BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition
Abstract
:1. Introduction
2. Results
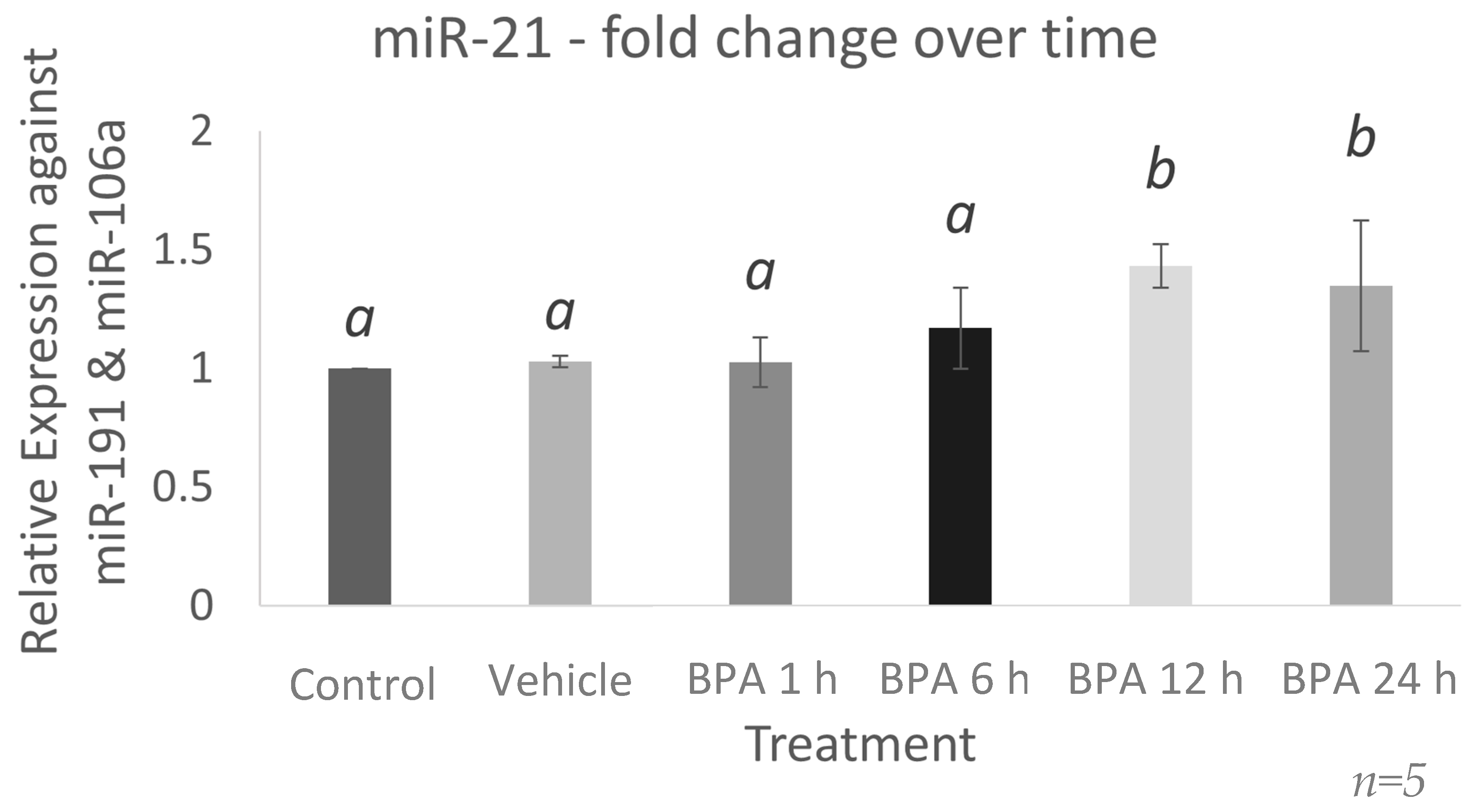
