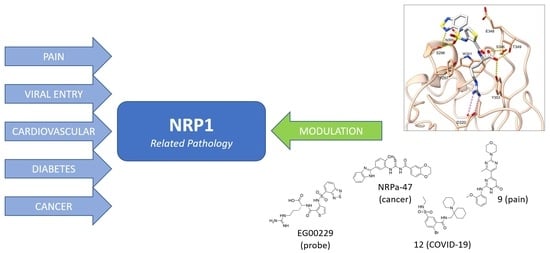
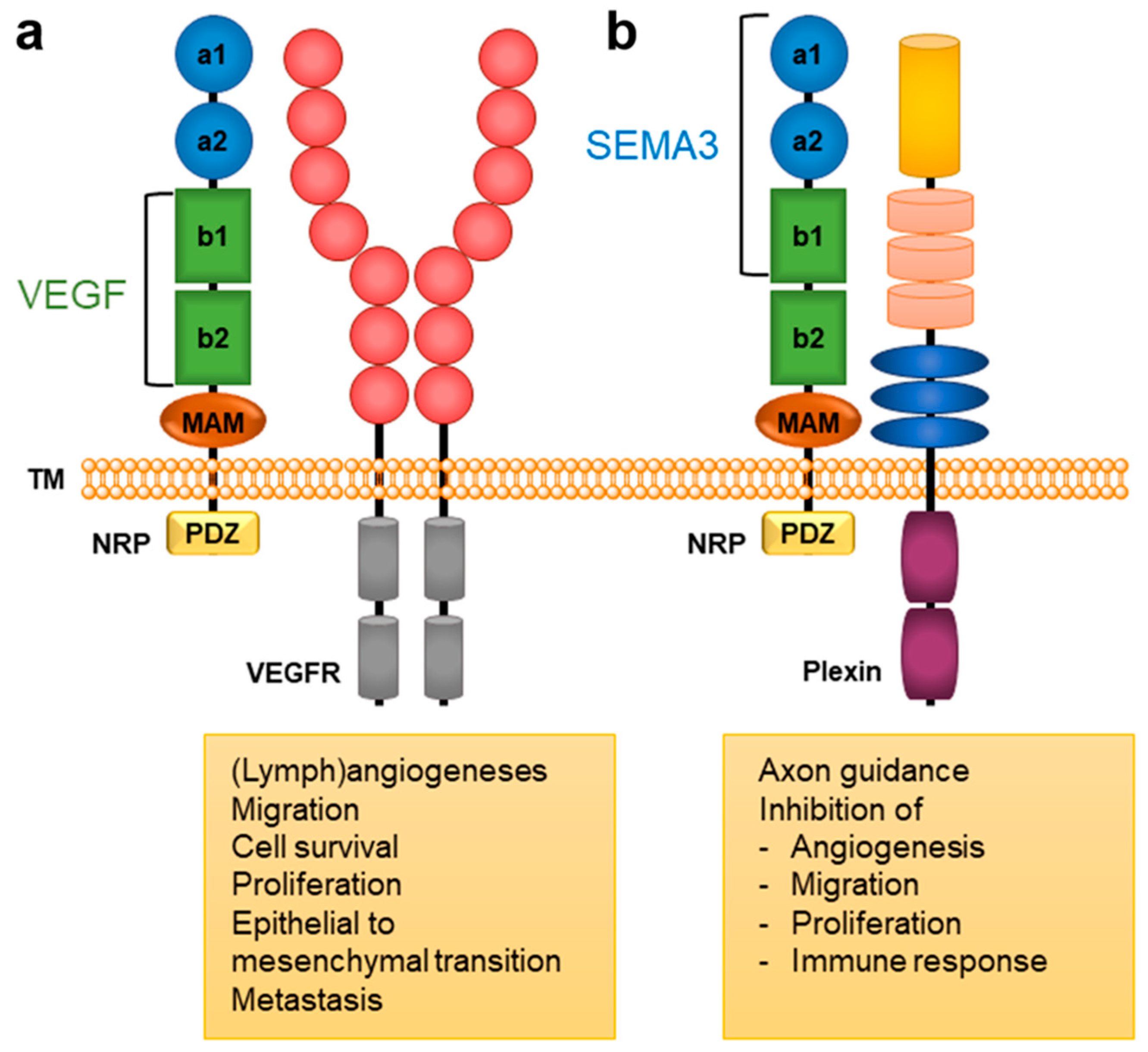
Neuropilin (NRPs) Related Pathological Conditions and Their Modulators
Abstract
:1. Introduction
2. NRP Binding of Endogenous Ligands

3. Neuropilin-Related Pathology
3.1. Pain
3.2. Viral Entry
3.3. Cardiovascular Diseases
3.4. Diabetes
3.5. Cancer
| Cancer Type | NRP1 | NRP2 | Reference |
|---|---|---|---|
| Leukaemia | x | [86,87,88,89,90,91] | |
| Breast cancer | x | x | [92,93,94,95,96,97,98,99,100,101,102] |
| Carcinoma | x | x | [103,104,105,106,107,108,109,110,111,112,113,114,115,116] |
| Colon & Colorectal cancer | x | x | [117,118,119,120,121,122,123,124,125,126] |
| Gastric cancer | x | [127,128,129,130,131,132,133,134,135,136,137,138] | |
| Lung cancer | x | x | [139,140,141,142,143,144,145,146,147,148,149] |
| Pancreatic cancer | x | [150,151,152,153,154] | |
| Prostate cancer | x | x | [155,156,157] |
| Melanoma | x | x | [158,159,160,161] |
| Glioma | x | [162,163,164,165,166,167,168] | |
| Liver cancer | x | [169] | |
| Mammary stem cells cancer | x | [170,171] | |
| Esophageal cancer | x | [172,173] | |
| Stem cell cancer | x | [174,175] | |
| Thyroid cancer | x | [176,177] | |
| Multiple myeloma | x | [51] | |
| Lymphoma | x | [178] | |
| Bladder cancer | x | [179,180] | |
| Tongue cancer | x | [181] | |
| Cervical cancer | x | [182,183] | |
| Gallbladder cancer | x | [184] | |
| Endometrium cancer | x | x | [185,186] |
4. Neuropilin-1 Modulators
4.1. Peptidomimetics
| Compound | Binding Mode | Inhibition of VEGF-A Binding [μM] | Ref. |
|---|---|---|---|
| Cancer | |||
 1 (EG00229) |  [a] [a] | IC50 = 3 | [35] |
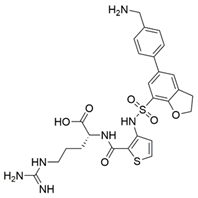 2 (EG01337) | 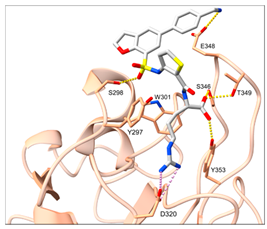 [b] [b] | IC50 = 0.6 | [188] |
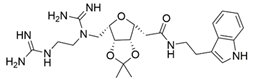 3 | NA | IC50 = 92 | [191] |
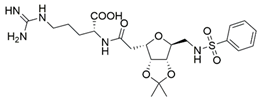 4 | NA | IC50 = 39 | [192] |
4.2. Small-Molecule Antagonists
| Compound | Binding Mode Prediction | Inhibition of VEGF-A Binding [μM] | Ref. |
|---|---|---|---|
| Cancer | |||
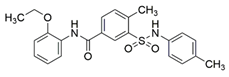
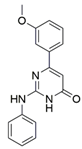
 5 (NRPa-47) | 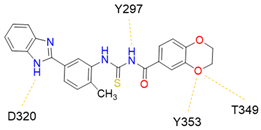 | IC50 = 34 | [193] |
 6 (NRPa-308) | 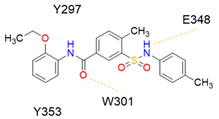 | 32% [a] | [194] |
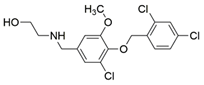 7 | 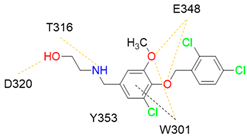 | Ki = 7.3 | [196] |
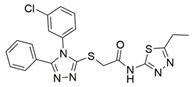 8 | 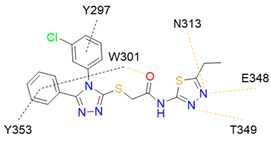 | IC50 = 19.1 | [197] |
| Neuropathic pain | |||
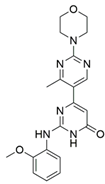 9 | 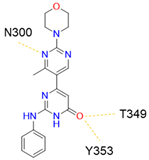 | IC50 = 0.000598 | [40] |
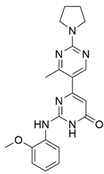 10 | NA | IC50 = 0.00314 | [40] |
| COVID-19 | SARS-CoV-2 inhibition [%] | ||
 11 | 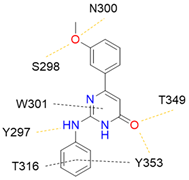 | >50 [b] | [40] |
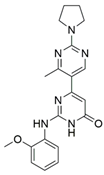 10 | NA | >50 [b] | [40] |
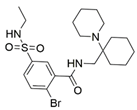 12 |  | 61.44 ± 2.48 [c] | [200] |
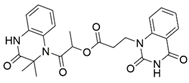 13 | 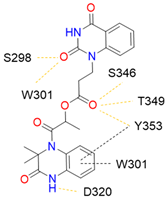 | 63.58 ± 1.27 [c] | [200] |
5. NRP2 Antagonists
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted role of neuropilins in the immune system: Potential targets for immunotherapy. Front. Immunol. 2017, 8, 1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niland, S.; Eble, J.A. Neuropilin: Handyman and power broker in the tumor microenvironment. Adv. Exp. Med. Biol. 2020, 1223, 31–67. [Google Scholar]
- Carmeliet, P.; Tessier-Lavigne, M. Common mechanisms of nerve and blood vessel wiring. Nature 2005, 436, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153. [Google Scholar] [CrossRef] [PubMed]
- Yelland, T.; Djordjevic, S. Crystal structure of the neuropilin-1 MAM domain: Completing the neuropilin-1 ectodomain picture. Structure 2016, 24, 2008–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, F.; Goshima, Y. Structural and functional relation of neuropilins. Neuropilin: From Nervous System to Vascular and Tumor Biol. Adv. Exp. Med. Bio 2002, 515, 55–69. [Google Scholar]
- Nasarre, P.; Gemmill, R.M.; Drabkin, H.A. The emerging role of class-3 semaphorins and their neuropilin receptors in oncology. Onco Targets Ther. 2014, 7, 1663. [Google Scholar] [PubMed] [Green Version]
- Prud’homme, G.J.; Glinka, Y. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget 2002, 3, 921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Tessier-Lavigne, M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 1997, 90, 739–751. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin, A.L.; Levengood, D.V.; Rowe, E.G.; Tai, Y.T.; Giger, R.J.; Ginty, D.D. Neuropilin is a semaphorin III receptor. Cell 1997, 90, 753–762. [Google Scholar] [CrossRef] [Green Version]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Chen, H.; Lu, L.; Wang, L.; Zhang, X.; Guo, X. New insights into the role of coreceptor neuropilins in tumour angiogenesis and lymphangiogenesis and targeted therapy strategies. J. Drug Target. 2021, 29, 155–167. [Google Scholar] [CrossRef]
- Takahashi, T.; Fournier, A.; Nakamura, F.; Wang, L.H.; Murakami, Y.; Kalb, R.G.; Fujisawa, H.; Strittmatter, S.M. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999, 99, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Tamagnone, L.; Artigiani, S.; Chen, H.; He, Z.; Ming, G.L.; Song, H.J.; Chedotal, A.; Winberg, M.L.; Goodman, C.S.; Poo, M.; et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999, 99, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Meyer, L.A.; Fritz, J.; Pierdant-Mancera, M.; Bagnard, D. Current drug design to target the Semaphorin/Neuropilin/Plexin complexes. Cell Adhes. Migr. 2016, 10, 700–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledano, S.; Nir-Zvi, I.; Engelman, R.; Kessler, O.; Neufeld, G. Class-3 semaphorins and their receptors: Potent multifunctional modulators of tumor progression. Int. J. Mol. Sci. 2019, 20, 556. [Google Scholar] [CrossRef] [Green Version]
- West, D.C.; Rees, C.G.; Duchesne, L.; Patey, S.J.; Terry, C.J.; Turnbull, J.E.; Delehedde, M.; Heegaard, C.W.; Allain, F.; Vanpouille, C.; et al. Interactions of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J. Biol. Chem. 2005, 280, 13457–13464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulpice, E.; Plouet, J.; Bergé, M.; Allanic, D.; Tobelem, G.; Merkulova-Rainon, T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood 2008, 111, 2036–2045. [Google Scholar] [CrossRef]
