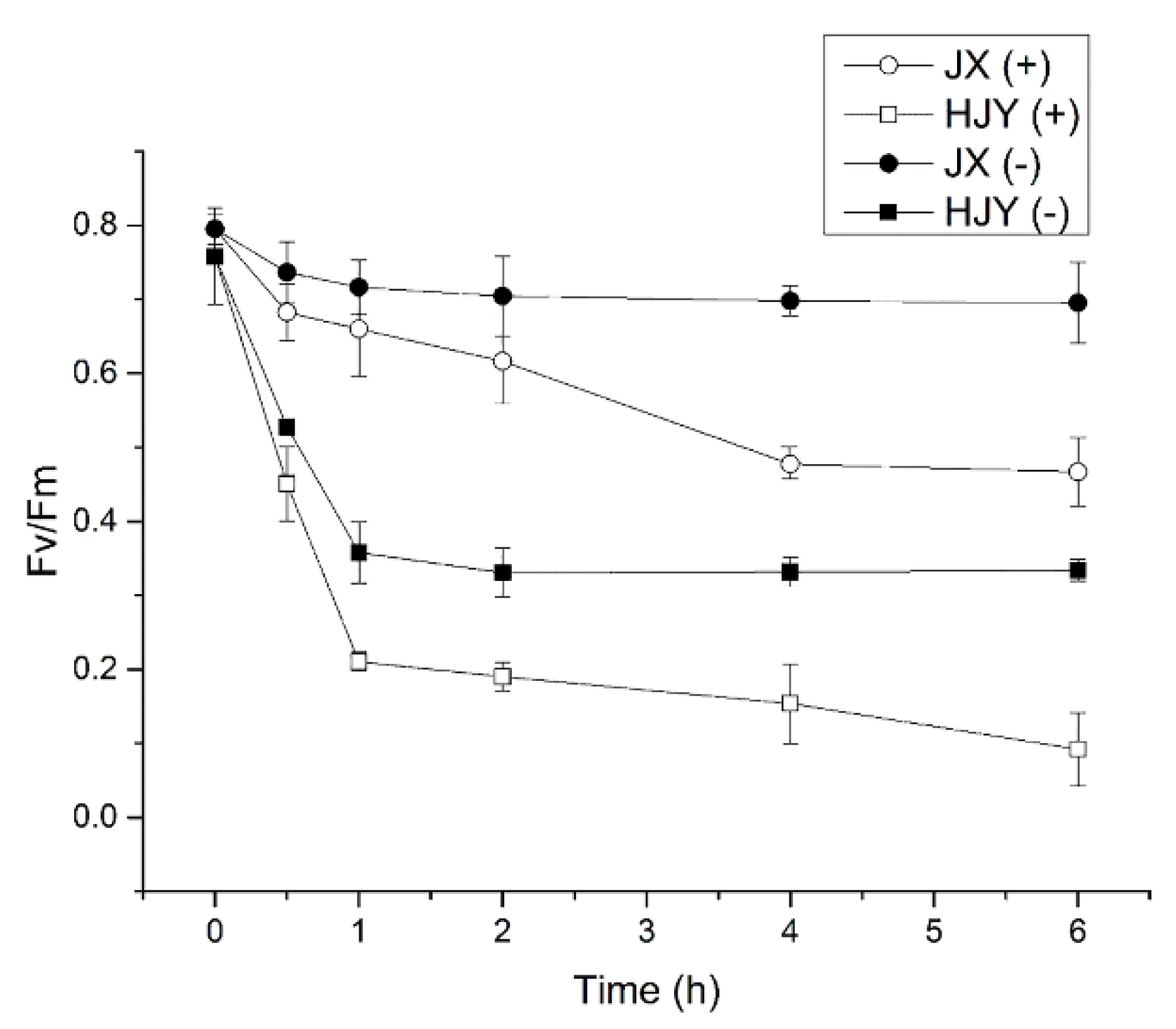
2.4. Effects of Light on PSII Activity
2.5. Effects of Inhibitor of Chloroplast-Encoded Protein Synthesis on PSII Activity
2.6. Sequence of Gene PsbC-Encoding CP43 Protein
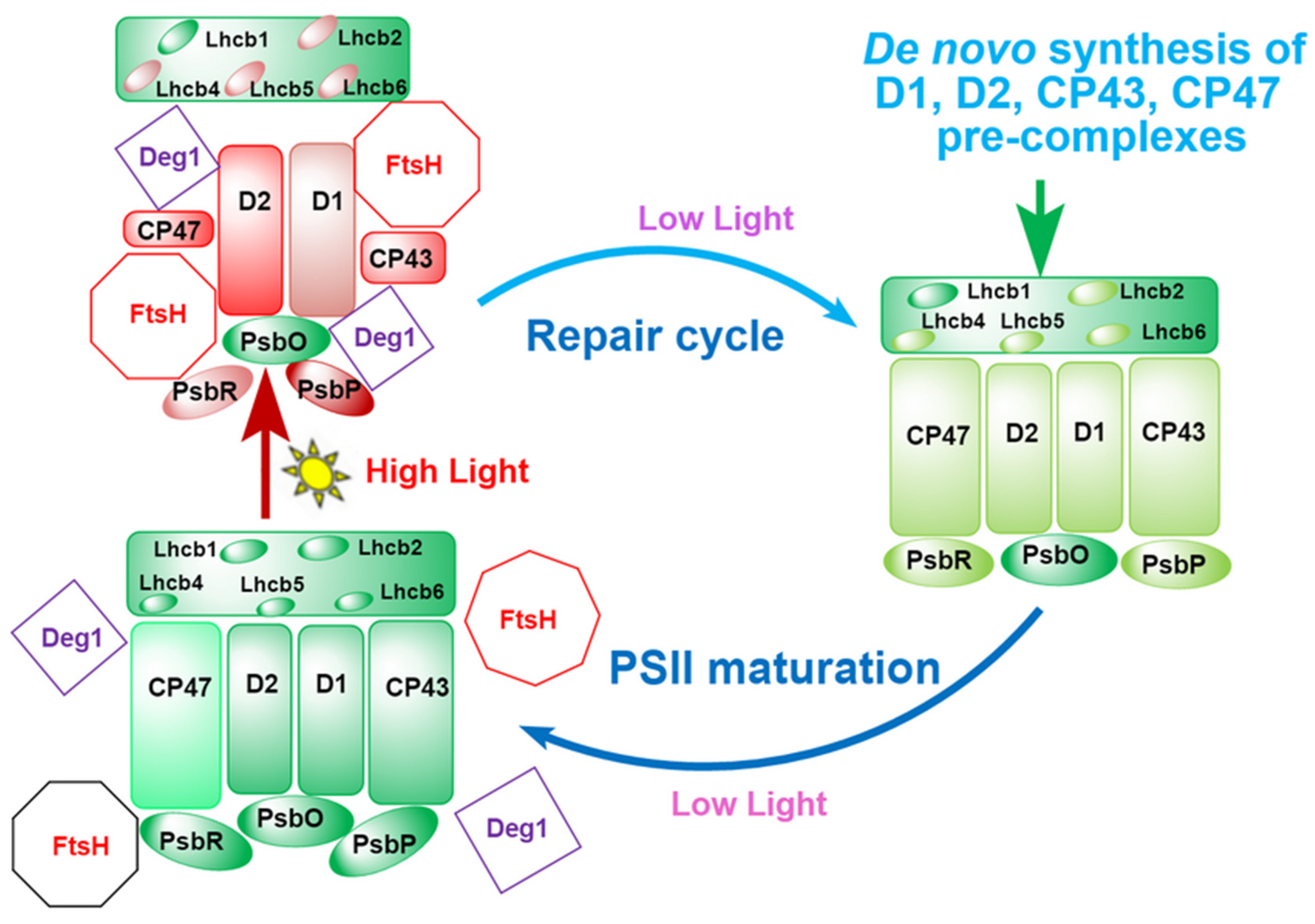
3. Discussion
3.1. Responses of PSII–LHCII SC Subunits in LS Albino Tea Cultivar to HL Stress
3.2. Photodamade of PSII and Albino Phenotype in LS Albino Tea Plant
3.3. Multicopy of Photosynthetic Proteins and Adaptation to HL Stress in LS Albino Tea Plant
3.4. HL-Induced Degradation of PSII Subunits and Crosslinking of Their Degradation Fragments
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Fv/Fm
4.3. Protein Preparation
4.4. SDS-PAGE and Immunoblotting Analysis
4.5. Cloning and Sequencing of PsbC
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interests
References
- Du, Y.Y.; Liang, Y.R.; Wang, H.; Wang, K.R.; Lu, J.L.; Zhang, G.H.; Lin, W.P.; Li, M.; Fang, Q.Y. A study on the chemical composition of albino tea cultivars. J. Hortic. Sci. Biotech. 2016, 81, 809–812. [Google Scholar] [CrossRef]
- Ma, Q.P.; Li, H.; Zou, Z.W.; Arkorful, E.; Lv, Q.R.; Zhou, Q.Q.; Chen, X.; Sun, K.; Li, X. Transcriptomic analyses identify albino-associated genes of a novel albino tea germplasm ‘Huabai 1’. Hortic. Res. 2018, 5, 54. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.H.; Yang, R.; Shi, Y.L.; Li, X.M.; Fu, Q.Y.; Lu, J.L.; Ye, J.H.; Wang, K.R.; Ma, S.C.; Zheng, X.Q.; et al. Light-sensitive albino tea plants and their characterization. Hortscience 2018, 53, 144–147. [Google Scholar] [CrossRef]
- Li, J.; Zeng, L.; Liao, Y.; Gu, D.; Tang, J.; Yang, Z. Influence of chloroplast defects on formation of jasmonic acid and characteristic aroma compounds in tea (Camellia sinensis) leaves exposed to postharvest stresses. Int. J. Mol. Sci. 2019, 20, 1044. [Google Scholar] [CrossRef]
- Cheng, S.H.; Fu, X.M.; Liao, Y.Y.; Xu, X.L.; Zeng, L.T.; Tang, J.C.; Li, J.; Lai, J.; Yang, Z. Differential accumulation of specialized metabolite L-theanine in green and albino-induced yellow tea (Camellia sinensis) leaves. Food Chem. 2018, 276, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Kambe, Y.; Ohshio, M.; Kunihiro, A.; Tanaka, Y.; Suzuki, T.; Nakamura, Y.; Morita, A.; Ikka, T. Integrated metabolome and transcriptome analyses reveal etiolation-induced metabolic changes leading to high amino acid contents in a light-sensitive Japanese albino tea cultivar. Front. Plant Sci. 2021, 11, 611140. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Tang, D.D.; Liu, M.Y.; Ruan, J.Y. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2018, 231, 272–281. [Google Scholar] [CrossRef]