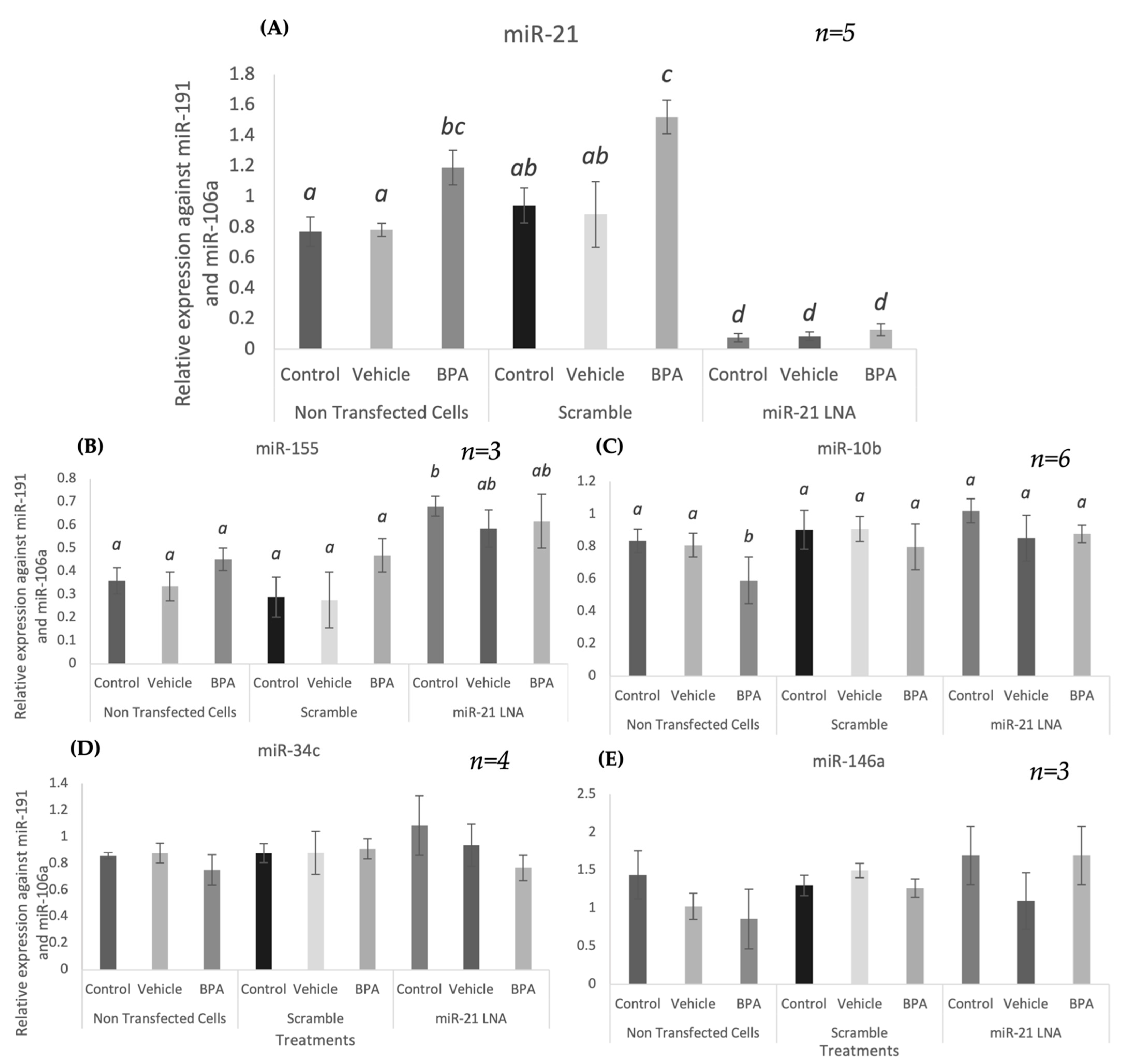
2.1. miR-21 Expression Increases 12 h after BPA Treatment in Bovine Granulosa Cells
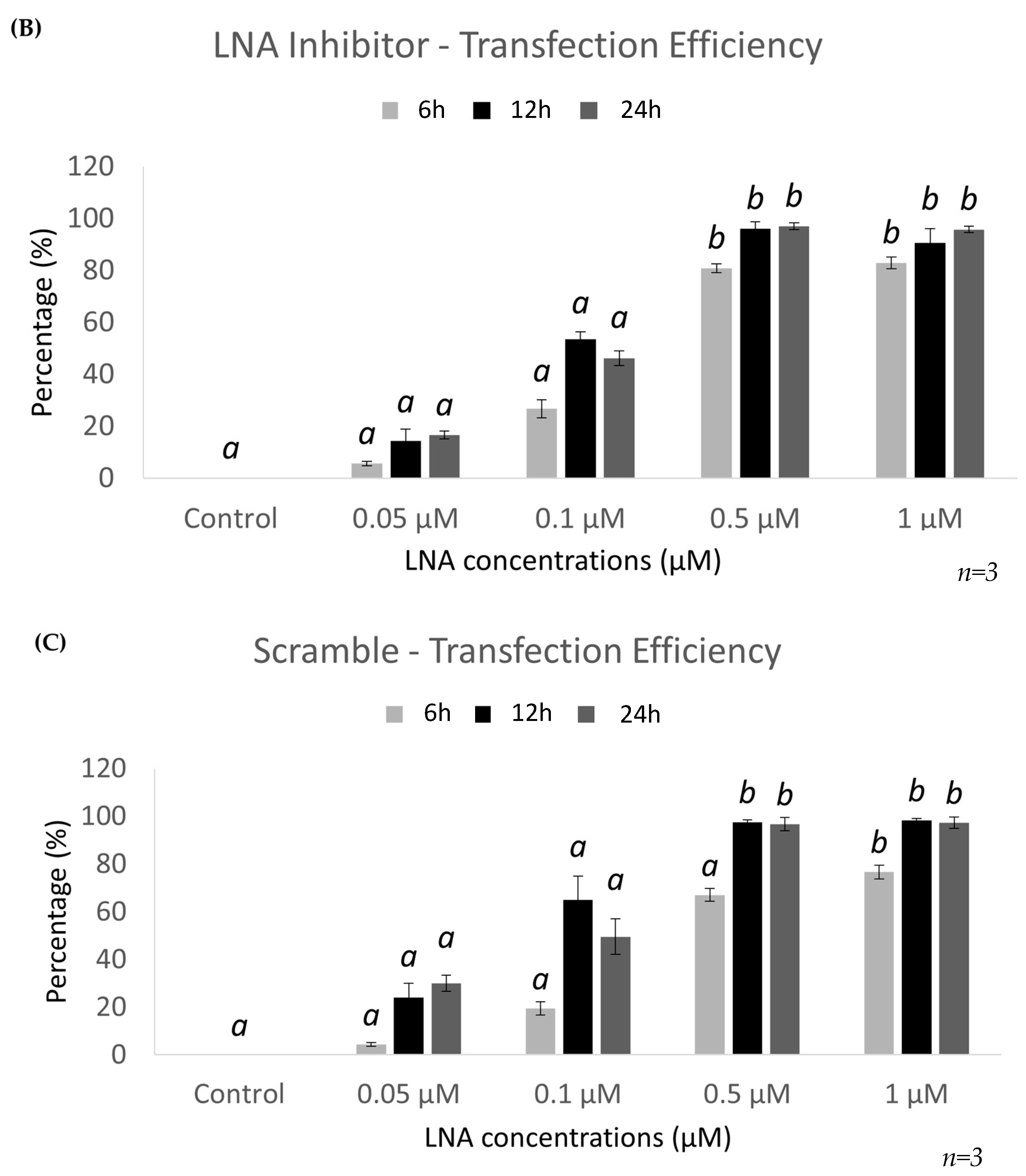