- Pellet-Many, C.; Mehta, V.; Fields, L.; Mahmoud, M.; Lowe, V.; Evans, I.; Ruivo, J.; Zachary, I. Neuropilins 1 and 2 mediate neointimal hyperplasia and re-endothelialisation following arterial injury. Cardiovasc. Res. 2015, 108, 288–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, J.L.; Sayers, J.; Chapman, C.; Pellet-Many, C. Emerging Roles for Neuropilin-2 in Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 5154. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, P.; Grubinger, M.; Gröger, C.; Huber, H.; Sieghart, W.; Peck-Radosavljevic, M.; Mikulits, W. Neuropilin-2 induced by transforming growth factor-β augments migration of hepatocellular carcinoma cells. BMC Cancer 2015, 15, 909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, Y.; Cho, J.G.; Park, J.Y.; Oh, S.; Park, D.; Yoo, K.H.; Lee, M.S.; Kwon, B.S.; Kim, J.; Yang, Y. MiR-146a Regulates Migration and Invasion by Targeting NRP2 in Circulating-Tumor Cell Mimicking Suspension Cells. Genes 2020, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Bai, Y.; Zhu, Q.; Hu, B.; Xu, Y. Targeting VEGF–neuropilin interactions: A promising antitumor strategy. Drug Discov. 2019, 24, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2019, 106, 16157–16162. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.Q.; Klagsbrun, M. Neuropilin is a mediator of angiogenesis. Cancer Metastasis Rev. 2000, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Lampropoulou, A.; Ruhrberg, C. Neuropilin regulation of angiogenesis. Biochem. Soc. Trans. 2014, 42, 1623–1628. [Google Scholar] [CrossRef]
- Staton, C.A.; Kumar, I.; Reed, M.W.R.; Brown, N.J. Neuropilins in physiological and pathological angiogenesis. J. Pathol. 2007, 212, 237–248. [Google Scholar] [CrossRef]
- Mei, B.; Chen, J.; Yang, N.; Peng, Y. The regulatory mechanism and biological significance of the Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and metastasis of gastric cancer. Cell Death Dis. 2020, 11, 241. [Google Scholar] [CrossRef]
- Hu, C.; Jiang, X. Role of NRP-1 in VEGF-VEGFR2-independent tumorigenesis. Target. Oncol. 2016, 11, 501–505. [Google Scholar] [CrossRef]
- Alghamdi, A.A.; Benwell, C.J.; Atkinson, S.J.; Lambert, J.; Johnson, R.T.; Robinson, S.D. NRP2 as an emerging angiogenic player; promoting endothelial cell adhesion and migration by regulating recycling of α5 integrin. Front. Cell Dev. Biol. 2020, 8, 395. [Google Scholar] [CrossRef]
- Iragavarapu-Charyulu, V.; Wojcikiewicz, E.; Urdaneta, A. Semaphorins in angiogenesis and autoimmune diseases: Therapeutic targets? Front. Immunol. 2020, 11, 346. [Google Scholar] [CrossRef]
- Caunt, M.; Mak, J.; Liang, W.C.; Stawicki, S.; Pan, Q.; Tong, R.K.; Plowman, G.; Kowalski, J.; Ho, C.; Reslan, H.B.; et al. Supplemental Data Blocking Neuropilin-2 Function Inhibits Tumor Cell Metastasis. Cancer Cell 2008, 13, 331–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvis, A.; Allerston, C.K.; Jia, H.; Herzog, B.; Garza-Garcia, A.; Winfield, N.; Ellard, K.; Aqil, R.; Lynch, R.; Chapman, C.; et al. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J. Med. Chem. 2010, 53, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Mota, F.; Fotinou, C.; Rana, R.R.; Chan, A.; Yelland, T.; Arooz, M.T.; O’Leary, A.P.; Hutton, J.; Frankel, P.; Zachary, I.; et al. Architecture and hydration of the arginine-binding site of neuropilin-1. FEBS J. 2018, 285, 1290–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Parker, M.W.; Xu, P.; Li, X.; Vander Kooi, C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012, 287, 11082–11089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llorián-Salvador, M.; González-Rodríguez, S. Painful understanding of VEGF. Front. Pharmacol. 2018, 9, 1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Miller, S.; Patek, M.; Moutal, A.; Duran, P.; Cabel, C.R.; Thorne, C.A.; Khanna, R. Novel compounds targeting neuropilin receptor 1 with potential to interfere with SARS-CoV-2 virus entry. ACS Chem. Neurosci. 2021, 12, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Hulse, R.P. Role of VEGF-A in chronic pain. Oncotarget 2017, 8, 10775. [Google Scholar] [CrossRef]
- Wang, H.B.; Zhang, H.; Zhang, J.P.; Li, Y.; Zhao, B.; Feng, G.K.; Zeng, M.S. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015, 6, 6240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghez, D.; Lepelletier, Y.; Lambert, S.; Fourneau, J.M.; Blot, V.; Janvier, S.; Hermine, O. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 2006, 80, 6844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, S.; Bouttier, M.; Vassy, R.; Seigneuret, M.; Petrow-Sadowski, C.; Janvier, S.; Pique, C. HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 2009, 113, 5176–5185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Lane, R.K.; Guo, H.; Fisher, A.D.; Diep, J.; Lai, Z.; Chen, Y.; Kaiser, W.J. Necroptosis-based CRISPR knockout screen reveals Neuropilin-1 as a critical host factor for early stages of murine cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 20109–20116. [Google Scholar] [CrossRef]
- Wang, H.C.; Huang, P.N.; Hung, H.C.; Tseng, S.N.; Chang, C.C.; Tsai, Y.R.; Hsu, J.T. Effect of a Neuropilin-1-Derived Virus Receptor Trap on Enterovirus A71 infection in vitro. Antimicrob. Agents Chemother. 2020, 65, e00695-20. [Google Scholar] [CrossRef]
- Raaben, M.; Jae, L.T.; Herbert, A.S.; Kuehne, A.I.; Stubbs, S.H.; Chou, Y.Y.; Whelan, S.P. NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 2017, 22, 688–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Martin, N.; Marcandalli, J.; Huang, C.S.; Arthur, C.P.; Perotti, M.; Foglierini, M.; Ciferri, C. An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 2018, 174, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Kusunoki, H.; Tanaka, T.; Kohno, T.; Matsuhashi, K.; Hosoda, K.; Wakamatsu, K.; Hamaguchi, I. A novel neuropilin-1–binding sequence in the human T-cell lymphotropic virus type 1 envelope glycoprotein. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 541–548. [Google Scholar] [CrossRef]
- Gu, Y.Y.; Luo, B.; Li, C.Y.; Huang, L.S.; Chen, G.; Feng, Z.B.; Peng, Z.G. Expression and clinical significance of neuropilin-1 in Epstein-Barr virus-associated lymphomas. Cancer Biomark. 2019, 25, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Sun, Y.; Carroll, C.R.; Usherwood, E.J. Neuropilin-1 regulates the secondary CD8 T cell response to virus infection. mSphere 2019, 4, e00221-19. [Google Scholar] [CrossRef] [Green Version]