- Xu, Y.X.; Chen, W.; Ma, C.L.; Shen, S.Y.; Zhou, Y.Y.; Zhou, L.Q.; Chen, L. Proteome and acetyl-proteome profiling of Camellia sinensis cv. ‘Anjin Baicha’ during periodic albinism reveals alterations in photosynthetic and secondary metabolite biosynthetic pathways. Front. Plant Sci. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Jiang, X.F.; Zhao, H.; Guo, F.; Shi, X.P.; Ye, C.; Yang, P.X.; Liu, B.; Ni, D. Transcriptomic analysis reveals mechanism of light-sensitive albinism in tea plant Camellia sinensis ‘Huangjinju’. BMC Plant Biol. 2020, 20, 216. [Google Scholar] [CrossRef] [PubMed]
- Song, L.B.; Ma, Q.P.; Zou, Z.W.; Sun, K.; Yao, Y.T.; Tao, J.H.; Kaleri, N.A.; Li, X. Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar ‘Huangjinya’. Front. Plant Sci. 2017, 8, 803. [Google Scholar] [CrossRef]
- Li, N.N.; Yang, Y.P.; Ye, J.H.; Lu, J.L.; Zheng, X.Q.; Liang, Y.R. Effects of sunlight on gene expression and chemical composition of light-sensitive albino tea plant. Plant Growth. Regul. 2016, 78, 253–262. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, J.D.; Wang, S.L.; Chen, L.; Ma, C.L.; Yao, M.Z. Repressed gene expression of photosynthetic antenna proteins associated with yellow leaf variation as revealed by bulked segregant RNA-seq in tea plant Camellia sinensis. J. Agric. Food Chem. 2020, 68, 8068–8079. [Google Scholar] [CrossRef] [PubMed]
- Drop, B.; Webber-Birungi, M.; Yadav, S.N.K.; Filipowicz-Szymanska, A.; Fusetti, F.; Boekema, E.J.; Croce, R. Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. BBA-Bioenergetics 2014, 1837, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.; Yocum, C.F. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [PubMed]
- Iwata, S.; Barber, J. Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr. Opin. Struc. Biol. 2004, 14, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Pokorska, B.; Zienkiewicz, M.; Powikrowska, M.; Drozak, A.; Romanowska, E. Differential turnover of the photosystem II reaction centre D1 protein in mesophyll and bundle sheath chloroplasts of maize. BBA-Bioenergetics 2009, 1787, 1161–1169. [Google Scholar] [CrossRef]
- Tyystjärvi, E.; Aro, E.M. The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc. Natl. Acad. Sci. USA 1996, 93, 2213–2218. [Google Scholar] [CrossRef]