2.2. Optimal Uptake Efficiencies of miR-21 and Scramble LNAs Occur after 12 h of Transfection at 0.5 μM Concentrations
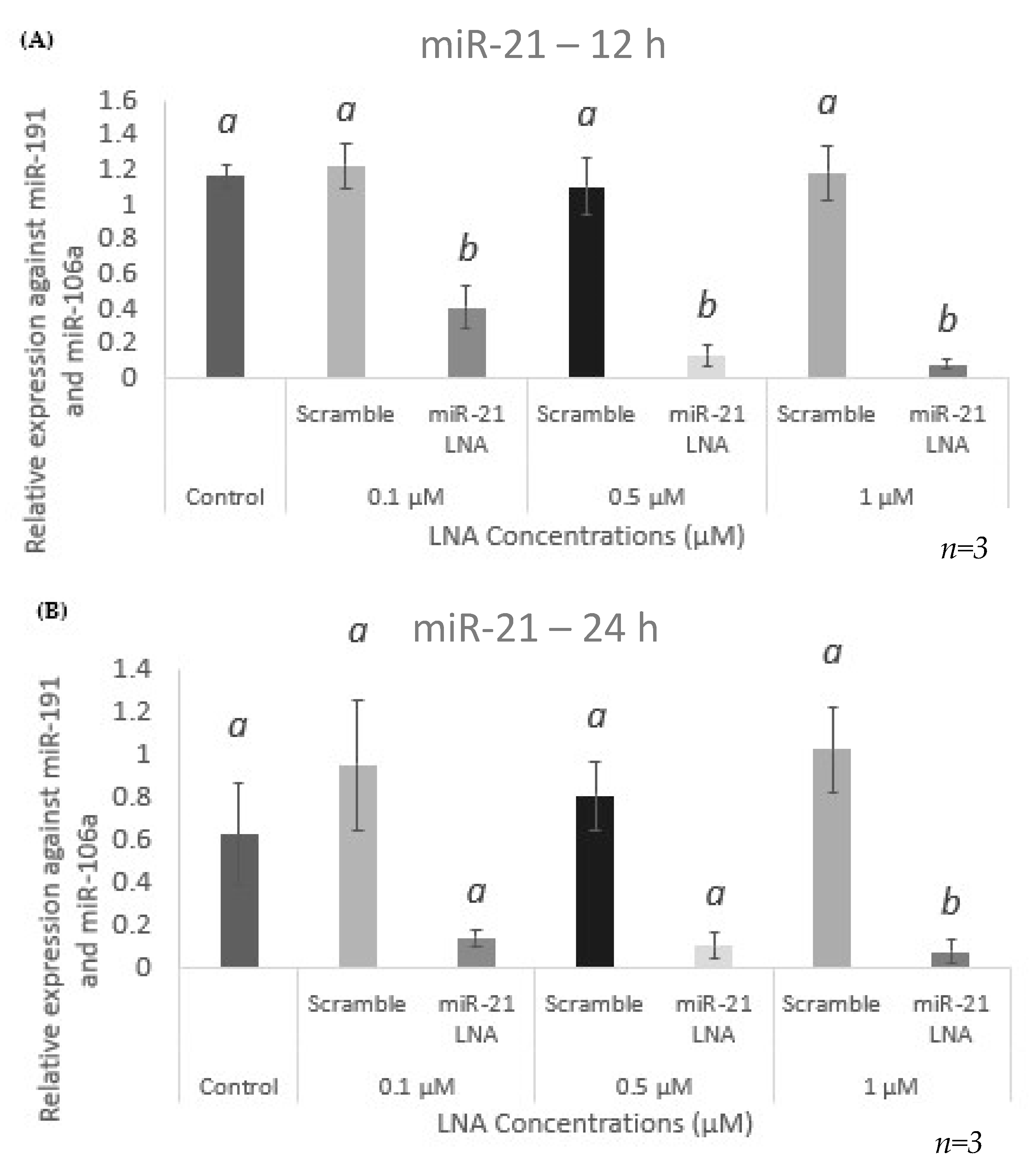