- Davies, J.; Randeva, H.S.; Chatha, K.; Hall, M.; Spandidos, D.A.; Karteris, E.; Kyrou, I. Neuropilin 1 as a new potential SARS-CoV-2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID 19. Mol. Med. Rep. 2020, 22, 4221–4226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 589, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Bruder, D.; Probst-Kepper, M.; Westendorf, A.M.; Geffers, R.; Beissert, S.; Loser, K.; von Boehmer, H.; Buer, J.; Hansen, W. Frontline: Neuropilin-1: A surface marker of regulatory T cells. Eur. J. Immunol. 2004, 34, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Louvet, C.; Davini, D.; Gardner, J.M.; Martinez-Llordella, M.; Bailey-Bucktrout, S.; Anthony, B.A.; Sverdrup, F.M.; Head, R.; Kuster, D.J.; et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 2012, 209, 1713–1722. [Google Scholar] [CrossRef] [Green Version]
- Delgoffe, G.M.; Woo, S.R.; Turnis, M.E.; Gravano, D.M.; Guy, C.; Overacre, A.E.; Bettini, M.L.; Vogel, P.; Finkelstein, D.; Bonnevier, J.; et al. Stability and function of regulatory T cells is maintained by a neuropilin-1–semaphorin-4a axis. Nature 2013, 501, 252–256. [Google Scholar] [CrossRef]
- Liu, C.; Somasundaram, A.; Manne, S.; Gocher, A.M.; Szymczak-Workman, A.L.; Vignali, K.M.; Scott, E.N.; Normolle, D.P.; Wherry, E.J.; Lipson, E.J.; et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat. Immunol. 2020, 21, 1010–1021. [Google Scholar] [CrossRef]
- Shen, X.R.; Geng, R.; Li, Q.; Chen, Y.; Li, S.F.; Wang, Q.; Min, J.; Yang, Y.; Li, B.; Jiang, R.D.; et al. ACE2-independent infection of T lymphocytes by SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 83. [Google Scholar] [CrossRef]
- Nishiura, H.; Kobayashi, T.; Miyama, T.; Suzuki, A.; Jung, S.M.; Hayashi, K.; Kinoshita, R.; Yang, Y.; Yuan, B.; Akhmetzhanov, A.R.; et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int. J. Infect. Dis. 2020, 94, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Moutal, A.; Martin, L.F.; Boinon, L.; Gomez, K.; Ran, D.; Zhou, Y.; Stratton, H.J.; Cai, S.; Luo, S.; Gonzalez, K.B.; et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain 2021, 162, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, Y.; Yamada, S.; Thirunavukkarasu, M.; Nin, V.; Joshi, M.; Rishi, M.T.; Bhattacharya, S.; Camacho-Pereira, J.; Sharma, A.K.; et al. Cardiomyopathy and Worsened Ischemic Heart Failure in SM22-α Cre-Mediated Neuropilin-1 Null Mice: Dysregulation of PGC1α and Mitochondrial Homeostasis. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1401–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilari, S.; Wang, Y.; Singh, A.; Graham, R.P.; Iyer, V.; Thompson, S.M.; Torbenson, M.S.; Mukhopadhyay, D.; Misra, S. Neuropilin-1 deficiency in vascular smooth muscle cells is associated with hereditary hemorrhagic telangiectasia arteriovenous malformations. JCI Insight 2022, 7, e155565. [Google Scholar] [CrossRef]
- Matilla, L.; Arrieta, V.; Jover, E.; Garcia-Peña, A.; Martinez-Martinez, E.; Sadaba, R.; Alvarez, V.; Navarro, A.; Fernandez-Celis, A.; Gainza, A.; et al. Soluble St2 Induces Cardiac Fibroblast Activation and Collagen Synthesis via Neuropilin-1. Cells 2020, 9, 1667. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, J.; Zhao, F.; Zhi, L.; Wang, X.; Liu, L.; Bi, Z.; Zhao, Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020, 109, 531–538. [Google Scholar] [CrossRef]
- Sahni, S.; Gupta, G.; Sarda, R.; Pandey, S.; Pandey, R.M.; Sinha, S. Impact of metabolic and cardiovascular disease on COVID-19 mortality: A systematic review and meta-analysis. Diabetes Metab. Syndr. 2021, 15, 102308. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, X.; Chen, M.; Feng, Y.; Xiong, C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020, 116, 1097–1100. [Google Scholar] [CrossRef] [Green Version]
- Nicin, L.; Abplanalp, W.T.; Mellentin, H.; Kattih, B.; Tombor, L.; John, D.; Schmitto, J.D.; Heineke, J.; Emrich, F.; Arsalan, M.; et al. Cell type-specific expression of the putative SARS-CoV-2 receptor ACE2 in human hearts. Eur. Heart J. 2020, 41, 1804–1806. [Google Scholar] [CrossRef] [Green Version]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef] [PubMed]
- Klagsbrun, M.; Takashima, S.; Mamluk, R. The role of neuropilin in vascular and tumor biology. In Neuropilin: From Nervous System to Vascular and Tumor Biology; Springer: Berlin/Heidelberg, Germany, 2002; pp. 33–48. [Google Scholar]
- Mourad, D.; Azar, N.S.; Azar, S.T. Diabetic nephropathy and COVID-19: The potential role of immune actors. Int. J. Mol. Sci. 2021, 22, 7762. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S.; Li, M.; Wu, D.; Liu, F.; Yang, R.; Ji, S.; Ji, A.; Li, Y. The neuropilin-1 inhibitor, ATWLPPR peptide, prevents experimental diabetes-induced retinal injury by preserving vascular integrity and decreasing oxidative stress. PLoS ONE 2015, 10, e0142571. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, I.; Rüster, C.; Franke, S.; Liebisch, M.; Wolf, G. Erythropoietin ameliorates podocyte injury in advanced diabetic nephropathy in the db/db mouse. Am. J. Physiol. Ren. Physiol. 2013, 305, F911–F918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondeva, T.; Wolf, G. Role of Neuropilin-1 in Diabetic Nephropathy. J. Clin. Med. 2015, 4, 1293–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramek, H.; Sarközi, R.; Lauterberg, C.; Kronbichler, A.; Pirklbauer, M.; Albrecht, R.; Noppert, S.-J.; Perco, P.; Rudnicki, M.; Strutz, F.M.; et al. Neuropilin-1 and neuropilin-2 are differentially expressed in human proteinuric nephropathies and cytokine-stimulated proximal tubular cells. Lab. Investig. 2009, 89, 1304–1316. [Google Scholar] [CrossRef] [Green Version]
- Bondeva, T.; Rüster, C.; Franke, S.; Hammerschmid, E.; Klagsbrun, M.; Cohen, C.D.; Wolf, G. Advanced glycation end-products suppress neuropilin-1 expression in podocytes. Kidney Int. 2009, 75, 605–616. [Google Scholar] [CrossRef] [Green Version]
- Steenblock, C.; Richter, S.; Berger, I.; Barovic, M.; Schmid, J.; Schubert, U.; Jarzebska, N.; von Mässenhausen, A.; Linkermann, A.; Schürmann, A.; et al. Viral infiltration of pancreatic islets in patients with COVID-19. Nat. Commun. 2021, 12, 3534. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021, 33, 1565–1576. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today/ (accessed on 24 June 2022).