- Cai, W.; Ma, J.; Guo, J.; Zhang, L. Function of ROC4 in the efficient repair of photodamaged photosystem II in Arabidopsis. Photochem. Photobiol. 2008, 84, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.R. The structure of Photosystem, II, and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 2015, 66, 23–48. [Google Scholar] [CrossRef]
- Kale, R.; Hebert, A.E.; Frankel, L.K.; Sallans, L.; Bricker, T.M.; Pospisil, P. Amino acid oxidation of the D1 and D2 proteins by oxygen radicals during photoinhibition of photosystem II. Proc. Natl. Acad. Sci. USA 2017, 114, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Barber, J. Photosystem II: The water splitting enzyme of photosynthesis and the origin of oxygen in our atmosphere. Q. Rev. Biophys. 2016, 49, e14. [Google Scholar] [CrossRef] [PubMed]
- Weisz, D.A.; Johnson, V.M.; Niedzwiedzki, D.M.; Shinn, M.K.; Liu, H.J.; Klitzke, C.F.; Gross, M.L.; Blankenship, R.E.; Lohman, T.M.; Pakrasi, H.B. A novel chlorophyll protein complex in the repair cycle of photosystem II. Proc. Natl. Acad. Sci. USA 2019, 116, 21907–21913. [Google Scholar] [CrossRef]
- Roose, J.L.; Frankel, L.K.; Mummadisetti, M.P.; Bricker, T.M. The extrinsic proteins of photosystem II: Update. Planta 2016, 243, 889–908. [Google Scholar] [CrossRef]
- Shimada, Y.; Suzuki, H.; Tsuchiya, T.; Tomo, T.; Noguchi, T.; Mimuro, M. Effect of a single-amino acid substitution of the 43 kDa chlorophyll protein on the oxygen-evolving reaction of the Cyanobacterium Synechocystis sp. PCC 6803: Analysis of the Glu354Gln mutation. Biochemistry 2009, 48, 6095–6103. [Google Scholar] [CrossRef] [PubMed]
- Weisz, D.A.; Gross, M.L.; Pakrasi, H.B. The use of advanced mass spectrometry to dissect the life-cycle of photosystem II. Front. Plant Sci. 2016, 7, 617. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef]
- Scharff, L.B.; Ehrnthaler, M.; Janowski, M.; Childs, L.H.; Hasse, C.; Gremmels, J.; Ruf, S.; Zoschke, R.; Bock, R. Shine-dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell 2017, 29, 3085–3101. [Google Scholar] [CrossRef]
- Gawronski, P.; Enroth, C.; Kindgren, P.; Marquardt, S.; Karpinski, S.; Leister, D.; Jensen, P.E.; Vinther, J.; Scharff, L.B. Light-dependent translation change of Arabidopsis psbA correlates with RNA structure alterations at the translation initiation region. Cells 2021, 10, 322. [Google Scholar] [CrossRef]
- Li, L.; Aro, E.M.; Millar, A.H. Mechanisms of photodamage and protein turnover in photoinhibition. Trends Plant Sci. 2018, 23, 667–676. [Google Scholar] [CrossRef]