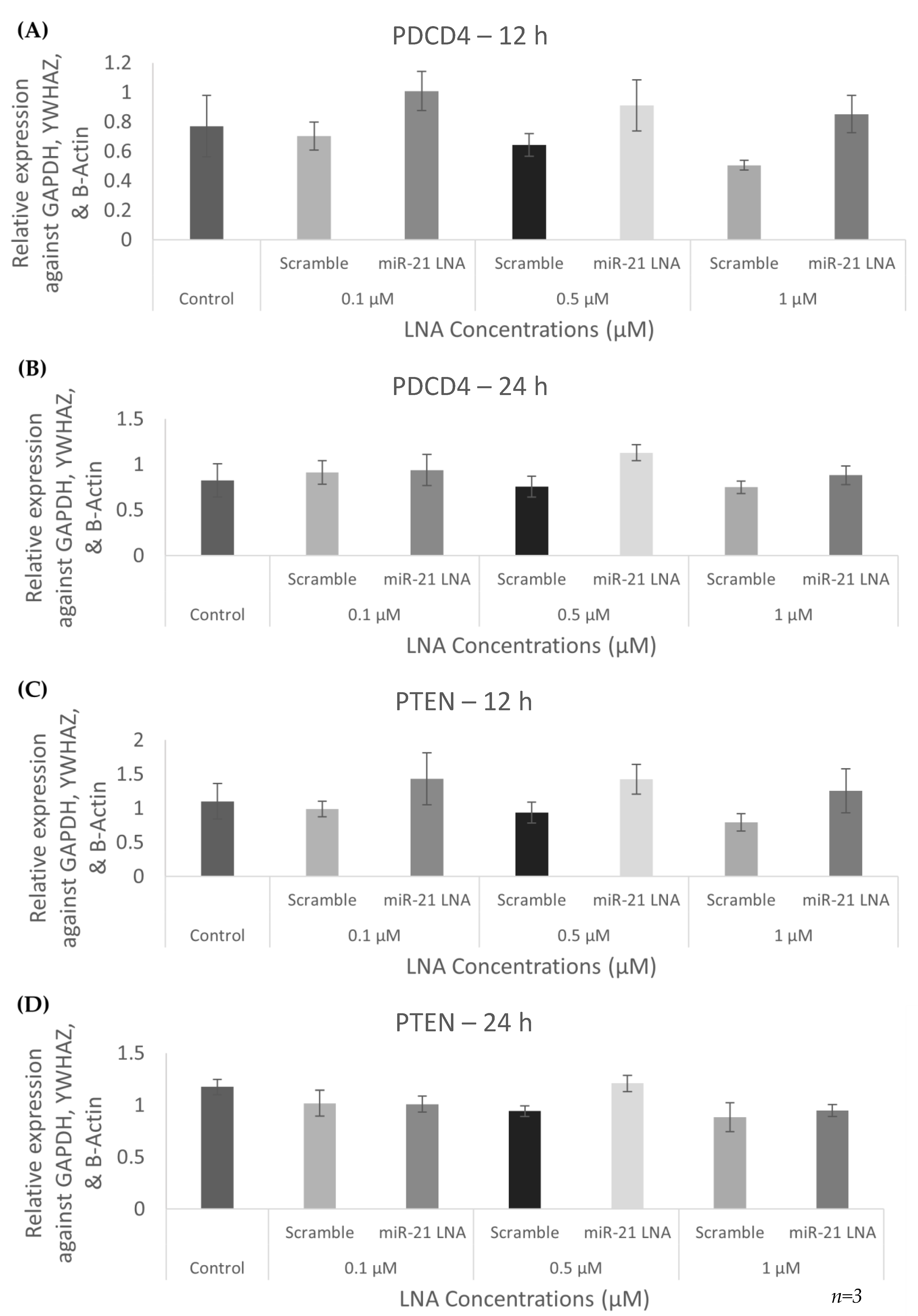
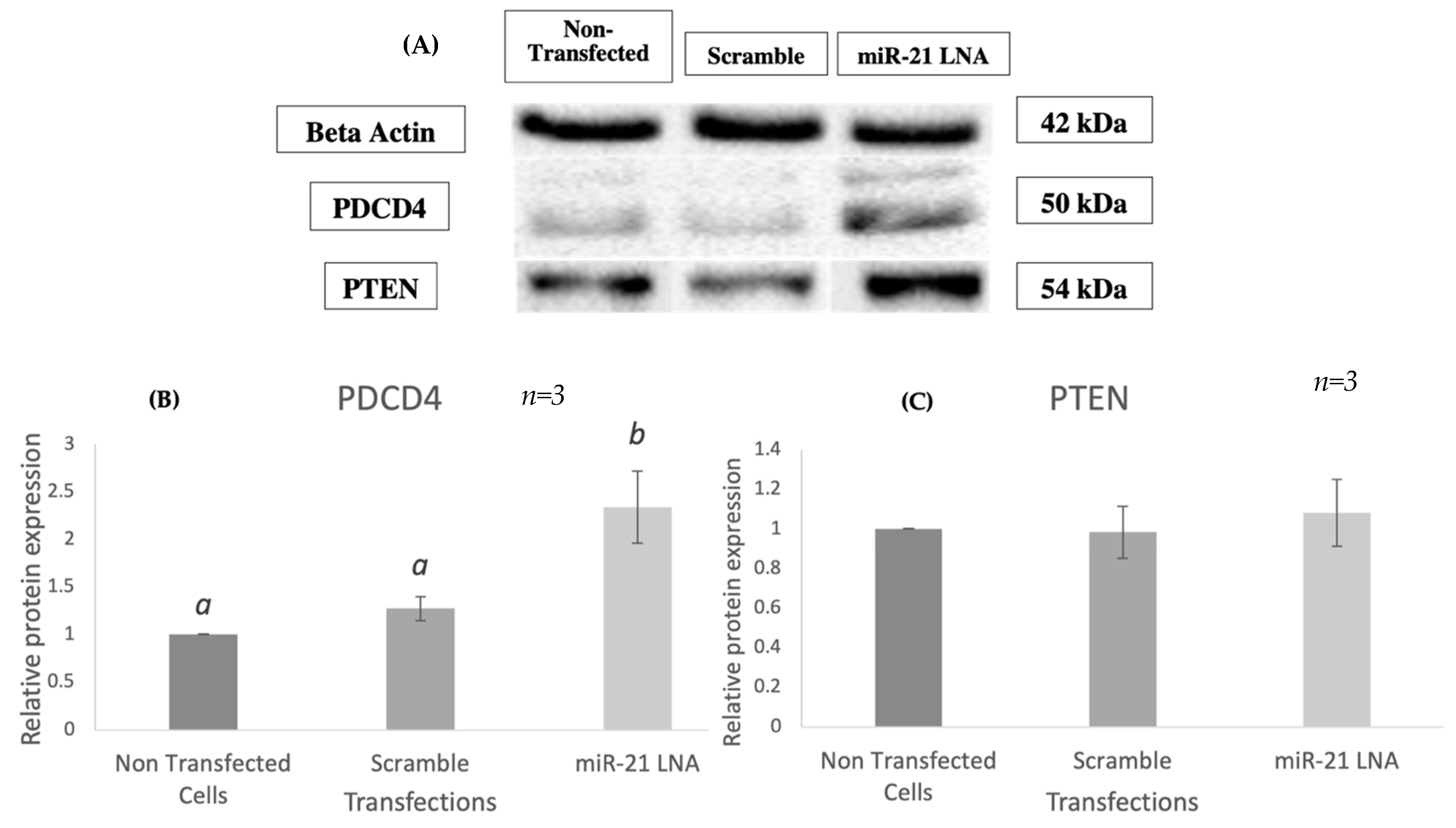
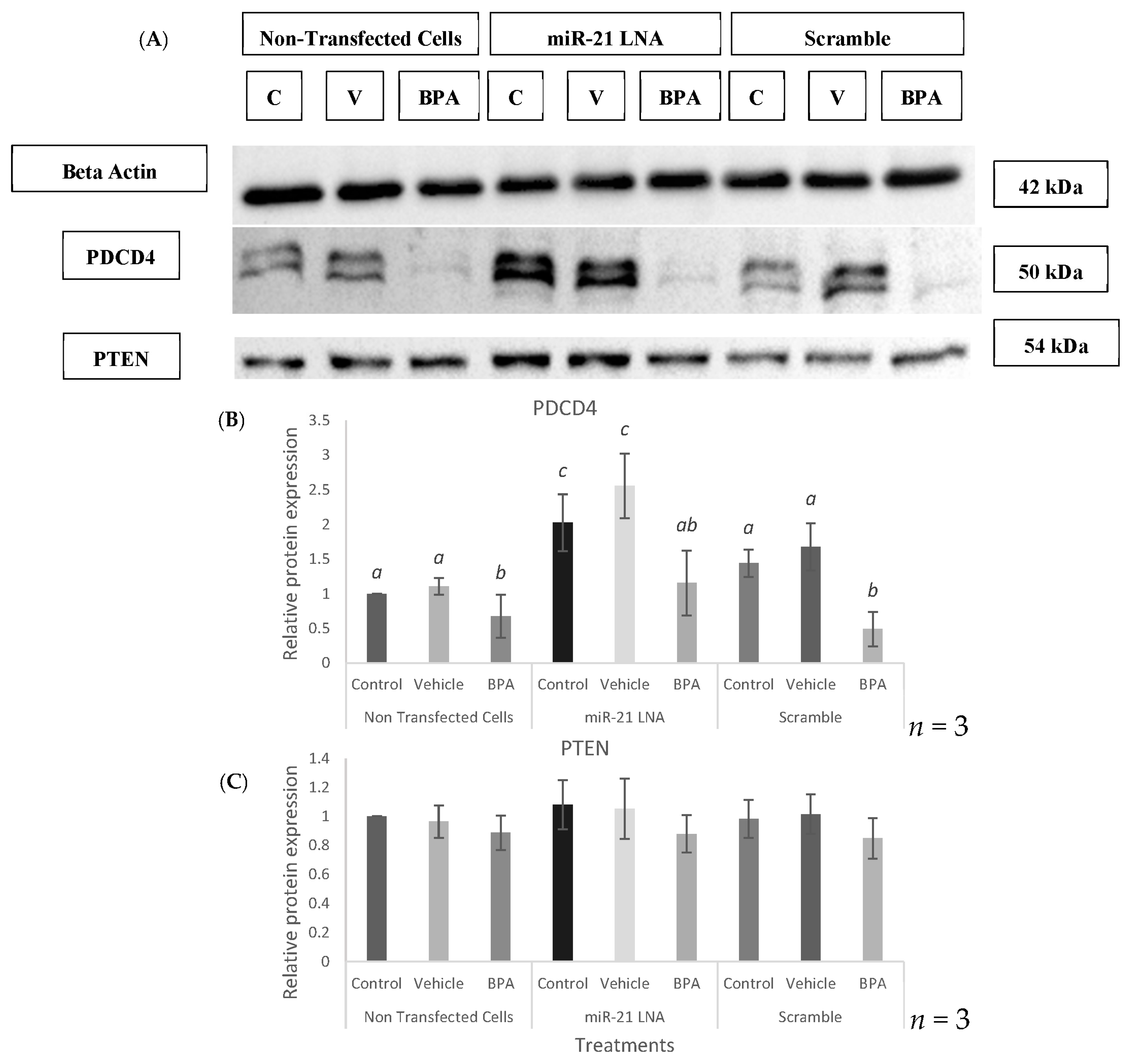