- Roy, S.; Bag, A.K.; Dutta, S.; Polavaram, N.S.; Islam, R.; Schellenburg, S.; Banwait, J.; Guda, C.; Ran, S.; Hollingsworth, M.A.; et al. Macrophage-Derived Neuropilin-2 Exhibits Novel Tumor-Promoting Functions. Cancer Res. 2018, 78, 5600–5617. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Vera, J.; Palmer, S.; Hernandez-Fernaud, J.R.; Dornier, E.; Mitchell, L.E.; Macpherson, I.; Edwards, J.; Zanivan, S.; Norman, J.C. A Proteomic Approach to Identify Endosomal Cargoes Controlling Cancer Invasiveness. J. Cell Sci. 2017, 78, 5600–5617. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Piechnik, A.; Dmoszynska, A.; Omiotek, M.; Mlak, R.; Kowal, M.; Stilgenbauer, S.; Bullinger, L.; Giannopoulos, K. The VEGF Receptor, Neuropilin-1, Represents a Promising Novel Target for Chronic Lymphocytic Leukemia Patients. Int. J. Cancer 2013, 133, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, L.; Xiao, Z.; Lu, S.; Yang, R.; Han, Z.C. Neuropilin-1 in Acute Myeloid Leukemia: Expression and Role in Proliferation and Migration of Leukemia Cells. Leuk. Lymphoma 2008, 49, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, W.; Ding, Y.; Xiu, B.; Li, P.; Dong, Y.; Zhu, Q.; Liang, A. Up-regulation of VEGF and its receptor in refractory leukemia cells. Int. J. Clin. Exp. Pathol. 2015, 8, 5282–5290. [Google Scholar] [PubMed]
- Palodetto, B.; da Silva Santos Duarte, A.; Rodrigues Lopes, M.; Adolfo Corrocher, F.; Roversi, F.M.; Soares Niemann, F.; Priscila Vieira Ferro, K.; Leda Figueiredo Longhini, A.; Melo Campos, P.; Favaro, P.; et al. SEMA3A Partially Reverses VEGF Effects through Binding to Neuropilin-1. Stem Cell Res. 2017, 22, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-G.; Wen, R.-T.; Qi, K.; Li, J.; Zheng, G.-X.; Wang, Y.-F.; Hong, Y.-G.; Zhang, Y.-M. The Neuropilin-1 Ligand, SEMA3A, Acts as a Tumor Suppressor in the Pathogenesis of Acute Leukemia. Anat. Rec. 2018, 302, 1127–1135. [Google Scholar] [CrossRef]
- Karjalainen, K.; Jaalouk, D.E.; Bueso-Ramos, C.E.; Zurita, A.J.; Kuniyasu, A.; Eckhardt, B.L.; Marini, F.C.; Lichtiger, B.; O’Brien, S.; Kantarjian, H.M.; et al. Targeting neuropilin-1 in human leukemia and lymphoma. Blood 2011, 117, 920–927. [Google Scholar] [CrossRef]
- Arpel, A.; Gamper, C.; Spenlé, C.; Fernandez, A.; Jacob, L.; Baumlin, N.; Laquerriere, P.; Orend, G.; Crémel, G.; Bagnard, D. Inhibition of Primary Breast Tumor Growth and Metastasis Using a Neuropilin-1 Transmembrane Domain Interfering Peptide. Oncotarget 2016, 7, 54723–54732. [Google Scholar] [CrossRef] [PubMed]
- Naik, A.; Al-Zeheimi, N.; Bakheit, C.S.; Al Riyami, M.; Al Jarrah, A.; Al Moundhri, M.S.; Al Habsi, Z.; Basheer, M.; Adham, S.A. Neuropilin-1 Associated Molecules in the Blood Distinguish Poor Prognosis Breast Cancer: A Cross-Sectional Study. Sci. Rep. 2017, 7, 3301. [Google Scholar] [CrossRef] [Green Version]
- Hellec, C.; Diawara, M.; Carpentier, M.; Denys, A.; Allain, F. The pro-Tumoral Activity of Heparan Sulfate 3-O-Sulfotransferase 3B (HS3ST3B) in Breast Cancer MDA-MB-231 Cells Is Dependent on the Expression of Neuropilin-1. Molecules 2018, 23, 2718. [Google Scholar] [CrossRef] [Green Version]
- Seifi-Alan, M.; Shams, R.; Bandehpour, M.; Mirfakhraie, R.; Ghafouri-Fard, S. Neuropilin-1 Expression Is Associated with Lymph Node Metastasis in Breast Cancer Tissues. Cancer Manag. Res. 2018, 10, 1969–1974. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, Y.; Wang, H.; Zheng, X.; Li, C.; Han, Z. Mir-376A Inhibits Breast Cancer Cell Progression by Targeting Neuropilin-1 NR. Onco Targets Ther. 2018, 11, 5293–5302. [Google Scholar] [CrossRef] [Green Version]
- Kiso, M.; Tanaka, S.; Saji, S.; Toi, M.; Sato, F. Long Isoform of VEGF Stimulates Cell Migration of Breast Cancer by Filopodia Formation via NRP1/ARHGAP17/cdc42 Regulatory Network. Int. J. Cancer 2018, 143, 2905–2918. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Y.; Li, C.; Liu, J.; Ren, H.; Li, L.; Zheng, X.; Wang, H.; Han, Z. RNA Binding Protein pum2 Promotes the Stemness of Breast Cancer Cells via Competitively Binding to Neuropilin-1 (NRP-1) Mrna with Mir-376a. Biomed. Pharm. 2019, 114, 108772. [Google Scholar] [CrossRef]
- Al-Zeheimi, N.; Naik, A.; Bakheit, C.S.; Al Riyami, M.; Al Ajarrah, A.; Al Badi, S.; Al Baimani, K.; Malik, K.; Al Habsi, Z.; Al Moundhri, M.S.; et al. Neoadjuvant Chemotherapy Alters Neuropilin-1, PLGF, and SNAI1 Expression Levels and Predicts Breast Cancer Patients Response. Front. Oncol. 2019, 25, 323. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, Y.N.; Xu, D.Q.; Huang, J.G.; Lv, D.; Shi, X.Y.; Liu, J.Y.; Ren, H.W.; Han, Z.X. Neuropilin1, a Novel Independent Prognostic Factor and Therapeutic Target in Triple-Negative Breast Cancer. Neoplasma 2021, 67, 1335–1345. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, J.; Wu, Q.; Cheng, Y.; Zheng, H.; Zhan, T.; Wang, H.; Yang, Y.; Wang, H.; Liu, Y.; et al. LINC-OIP5 in the Breast Cancer Cells Regulates Angiogenesis of Human Umbilical Vein Endothelial Cells through Yap1/Notch/NRP1 Signaling Circuit at a Tumor Microenvironment. Biol. Res. 2020, 53, 5. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, M.; Tao, Z.; Cao, J.; Wang, L.; Hu, X. Mir-331-3p Suppresses Cell Proliferation in TNBC Cells by Downregulating NRP2. Technol. Cancer Res. Treat. 2020, 19, 153303382090582. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, P.; Jiang, Y.; Dou, X.; Yan, J.; Ma, C.; Fan, Q.; Wang, W.; Su, F.; Tang, H.; et al. High Expression of Neuropilin-1 Associates with Unfavorable Clinicopathological Features in Hepatocellular Carcinoma. Pathol. Oncol. Res. 2015, 22, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.M.; Ng, K.Y.; Tong, M.; Chen, J.-N.; Chai, S.; Chan, K.-T.; Law, S.; Lee, N.P.; Choi, M.Y.; Li, B.; et al. Neuropilin-2 Promotes Tumourigenicity and Metastasis in Oesophageal Squamous Cell Carcinoma through Erk-MAPK-ETV4-MMP-E-Cadherin Deregulation. J. Pathol. 2016, 239, 309–319. [Google Scholar] [CrossRef]
- Dong, X.; Guo, W.; Zhang, S.; Wu, T.; Sun, Z.; Yan, S.; Zheng, S. Elevated Expression of Neuropilin-2 Associated with Unfavorable Prognosis in Hepatocellular Carcinoma. Onco Targets Ther. 2017, 10, 3827–3833. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Zhang, Y.; Wu, J.; Li, L.; Chen, N.; Ni, P.; Song, L.; Liu, X. Neuropilin 1 (NRP1) Is a Novel Tumor Marker in Hepatocellular Carcinoma. Clin. Chim. Acta 2018, 485, 158–165. [Google Scholar] [CrossRef]
- Stasikowska-Kanicka, O.; Wągrowska-Danilewicz, M.; Danilewicz, M. Immunohistochemical Study on Neuropilin 1 (NRP1) Immunoexpression in Oral Squamous Cell Carcinoma. Folia Histochem. Cytobiol. 2018, 56, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Cheng, C.; Xiong, H.; Wang, Y.; Chen, K.K.; Yang, J.; Xiao, B.; Zhang, R.; Li, S.; Sang, Y. NRP1 promotes cell migration and invasion and serves as a therapeutic target in nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 2460–2469. [Google Scholar]
- Adil, A.A.; Bommanabonia, A.K.; Vaithy, A.; Kumar, S.; Waseem, M.; Jamal, S.; Ahmed, N. Differential Expression of Helios, Neuropilin-1 and Foxp3 in Head and Neck Squamous Cell Carcinoma (HNSCC) Patients. 3 Biotech. 2019, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Santoro, A.; Kelley, R.K.; Gane, E.; Paradis, V.; Cleverly, A.; Smith, C.; Estrem, S.T.; Man, M.; Wang, S.; et al. Correction: Biomarkers and Overall Survival in Patients with Advanced Hepatocellular Carcinoma Treated with TGF-ΒRI Inhibitor Galunisertib. PLoS ONE 2021, 16, e0253671. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, C.; Song, A.; Jiang, F.; Zhou, J.; Li, G.; Zhang, W.; Ye, J.; Ding, X.; Zhang, W.; et al. CMTM6 promotes cell proliferation and invasion in oral squamous cell carcinoma by interacting with NRP1. Am. J. Cancer Res. 2020, 10, 1691–1709. [Google Scholar]
- Wu, Y.-N.; He, L.-H.; Bai, Z.-T.; Li, X. NRP1 Is a Prognostic Factor and Promotes the Growth and Migration of Cells in Intrahepatic Cholangiocarcinoma. Cancer Manag. Res. 2020, 12, 7021–7032. [Google Scholar] [CrossRef]