- Fu, H.Y.; Ghandour, R.; Ruf, S.; Zoschke, R.; Bock, R.; Schottler, M.A. The availability of neither D2 nor CP43 limits the biogenesis of photosystem II in tobacco. Plant Physiol. 2021, 185, 1111–1130. [Google Scholar] [CrossRef]
- Shi, Y.; Che, Y.; Wang, Y.; Luan, S.; Hou, X. Loss of mature D1 leads to compromised CP43 assembly in Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.S.; Zhang, L.; Tu, W.F.; Sun, R.X.; Li, F.; Lin, Y.J.; Zhang, Y.; Liu, C.; Yang, C. Photosynthetic inner antenna CP47 plays important roles in ephemeral plants in adapting to high light stress. J. Plant Physiol. 2020, 251, 153189. [Google Scholar] [CrossRef]
- Che, Y.F.; Fu, A.G.; Hou, X.; McDonald, K.; Buchanan, B.B.; Huang, W.D.; Luan, S. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 16247–16252. [Google Scholar] [CrossRef]
- Komenda, J.; Reisinger, V.; Muller, B.C.; Dobakova, M.; Granvogl, B.; Eichacker, L.A. Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 2004, 279, 48620–48629. [Google Scholar] [CrossRef] [PubMed]
- Rokka, A.; Suorsa, M.; Saleem, A.; Battchikova, N.; Aro, E.M. Synthesis and assembly of thylakoid protein complexes: Multiple assembly steps of photosystem II. Biochem. J. 2005, 388, 159–168. [Google Scholar] [CrossRef]
- Boehm, M.; Romero, E.; Reisinger, V.; Yu, J.F.; Komenda, J.; Eichacker, L.A.; Dekker, J.P.; Nixon, P.J. Investigating the early stages of photosystem ii assembly in Synechocystis sp PCC 6803 isolation of CP47 and CP43 complexes. J. Biol. Chem. 2011, 286, 14812–14819. [Google Scholar] [CrossRef] [PubMed]
- Adir, N.; Zer, H.; Shochat, S.; Ohad, I. Photoinhibition—A historical perspective. Photosynth. Res. 2003, 76, 343–370. [Google Scholar] [CrossRef] [PubMed]
- Chotewutmontri, P.; Barkan, A. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 2016, 12, e1006106. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Aro, E.M. Photosynthetic responses of plants to excess light: Mechanisms and conditions for photoinhibition, excess energy dissipation and repair. Photosynthesis 2012, 34, 275–297. [Google Scholar] [CrossRef]
- Kouril, R.; Nosek, L.; Opatikova, M.; Arshad, R.; Semchonok, D.A.; Chamrad, I. Unique organization of photosystem II supercomplexes and megacomplexes in Norway spruce. Plant J. 2020, 104, 215–225. [Google Scholar] [CrossRef]
- Jansson, S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef]