2.3. Knockdown of miR-21 Confirmed by qPCR Leads to Functional Differences on the Protein Level but Not at the Transcript Level of Known miR-21 Targets
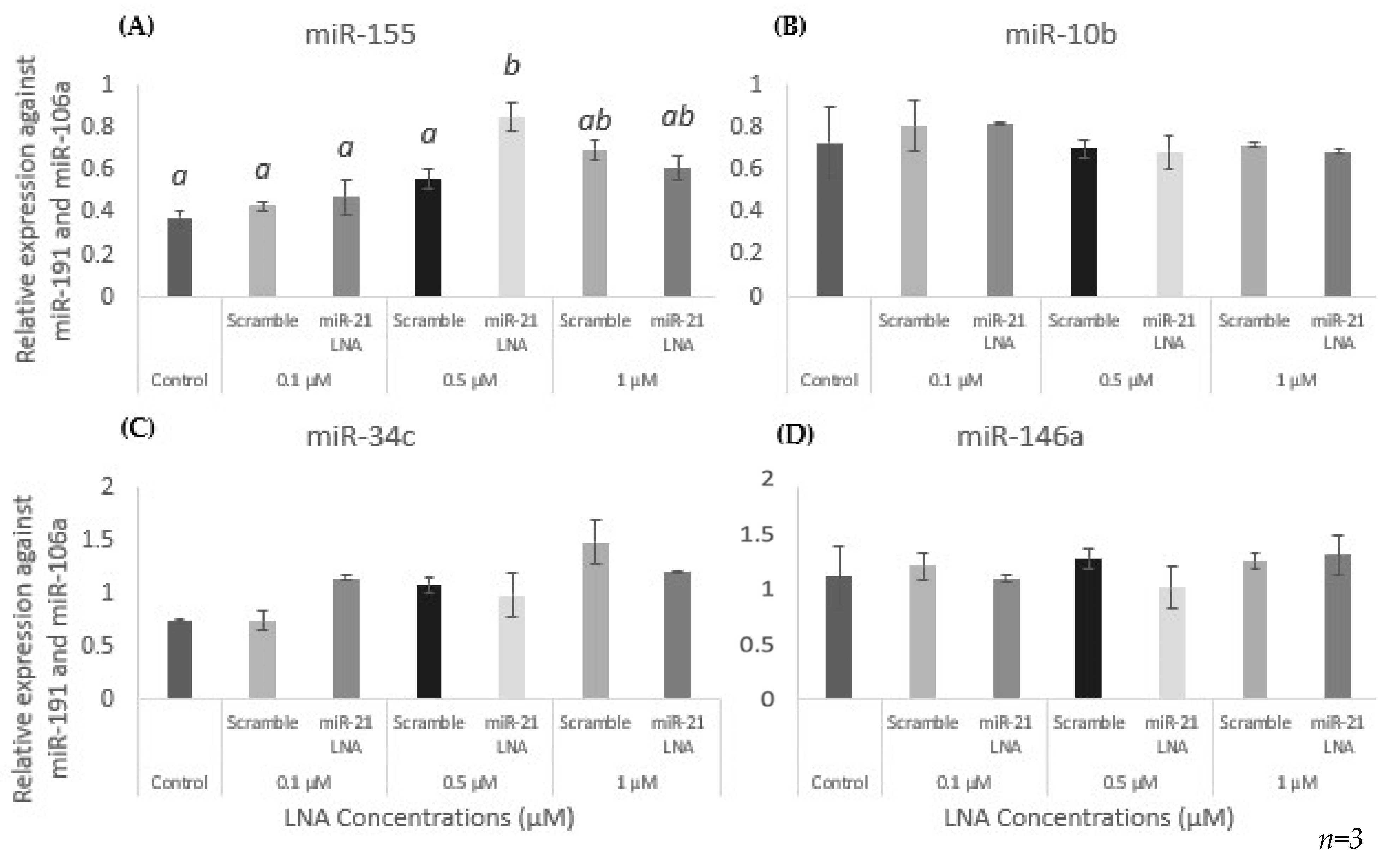