- Morin, E.; Lindskog, C.; Johansson, M.; Egevad, L.; Sandström, P.; Harmenberg, U.; Claesson-Welsh, L.; Sjöberg, E. Perivascular Neuropilin-1 Expression Is an Independent Marker of Improved Survival in Renal Cell Carcinoma. J. Pathol. 2020, 250, 387–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.-C.; Chen, T.-Y.; Liao, L.-T.; Chen, T.; Li, Q.-L.; Xu, J.-X.; Hu, J.-W.; Zhou, P.-H.; Zhang, Y.-Q. NETO2 Promotes Esophageal Cancer Progression by Inducing Proliferation and Metastasis via PI3K/Akt and Erk Pathway. Int. J. Biol. Sci. 2021, 17, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Dumond, A.; Brachet, E.; Durivault, J.; Vial, V.; Puszko, A.K.; Lepelletier, Y.; Montemagno, C.; Pagnuzzi-Boncompagni, M.; Hermine, O.; Garbay, C.; et al. Neuropilin 1 and Neuropilin 2 Gene Invalidation or Pharmacological Inhibition Reveals Their Relevance for the Treatment of Metastatic Renal Cell Carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 33. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Park, K.C.; Kim, J.G.; Moon, S.J.; Kang, S.B.; Lee, D.S.; Sul, H.J.; Ji, J.S.; Jeong, H.Y. Dysregulation of MicroRNA-196B-5p and MicroRNA-375 in Gastric Cancer. J. Gastric Cancer 2016, 16, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.; Liu, C.; Li, M.; Hu, Y.; Shi, N.; Yu, H.; Liu, H. Mir-486-5p Attenuates Tumor Growth and Lymphangiogenesis by Targeting Neuropilin-2 in Colorectal Carcinoma. Onco Targets Ther. 2016, 9, 2865–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.-P.; Wei, J.-C.; Wang, Q.; Yang, P.; Li, W.-L.; He, F.; Chen, H.C.; Hu, H.; Zhong, J.-B.; Cao, J. Long Non-Coding RNA 00152 Functions as a Competing Endogenous RNA to Regulate NRP1 Expression by Sponging with MIRNA-206 in Colorectal Cancer. Int. J. Oncol. 2018, 53, 1227–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomida, C.; Yamagishi, N.; Nagano, H.; Uchida, T.; Ohno, A.; Hirasaka, K.; Nikawa, T.; Teshima-Kondo, S. VEGF Pathway-Targeting Drugs Induce Evasive Adaptation by Activation of Neuropilin-1/CMET in Colon Cancer Cells. Int. J. Oncol. 2018, 52, 1350–1362. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.; Liu, L.; Lu, H. LncRNA Xist Facilitates Proliferation and Epithelial–Mesenchymal Transition of Colorectal Cancer Cells through Targeting Mir-486-5p and Promoting Neuropilin-2. J. Cell Physiol. 2019, 234, 13747–13761. [Google Scholar] [CrossRef] [PubMed]
- Bollard, J.; Patte, C.; Radkova, K.; Massoma, P.; Chardon, L.; Valantin, J.; Gadot, N.; Goddard, I.; Vercherat, C.; Hervieu, V.; et al. Neuropilin-2 Contributes to Tumor Progression in Preclinical Models of Small Intestinal Neuroendocrine Tumors. J. Pathol. 2019, 249, 343–355. [Google Scholar] [CrossRef]
- Huang, X.; Ye, Q.; Chen, M.; Li, A.; Mi, W.; Fang, Y.; Zaytseva, Y.Y.; O’Connor, K.L.; Vander Kooi, C.W.; Liu, S.; et al. N-Glycosylation-Defective Splice Variants of Neuropilin-1 Promote Metastasis by Activating Endosomal Signals. Nat. Commun. 2019, 10, 3708. [Google Scholar] [CrossRef]
- Ding, G.Y.; Xu, W.G.; Guo, S.; Zhan, Q.Q.; Yang, X.; Jia, C.L. Effect of RNA interference targeting neuropilin-2 gene on proliferation and apoptosis of colon cancer cell line HCT-8. Zhonghua Yi Xue Za Zhi 2020, 100, 3879–3883. [Google Scholar] [PubMed]
- De Vlaeminck, Y.; Bonelli, S.; Awad, R.M.; Dewilde, M.; Rizzolio, S.; Lecocq, Q.; Bolli, E.; Santos, A.R.; Laoui, D.; Schoonooghe, S.; et al. Targeting Neuropilin-1 with Nanobodies Reduces Colorectal Carcinoma Development. Cancers 2020, 12, 3582. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, S.; Park, S. IRGD Peptide as a Tumor-Penetrating Enhancer for Tumor-Targeted Drug Delivery. Polymers 2020, 12, 1906. [Google Scholar] [CrossRef] [PubMed]
- Akagi, M.; Kawaguchi, M.; Liu, W.; McCarty, M.F.; Takeda, A.; Fan, F.; Stoeltzing, O.; Parikh, A.A.; Jung, Y.D.; Bucana, C.D.; et al. Induction of Neuropilin-1 and Vascular Endothelial Growth Factor by Epidermal Growth Factor in Human Gastric Cancer Cells. Br. J. Cancer 2003, 88, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jiang, X.; Zhang, Q.; Dong, X.; Gao, Y.; He, Y.; Qiao, H.; Xie, F.; Xie, X.; Sun, X. Neuropilin-1 Is Associated with Clinicopathology of Gastric Cancer and Contributes to Cell Proliferation and Migration as Multifunctional Coreceptors. J. Exp. Clin. Cancer Res. 2016, 35, 16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xing, Y.; Gao, Q.; Sun, X.; Zhang, D.; Cao, G. Combination of NRP1-Mediated Irgd with 5-Fluorouracil Suppresses Proliferation, Migration and Invasion of Gastric Cancer Cells. Biomed. Pharmacother. 2017, 93, 1136–1143. [Google Scholar] [CrossRef]
- Wang, X.; Hu, H.; Liu, H. RNA Binding Protein lin28b Confers Gastric Cancer Cells Stemness via Directly Binding to NRP-1. Biomed. Pharmacother. 2018, 104, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Shi, B.; Fu, Y.; Zhao, S.; Qu, K.; Guo, Q.; Li, K.; She, J. Hypomethylated Gene NRP1 Is Co-Expressed with PDGFRB and Associated with Poor Overall Survival in Gastric Cancer Patients. Biomed. Pharmacother. 2019, 111, 1334–1341. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Y.J.; Shi, Y.; Wu, T. NRP-1 and KDR Polymorphisms Are Associated with Survival Time in Patients with Advanced Gastric Cancer. Oncol. Lett. 2019, 18, 4629–4638. [Google Scholar] [CrossRef]
- Pang, W.; Zhai, M.; Wang, Y.; Li, Z. Long Noncoding RNA SNHG16 Silencing Inhibits the Aggressiveness of Gastric Cancer via Upregulation of MicroRNA-628-3P and Consequent Decrease of nrp1. Cancer Manag. Res. 2019, 11, 7263–7277. [Google Scholar] [CrossRef] [Green Version]
- Hang, C.; Yan, H.S.; Gong, C.; Gao, H.; Mao, Q.H.; Zhu, J.X. MicroRNA-9 Inhibits Gastric Cancer Cell Proliferation and Migration by Targeting Neuropilin-1. Exp. Ther. Med. 2019, 18, 2524–2530. [Google Scholar] [CrossRef]
- Wu, C.; Zeng, M.-H.; Liao, G.; Qian, K.; Li, H. Neuropilin-1 Interacts with Fibronectin-1 to Promote Epithelial–Mesenchymal Transition Progress in Gastric Cancer. Onco Targets Ther. 2020, 13, 10677–10687. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Guo, S.; Tang, J.; Wen, J.; Wang, H.; Hu, X.; Gu, Q. MicroRNA-19B-3P Suppresses Gastric Cancer Development by Negatively Regulating Neuropilin-1. Cancer Cell Int. 2020, 25, 193. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Li, Y.; Huang, L.; Bai, B.; Xu, Z. Clinicopathological Significance of Neuropilin 1 Expression in Gastric Cancer: A Meta-Analysis. Dis. Markers 2020, 2020, 4763492. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Hyeon, J.; Song, I.H.; Lee, H.H. Relationship between Neuropilin-1 Expression and Prognosis, According to Gastric Cancer Histology. J. Mol. Histol. 2020, 51, 199–208. [Google Scholar] [CrossRef]
- Dong, J.C.; Gao, H.; Zuo, S.Y.; Zhang, H.Q.; Zhao, G.; Sun, S.L.; Han, H.L.; Jin, L.L.; Shao, L.H.; Wei, W.; et al. Neuropilin 1 Expression Correlates with the Radio-Resistance of Human Non-Small-Cell Lung Cancer Cells. J. Cell Mol. Med. 2015, 19, 2286–2295. [Google Scholar] [CrossRef]