- Dunsmuir, P. The petunia chlorophyll a/b binding protein genes: A comparison of Cab genes from different gene families. Nucleic Acids Res. 1985, 13, 2503–2518. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zolla, L.; Timperio, A.M.; Walcher, W.; Huber, C.G. Proteomics of light-harvesting proteins in different plant species. Analysis and comparison by liquid chromatography-electrospray ionization mass spectrometry. Photosystem II. Plant Physiol. 2003, 131, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.F.; Yan, H.C.; Wang, K.B.; Kuang, T.Y.; Zhang, J.P.; Gui, L.L.; An, X.; Chang, W. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 2004, 428, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Zhu, Q.D.; Pei, X.X.; Xing, G.Z.; Ou, X.Q.; Li, H. Comparative analysis of the photosynthetic physiology and transcriptome of a high-yielding wheat variety and its parents. Crop J. 2020, 8, 1037–1048. [Google Scholar] [CrossRef]
- Rea, G.; Volpicella, M.; De Leo, F.; Zolla, L.; Gallerani, R.; Ceci, L.R. Characterization of three members of the multigene family coding for isoforms of the chlorophyll-a/b-binding protein Lhcb1 in spinach. Physiol. Plantarum. 2007, 130, 167–176. [Google Scholar] [CrossRef]
- Hess, W.R.; Rocap, G.; Ting, C.S.; Larimer, F.; Stilwagen, S.; Lamerdin, J.; Chisholm, S.W. The photosynthetic apparatus of Prochlorococcus: Insights through comparative genomics. Photosynth. Res. 2001, 70, 53–71. [Google Scholar] [CrossRef]
- Bibby, T.S.; Mary, I.; Nield, J.; Partensky, F.; Barber, J. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 2003, 424, 1051–1054. [Google Scholar] [CrossRef]
- Yamatani, H.; Kohzuma, K.; Nakano, M.; Takami, T.; Kato, Y.; Hayashi, Y.; Monden, Y.; Okumoto, Y.; Abe, T.; Kumamaru, T.; et al. Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J. Exp. Bot. 2018, 69, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Rintamaki, E.; Kettunen, R.; Aro, E.M. Differential D1 dephosphorylation in functional and photodamaged photosystem II centers—Dephosphorylation is a prerequisite for degradation of damaged D1. J. Biol. Chem. 1996, 271, 14870–14875. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Akasaka, T. Degradation of antenna chlorophyll-binding protein CP43 during photoinhibition of photosystem II. Biochemistry 1995, 34, 9038–9045. [Google Scholar] [CrossRef] [PubMed]