- Gemmill, R.M.; Nasarre, P.; Nair-Menon, J.; Cappuzzo, F.; Landi, L.; D’Incecco, A.; Uramoto, H.; Yoshida, T.; Haura, E.B.; Armeson, K.; et al. The Neuropilin 2 Isoform nrp2b Uniquely Supports TGFΒ-Mediated Progression in Lung Cancer. Sci. Signal. 2017, 10, eaag0528. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhu, P.; Xia, Y.; Hui, K.; Wang, M.; Jiang, X. Role of the NRP-1-Mediated VEGFR2-Independent Pathway on Radiation Sensitivity of Non-Small Cell Lung Cancer Cells. J. Cancer Res. Clin. Oncol. 2018, 144, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Hernandez, L.; Vazquez-Santillan, K.; Castro-Oropeza, R.; Martinez-Ruiz, G.; Muñoz -Galindo, L.; Gonzalez-Torres, C.; Cortes-Gonzalez, C.; Victoria-Acosta, G.; Melendez-Zajgla, J.; Maldonado, V. NRP1-Positive Lung Cancer Cells Possess Tumor-Initiating Properties. Oncol. Rep. 2017, 39, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Xiong, K.; Shao, L.H.; Zhang, H.Q.; Jin, L.; Wei, W.; Dong, Z.; Zhu, Y.Q.; Wu, N.; Jin, S.Z.; Xue, L.X. MicroRNA-9 Functions as a Tumor Suppressor and Enhances Radio-Sensitivity in Radio-Resistant A549 Cells by Targeting Neuropilin 1. Oncol. Lett. 2017, 15, 2863–2870. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Du, W.; Lei, Z.; Zhang, Y.; Zhu, J.; Zeng, Y.; Wang, S.; Zheng, Y.; Liu, Z.; Huang, J.A. Neuropilin 1 Modulates Tgf-β1-Induced Epithelial-Mesenchymal Transition in Non-Small Cell Lung Cancer. Int. J. Oncol. 2019, 15, 2863–2870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Zhu, J.; Zeng, Y.; Du, W.; Zhang, Y.; Tang, H.; Zheng, Y.; Qin, H.; Liu, Z.; Huang, J. The Regulation of Neuropilin 1 Expression by Mir-338-3p Promotes Non-Small Cell Lung Cancer via Changes in EGFR Signaling. Mol. Carcinog. 2019, 58, 1019–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Zhang, H.; Gong, X.; Wei, W.; Lv, Y.; Chen, Z.; Wang, R.; Yi, J.; Shen, Y.; Jin, S. The Role of the Tumor Microenvironment in Neuropilin 1-Induced Radiation Resistance in Lung Cancer Cells. J. Cancer 2019, 10, 4017–4030. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Baek, D.-S.; Lee, S.; Park, D.; Kang, H.N.; Cho, B.C.; Kim, Y.-S. Dual-Targeting of EGFR and Neuropilin-1 Attenuates Resistance to EGFR-Targeted Antibody Therapy in KRAS-Mutant Non-Small Cell Lung Cancer. Cancer Lett. 2019, 466, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Liu, L.; Hu, C.; Liu, Y.; Qiao, Y.; Jiang, X. Expression of VEGFR2 and NRP-1 in non-small cell lung cancer and their clinical significance. Chin. J. Cancer Res. 2014, 26, 669–677. [Google Scholar]
- Dimou, A.; Nasarre, C.; Peterson, Y.K.; Pagano, R.; Gooz, M.; Nasarre, P.; Drabkin, H.A.; Armeson, K.E.; Gibney, B.C.; Gemmill, R.M.; et al. Neuropilin-2B Facilitates Resistance to Tyrosine Kinase Inhibitors in Non–Small Cell Lung Cancer. J. Thorac. Cardiovasc. Surg. 2021, 162, 463–473. [Google Scholar] [CrossRef]
- Matkar, P.N.; Singh, K.K.; Rudenko, D.; Kim, Y.J.; Kuliszewski, M.A.; Prud’homme, G.J.; Hedley, D.W.; Leong-Poi, H. Novel Regulatory Role of Neuropilin-1 in Endothelial-to-Mesenchymal Transition and Fibrosis in Pancreatic Ductal Adenocarcinoma. Oncotarget 2016, 7, 69489–69506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Lin, P.; Perrett, I.; Lin, J.; Liao, Y.-P.; Chang, C.H.; Jiang, J.; Wu, N.; Donahue, T.; Wainberg, Z.; et al. Tumor-Penetrating Peptide Enhances Transcytosis of Silicasome-Based Chemotherapy for Pancreatic Cancer. J. Clin. Investig. 2017, 127, 2007–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, E.; Sjöberg, E.; Tjomsland, V.; Testini, C.; Lindskog, C.; Franklin, O.; Sund, M.; Öhlund, D.; Kiflemariam, S.; Sjöblom, T.; et al. VEGF Receptor-2/Neuropilin 1 Trans.-Complex Formation between Endothelial and Tumor Cells Is an Independent Predictor of Pancreatic Cancer Survival. J. Pathol. 2018, 246, 311–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Ma, L.; Zhai, B.; Zhu, H.; Li, W.; Jiang, W.; Lei, L.; Zhang, S.; Qiao, H.; Jiang, X.; Sun, X. The Mir-141/Neuropilin-1 Axis Is Associated with the Clinicopathology and Contributes to the Growth and Metastasis of Pancreatic Cancer. Cancer Cell Int. 2019, 19, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Fan, S.; Pan, X.; Xiaokaiti, Y.; Duan, J.; Shi, Y.; Pan, Y.; Tie, L.; Wang, X.; Li, Y.; et al. Nordihydroguaiaretic Acid Impairs Prostate Cancer Cell Migration and Tumor Metastasis by Suppressing Neuropilin 1. Oncotarget 2016, 7, 86225–86238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tse, B.W.; Volpert, M.; Ratther, E.; Stylianou, N.; Nouri, M.; McGowan, K.; Lehman, M.L.; McPherson, S.J.; Roshan-Moniri, M.; Butler, M.S.; et al. Neuropilin-1 Is Upregulated in the Adaptive Response of Prostate Tumors to Androgen-Targeted Therapies and Is Prognostic of Metastatic Progression and Patient Mortality. Oncogene 2017, 36, 3417–3427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borkowetz, A.; Froehner, M.; Rauner, M.; Conrad, S.; Erdmann, K.; Mayr, T.; Datta, K.; Hofbauer, L.C.; Baretton, G.B.; Wirth, M.; et al. Neuropilin-2 Is an Independent Prognostic Factor for Shorter Cancer-Specific Survival in Patients with Acinar Adenocarcinoma of the Prostate. Int. J. Cancer 2019, 146, 2619–2627. [Google Scholar] [CrossRef] [Green Version]
- Moriarty, W.F.; Kim, E.; Gerber, S.A.; Hammers, H.; Alani, R.M. Neuropilin-2 Promotes Melanoma Growth and Progression in Vivo. Melanoma Res. 2016, 26, 321–328. [Google Scholar] [CrossRef]
- Dou, X.; Yan, J.; Zhang, Y.; Liu, P.; Jiang, Y.; Lv, S.; Zeng, F.; Chen, X.; Wang, S.; Zhang, H.; et al. SPECT Imaging of Neuropilin Receptor Type-1 Expression with 131i-Labeled Monoclonal Antibody. Int. J. Oncol. 2016, 49, 961–970. [Google Scholar] [CrossRef] [Green Version]
- Eisenstein, A.; Panova, I.P.; Chung, H.J.; Goldberg, L.J.; Zhang, Q.; Lazova, R.; Bhawan, J.; Busam, K.J.; Symanowski, J.T.; Alani, R.M.; et al. Quantitative Assessment of Neuropilin-2 as a Simple and Sensitive Diagnostic Assay for SPITZOID Melanocytic Lesions. Melanoma Res. 2018, 28, 71–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzolio, S.; Cagnoni, G.; Battistini, C.; Bonelli, S.; Isella, C.; Van Ginderachter, J.A.; Bernards, R.; Di Nicolantonio, F.; Giordano, S.; Tamagnone, L. Neuropilin-1 Upregulation Elicits Adaptive Resistance to Oncogene-Targeted Therapies. J. Clin. Investig. 2018, 128, 3976–3990. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, L.; Sun, K.; Khan, A.A.; Yan, J.; Liu, H.; Lu, A.; Gu, N. Neuropilin-1 (NRP-1)/GIPC1 Pathway Mediates Glioma Progression. Tumor Biol. 2016, 37, 13777–13788. [Google Scholar] [CrossRef]
- Kwiatkowski, S.C.; Guerrero, P.A.; Hirota, S.; Chen, Z.; Morales, J.E.; Aghi, M.; McCarty, J.H. Neuropilin-1 Modulates TGFΒ Signaling to Drive Glioblastoma Growth and Recurrence after Anti-Angiogenic Therapy. PLoS ONE 2017, 12, e0185065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caponegro, M.D.; Moffitt, R.A.; Tsirka, S.E. Expression of Neuropilin-1 Is Linked to Glioma Associated Microglia and Macrophages and Correlates with Unfavorable Prognosis in High Grade Gliomas. Oncotarget 2018, 9, 35655–35665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyauchi, J.T.; Caponegro, M.D.; Chen, D.; Choi, M.K.; Li, M.; Tsirka, S.E. Deletion of Neuropilin 1 from Microglia or Bone Marrow–Derived Macrophages Slows Glioma Progression. Cancer Res. 2017, 78, 685–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Chen, L.; Khan, A.A.; Li, B.; Gu, B.; Lin, F.; Su, X.; Yan, J. MIRNA-124-3p/Neuropilin-1(NRP-1) Axis Plays an Important Role in Mediating Glioblastoma Growth and Angiogenesis. Int. J. Cancer 2018, 143, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, C.; Almasoud, A.; Pellegrini-Moïse, N.; Pinel, S.; Barberi-Heyob, M.; Chastagner, P.; Boura, C. PO-013 a Novel Peptidomimetic Targeting NRP1 Increases Radiosensitivity of Medulloblastoma Stem Cells. ESMO Open 2018, 3, A232. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Valduga, J.; Chateau, A.; Richard, M.; Pellegrini-Moïse, N.; Barberi-Heyob, M.; Chastagner, P.; Boura, C. Stimulation of Medulloblastoma Stem Cells Differentiation by a Peptidomimetic Targeting Neuropilin-1. Oncotarget 2018, 9, 15312–15325. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.C.; Shen, H.X.; Chen, C.; Ma, L.; Li, W.Z.; Wang, L.; Geng, Z.M. Neuropilin-1 Promotes Primary Liver Cancer Progression by Potentiating the Activity of Hepatic Stellate Cells. Oncol. Lett. 2017, 15, 2245–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grun, D.; Adhikary, G.; Eckert, R.L. VEGF-A Acts via Neuropilin-1 to Enhance Epidermal Cancer Stem Cell Survival and Formation of Aggressive and Highly Vascularised Tumors. Oncogene 2016, 35, 4379–4387. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wu, T.; Dong, X.; Zeng, Y.A. Neuropilin-1 Is Upregulated by Wnt/β-Catenin Signaling and Is Important for Mammary Stem Cells. Sci. Rep. 2017, 7, 10941. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.H.; Rockstroh, A.; Sokolowski, K.A.; Lynam, L.R.; Lehman, M.; Thompson, E.W.; Gregory, P.A.; Nelson, C.C.; Volpert, M.; Hollier, B.G. Neuropilin-1 is over-expressed in claudin-low breast cancer and promotes tumor progression through acquisition of stem cell characteristics and RAS/MAPK pathway activation. Breast Cancer Res. 2022, 24, 8. [Google Scholar] [CrossRef]
- Shi, F.; Shang, L.; Yang, L.-Y.; Jiang, Y.-Y.; Wang, X.-M.; Hao, J.-J.; Zhang, Y.; Huang, D.-K.; Cai, Y.; Xu, X.; et al. Neuropilin-1 Contributes to Esophageal Squamous Cancer Progression via Promoting p65-Dependent Cell Proliferation. Oncogene 2018, 37, 935–943. [Google Scholar] [CrossRef]
- Grun, D.; Adhikary, G.; Eckert, R.L. NRP-1 Interacts with GIPC1 and SYX to Activate p38 MAPK Signaling and Cancer Stem Cell Survival. Mol. Carcinog. 2018, 58, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Grun, D.; Adhikary, G.; Eckert, R.L. NRP-1 Interacts with GIPC1 and α6/Β4-Integrins to Increase yap1/∆np63α-Dependent Epidermal Cancer Stem Cell Survival. Oncogene 2019, 37, 4711–4722. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.-G.; Chang, W.-W.; Jan, M.-S.; Tu, C.-W.; Lu, Y.-C.; Tai, C.-K. Promotion of Metastasis of Thyroid Cancer Cells via NRP-2-Mediated Induction. Oncol. Lett. 2016, 12, 4224–4230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.; Kang, Y.E.; Oh, C.; Liu, L.; Jin, Y.; Lim, M.A.; Won, H.-R.; Chang, J.W.; Koo, B.S. Neuropilin-2 Promotes Growth and Progression of Papillary Thyroid Cancer Cells. Auris Nasus Larynx 2020, 47, 870–880. [Google Scholar] [CrossRef]
- Karczmarczyk, A.; Bilska, S.; Korpysz, M.; Purkot, J.; Grząśko, N.; Hus, M.; Giannopoulos, K. Expression and Clinical Significance of Neuropilin-1 in Patients with Multiple Myeloma. Anticancer Res. 2020, 40, 5437–5443. [Google Scholar] [CrossRef]
- Keck, B.; Wach, S.; Taubert, H.; Zeiler, S.; Ott, O.J.; Kunath, F.; Hartmann, A.; Bertz, S.; Weiss, C.; Hönscheid, P.; et al. Neuropilin-2 and Its Ligand VEGF-C Predict Treatment Response after Transurethral Resection and Radiochemotherapy in Bladder Cancer Patients. Int. J. Cancer 2014, 136, 443–451. [Google Scholar] [CrossRef]
- Schulz, A.; Gorodetska, I.; Behrendt, R.; Fuessel, S.; Erdmann, K.; Foerster, S.; Datta, K.; Mayr, T.; Dubrovska, A.; Muders, M.H. Linking NRP2 with EMT and Chemoradioresistance in Bladder Cancer. Front. Oncol. 2020, 9, 1461. [Google Scholar] [CrossRef]
- Ong, H.S.; Gokavarapu, S.; Xu, Q.; Tian, Z.; Li, J.; Ji, T.; Zhang, C.P. Cytoplasmic Neuropilin 2 Is Associated with Metastasis and a Poor Prognosis in Early Tongue Cancer Patients. Int. J. Oral Maxillofac. Surg. 2017, 46, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Shimada, K.; Asano, A.; Tatsumi, Y.; Yamaguchi, N.; Yamazaki, M.; Konishi, N. MicroRNA-331-3p Suppresses Cervical Cancer Cell Proliferation and E6/E7 Expression by Targeting NRP2. Int. J. Mol. Sci. 2016, 17, 1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Liu, L.; Zhu, Y.; Wang, B.-B.; Chen, Y.-N.; Zhang, F.; Zhang, X.-A.; Ren, C.-C. Neuropilin-1 Is Associated with the Prognosis of Cervical Cancer in Henan Chinese Population. Onco Targets Ther. 2019, 12, 2911–2920. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhang, R.; Ma, L.; Li, Q.; Zhao, Y.; Zhang, G.; Zhang, D.; Li, W.; Cao, S.; Wang, L.; et al. Neuropilin-1 Is up-Regulated by Cancer-Associated Fibroblast-Secreted Il-8 and Associated with Cell Proliferation of Gallbladder Cancer. J. Cell Mol. Med. 2020, 24, 12608–12618. [Google Scholar] [CrossRef] [PubMed]
- Oplawski, M.; Dziobek, K.; Grabarek, B.; Zmarzły, N.; Dąbruś, D.; Januszyk, P.; Brus, R.; Tomala, B.; Boroń, D. Expression of NRP-1 and NRP-2 in Endometrial Cancer. Curr. Pharm. Biotechnol. 2019, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Xu, L.; Qiu, Y.; Zhan, J.; Chen, X. Down-Regulated Mir-124-3p Enhanced the Migration and Epithelial-Stromal Transformation of Endometrial Stromal Cells Extracted from Eutopic Endometrium in Subjects with Adenomyosis by up-Regulating Neuropilin 1. Tissue Cell 2021, 69, 101474. [Google Scholar] [CrossRef]
- Miyauchi, J.T.; Chen, D.; Choi, M.; Nissen, J.C.; Shroyer, K.R.; Djordevic, S.; Zachary, I.C.; Selwood, D.; Tsirka, S.E. Ablation of Neuropilin 1 from glioma-associated microglia and macrophages slows tumor progression. Oncotarget 2016, 7, 9801–9814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, J.; Mota, F.; Steadman, D.; Soudy, C.; Miyauchi, J.T.; Crosby, S.; Jarvis, A.; Reisinger, T.; Winfield, N.; Evans, G.; et al. Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGFβ) Production in Regulatory T-Cells. J. Med. Chem. 2018, 61, 4135–4154. [Google Scholar] [CrossRef] [PubMed]
- Klaewkla, M.; Charoenwongpaiboon, T.; Mahalapbutr, P. Molecular basis of the new COVID-19 target neuropilin-1 in complex with SARS-CoV-2 S1 C-end rule peptide and small-molecule antagonists. J. Mol. Liq. 2021, 335, 116537. [Google Scholar] [CrossRef] [PubMed]
- Binetruy-Tournaire, R.; Demangel, C.; Malavaud, B.; Vassy, R.; Rouyre, S.; Kraemer, M.; Mazie, J.C. Identification of a peptide blocking vascular endothelial growth factor (VEGF)-mediated angiogenesis. EMBO J. 2020, 19, 1525–1533. [Google Scholar] [CrossRef] [Green Version]
- Perret, G.Y.; Starzec, A.; Hauet, N.; Vergote, J.; Le Pecheur, M.; Vassy, R.; Moretti, J.L. In vitro evaluation and biodistribution of a 99mTc-labeled anti-VEGF peptide targeting neuropilin-1. Nucl. Med. Biol. 2004, 31, 575–581. [Google Scholar] [CrossRef]
- Novoa, A.; Pellegrini-Moïse, N.; Bechet, D.; Barberi-Heyob, M.; Chapleur, Y. Sugar-based peptidomimetics as potential inhibitors of the vascular endothelium growth factor binding to neuropilin-1. Bioorg. Med. Chem. 2010, 18, 3285–3298. [Google Scholar] [CrossRef]
- Richard, M.; Chateau, A.; Jelsch, C.; Didierjean, C.; Manival, X.; Charron, C.; Pellegrini-Moise, N. Carbohydrate-based peptidomimetics targeting neuropilin-1: Synthesis, molecular docking study and in vitro biological activities. Bioorg. Med. Chem. 2016, 24, 5315–5325. [Google Scholar] [CrossRef]
- Borriello, L.; Montès, M.; Lepelletier, Y.; Leforban, B.; Liu, W.Q.; Demange, L.; Raynaud, F. Structure-based discovery of a small non-peptidic Neuropilins antagonist exerting in vitro and in vivo anti-tumor activity on breast cancer model. Cancer Lett. 2014, 349, 120–127. [Google Scholar] [CrossRef]
- Liu, W.Q.; Lepelletier, Y.; Montès, M.; Borriello, L.; Jarray, R.; Grépin, R.; Demange, L. NRPa-308, a new neuropilin-1 antagonist, exerts in vitro anti-angiogenic and anti-proliferative effects and in vivo anticancer effects in a mouse xenograft model. Cancer Lett. 2018, 414, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Brachet, E.; Dumond, A.; Liu, W.Q.; Fabre, M.; Selkti, M.; Raynaud, F.; Demange, L. Synthesis, 3D-structure and stability analyses of NRPa-308, a new promising anticancer agent. Bioorg. Med. Chem. Lett. 2019, 29, 126710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starzec, A.; Miteva, M.A.; Ladam, P.; Villoutreix, B.O.; Perret, G.Y. Discovery of novel inhibitors of vascular endothelial growth factor-A–Neuropilin-1 interaction by structure-based virtual screening. Bioorg. Med. Chem. 2014, 22, 4042–4048. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Li, Y.; Bai, Y.; Jiang, T.; Sun, H.; Zhu, Q.; Xu, Y. Discovery of novel nonpeptide small-molecule NRP1 antagonists: Virtual screening, molecular simulation and structural modification. Bioorg. Med. Chem. 2020, 28, 115183. [Google Scholar] [CrossRef] [PubMed]
- Seadawy, M.; Shamel, M.; Ahmed, A.; Zekri, A.R.N. In Silico Docking for Inhibition Neuropilin-1 (SARS-CoV-2 Receptor) by Some Natural and Approved Drugs. Available online: https://ssrn.com/abstract=3735823 (accessed on 26 July 2022).