- Lundin, B.; Thuswaldner, S.; Shutova, T.; Eshaghi, S.; Samuelsson, G.; Barber, J.; Andersson, B.; Spetea, C. Subsequent events to GTP binding by the plant PsbO protein: Structural changes, GTP hydrolysis and dissociation from the photosystem II complex. BBA-Bioenergetics 2007, 1767, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Lundin, B.; Hansson, M.; Schoefs, B.; Vener, A.V.; Spetea, C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 2007, 49, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Petersson, U.A.; Haas, B.J.; Funk, C.; Schroder, W.P.; Kieselbach, T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 8354–8365. [Google Scholar] [CrossRef] [PubMed]
- Krynicka, V.; Shao, S.; Nixon, P.J.; Komenda, J. Accessibility controls selective degradation of photosystem II subunits by FtsH protease. Nat. Plants 2015, 1, 15168. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Ozawa, S.I.; Takahashi, Y.; Sakamoto, W. D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynth. Res. 2015, 126, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.G.; Wang, X.L.; Kuang, T.Y. Light stability of the isolated CP43 studied by infrared and terahertz spectroscopy. Photosynthetica 2020, 58, 755–761. [Google Scholar] [CrossRef]
- Chotewutmontri, P.; Barkan, A. Light-induced psbA translation in plants is triggered by photosystem II damage via an assembly-linked autoregulatory circuit. Proc. Natl. Acad. Sci. USA 2020, 117, 21775–21784. [Google Scholar] [CrossRef]
- Gallo-Oller, G.; Ordonez, R.; Dotor, J. A new background subtraction method for Western blot densitometry band quantification through image analysis software. J. Immunol. Methods 2018, 457, 1–5. [Google Scholar] [CrossRef]
- Lee, M.; Park, J.; Lee, H.; Sohn, S.H.; Lee, J. Complete chloroplast genomic sequence of Citrus platymamma determined by combined analysis of Sanger and NGS data. Hortic. Environ. Biotechnol. 2015, 56, 704–711. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, W.-H.; Zheng, X.-Q.; Liang, Y.-R. High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants. Int. J. Mol. Sci. 2022, 23, 8522. https://doi.org/10.3390/ijms23158522
Cai W-H, Zheng X-Q, Liang Y-R. High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants. International Journal of Molecular Sciences. 2022; 23(15):8522. https://doi.org/10.3390/ijms23158522
Chicago/Turabian StyleCai, Wen-He, Xin-Qiang Zheng, and Yue-Rong Liang. 2022. "High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants" International Journal of Molecular Sciences 23, no. 15: 8522. https://doi.org/10.3390/ijms23158522
APA StyleCai, W.-H., Zheng, X.-Q., & Liang, Y.-R. (2022). High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants. International Journal of Molecular Sciences, 23(15), 8522. https://doi.org/10.3390/ijms23158522