- Vique-Sánchez, J.L. Potential inhibitors interacting in Neuropilin-1 to develop an adjuvant drug against COVID-19, by molecular docking. Bioorg. Med. Chem. 2021, 33, 116040. [Google Scholar] [CrossRef]
- Kolarič, A.; Jukič, M.; Bren, U. Novel Small-Molecule Inhibitors of the SARS-CoV-2 Spike Protein Binding to Neuropilin 1. Pharmaceuticals 2022, 15, 165. [Google Scholar] [CrossRef] [PubMed]
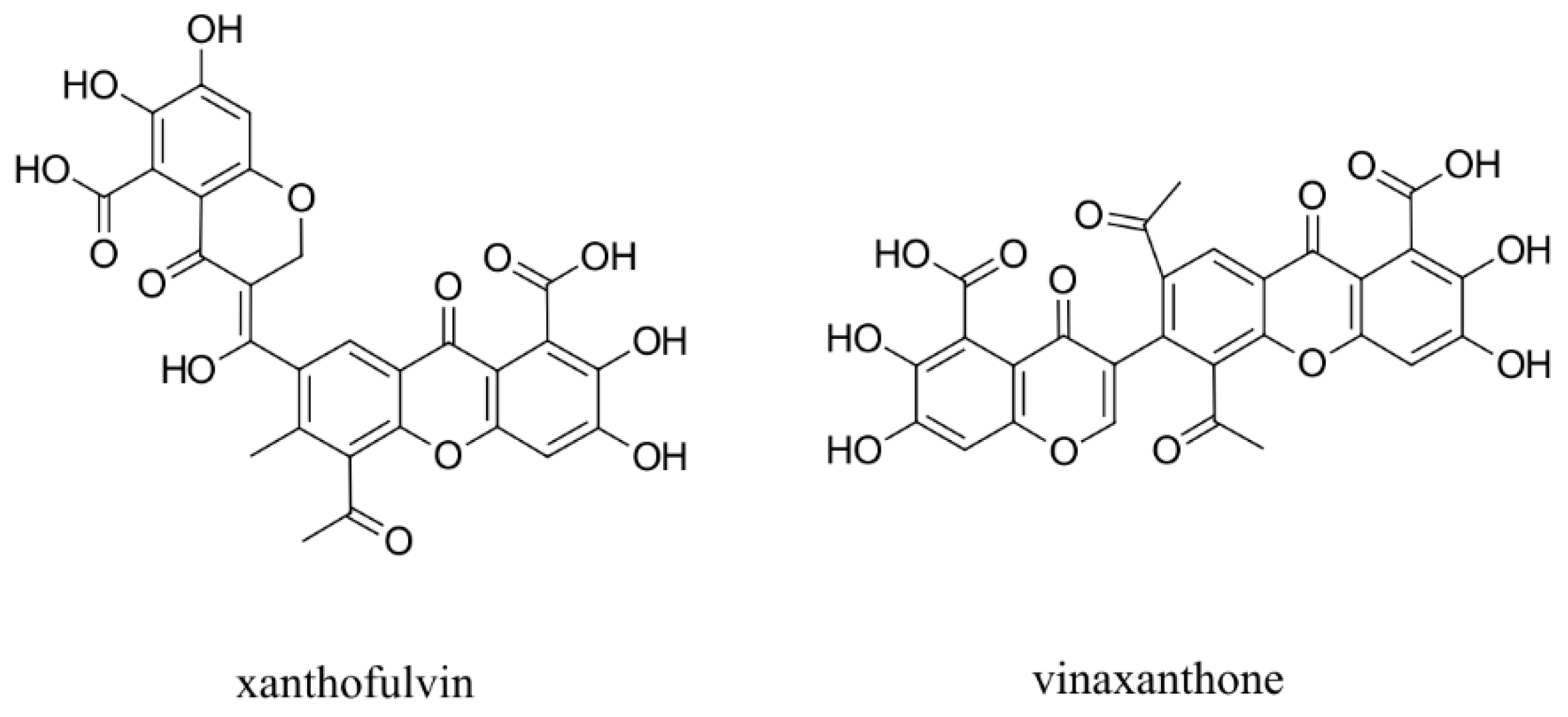
- Kumagai, K.; Hosotani, N.; Kikuchi, K.; Kimura, T.; Saji, I. Xanthofulvin, a novel semaphorin inhibitor produced by a strain of Penicillium. J. Antibiot. 2003, 56, 610–616. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, K.; Kishino, A.; Konishi, O.; Kumagai, K.; Hosotani, N.; Saji, I.; Kimura, T. In vitro and in vivo characterisation of a novel semaphorin 3A inhibitor, SM-216289 or xanthofulvin. J. Biol. Chem. 2003, 278, 42985–42991. [Google Scholar] [CrossRef] [Green Version]
- Said, A.M.; Parker, M.W.; Vander Kooi, C.W. Design, synthesis, and evaluation of a novel benzamidine-based inhibitor of VEGF-C binding to Neuropilin-2. Bioorg. Chem. 2020, 100, 103856. [Google Scholar] [CrossRef] [PubMed]
| Endogenous Ligand | Preferences for NRP Binding | Reference | |
|---|---|---|---|
| NRP1 | NRP2 | ||
| SEMA | SEMA3A SEMA3B SEMA3C SEMA3D SEMA3F | SEMA3B SEMA3C SEMA3D SEMA3F SEMA3G | [2,9] |
| VEGF | VEGF-A VEGF-A165 VEGF-A189 VEGF-B VEGF-C VEGF-D VEGF-E PIGF | VEGF-A VEGF-A145 VEGF-A165 VEGF-C VEGF-D PIGF | [3,9] |
| FGF | FGF-1 FGF-2 FGF-4 FGF-7 | FGF-2 | [18] |
| HGF | x | x | [19] |
| PDGF | PDGF-BB PDGF-C PDGF-D | PDGF-BB | [3,20] |
| TGF-β | TGF-β1 | TGF-β1 | [21,22] |
| miRNAs | x | x | [2,23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broz, M.; Kolarič, A.; Jukič, M.; Bren, U. Neuropilin (NRPs) Related Pathological Conditions and Their Modulators. Int. J. Mol. Sci. 2022, 23, 8402. https://doi.org/10.3390/ijms23158402
Broz M, Kolarič A, Jukič M, Bren U. Neuropilin (NRPs) Related Pathological Conditions and Their Modulators. International Journal of Molecular Sciences. 2022; 23(15):8402. https://doi.org/10.3390/ijms23158402
Chicago/Turabian StyleBroz, Matic, Anja Kolarič, Marko Jukič, and Urban Bren. 2022. "Neuropilin (NRPs) Related Pathological Conditions and Their Modulators" International Journal of Molecular Sciences 23, no. 15: 8402. https://doi.org/10.3390/ijms23158402
APA StyleBroz, M., Kolarič, A., Jukič, M., & Bren, U. (2022). Neuropilin (NRPs) Related Pathological Conditions and Their Modulators. International Journal of Molecular Sciences, 23(15), 8402. https://doi.org/10.3390/ijms23158402