2.4. Specificity of miR-21 LNAs Shown through No Effects on Other miRNAs except for the Significantly Increased Expression of miR-155
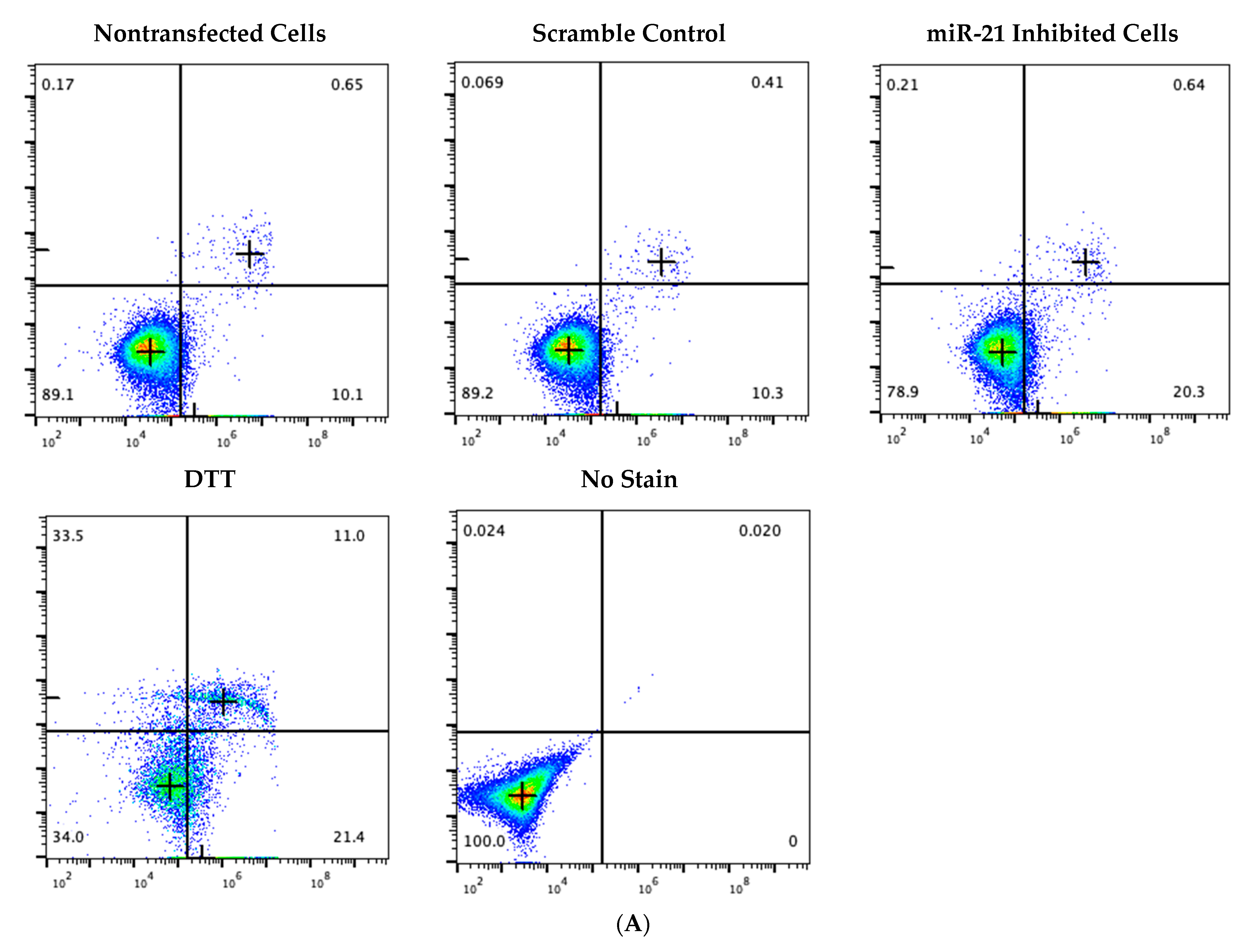
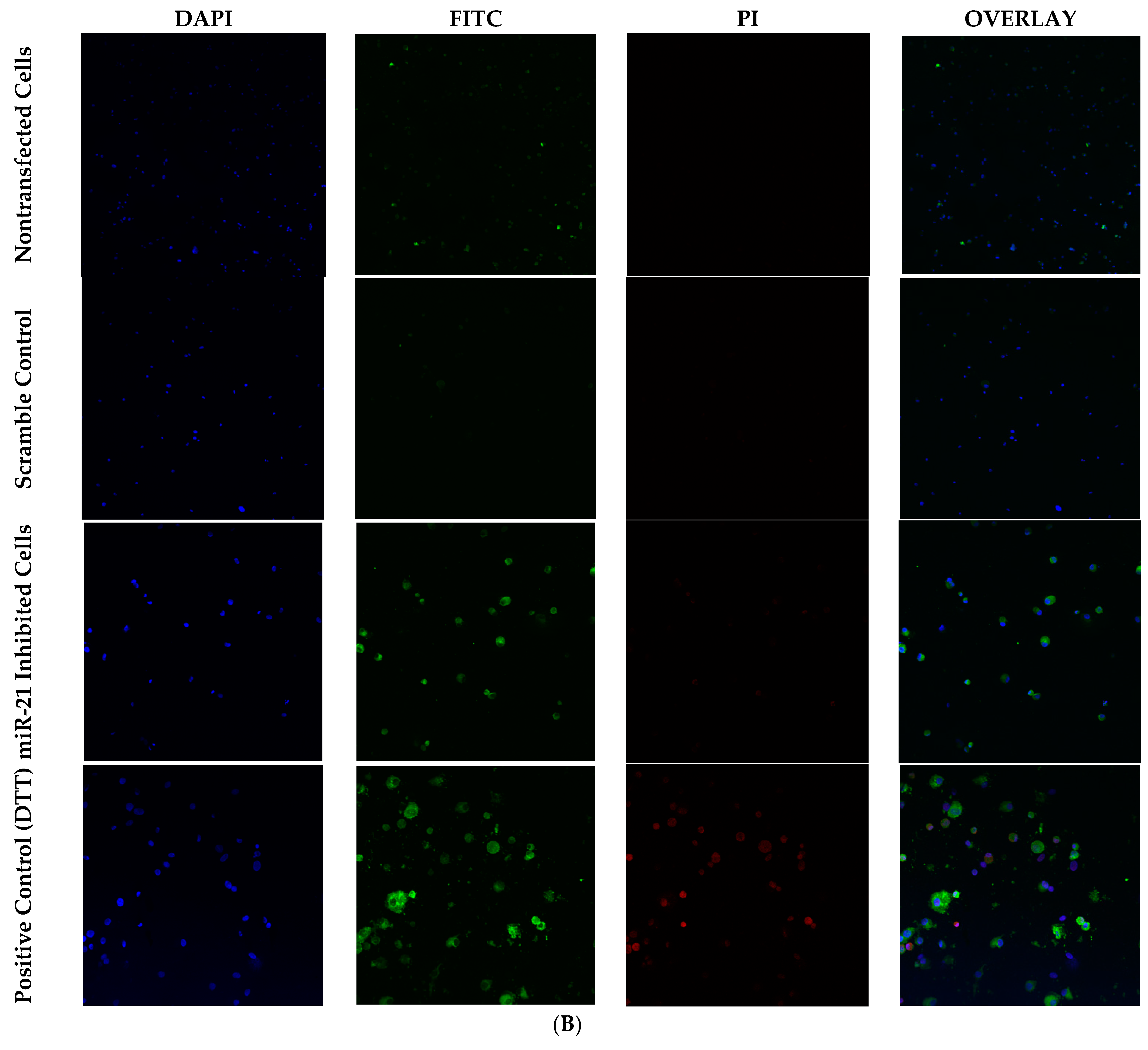
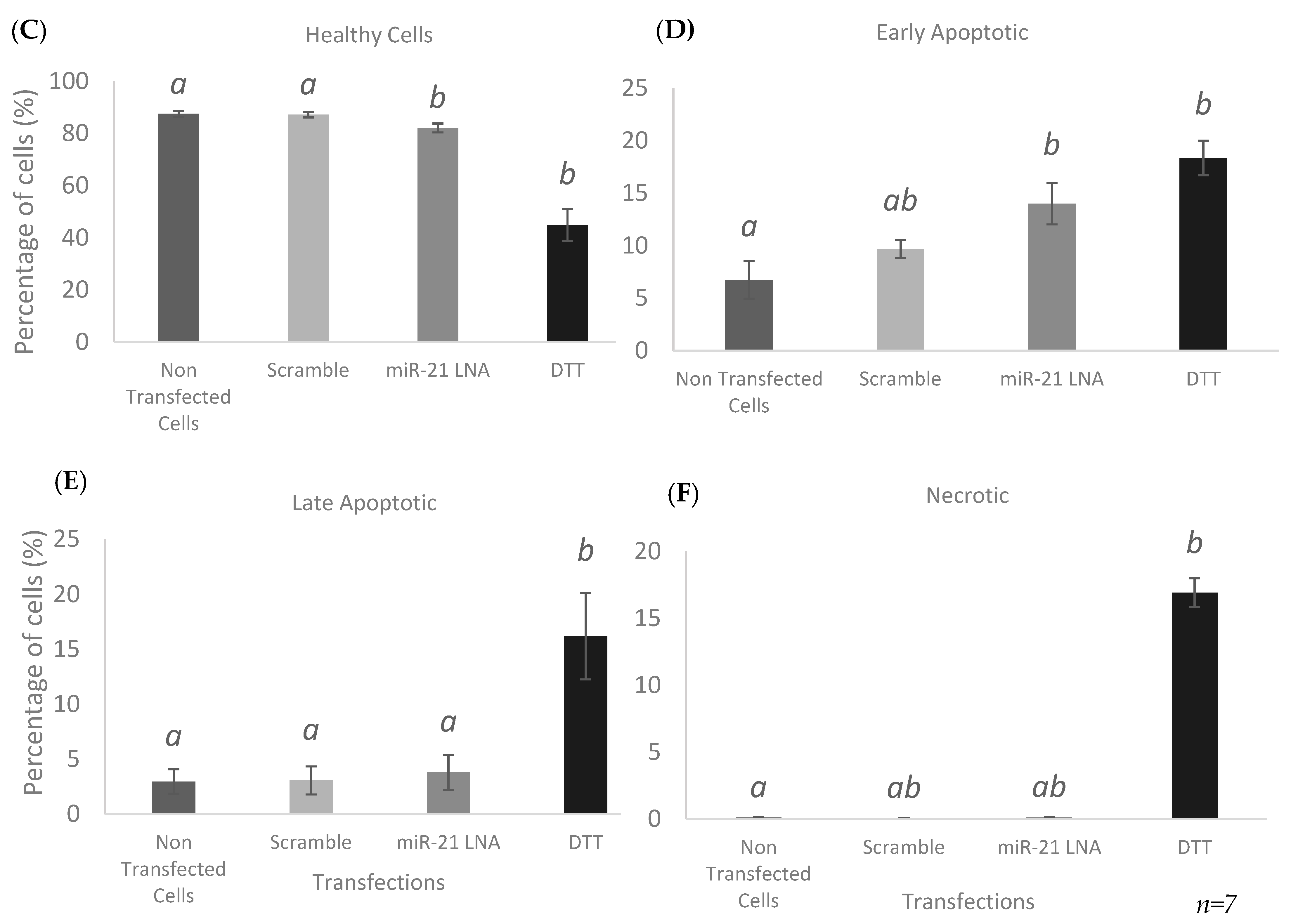
2.5. Knockdown of miR-21 Induces Apoptosis in Granulosa Cells through Positive Annexin Staining
2.6. BPA Affects miR-10b in Addition to miR-21, but LNA-21 Treatment Affects miR-155
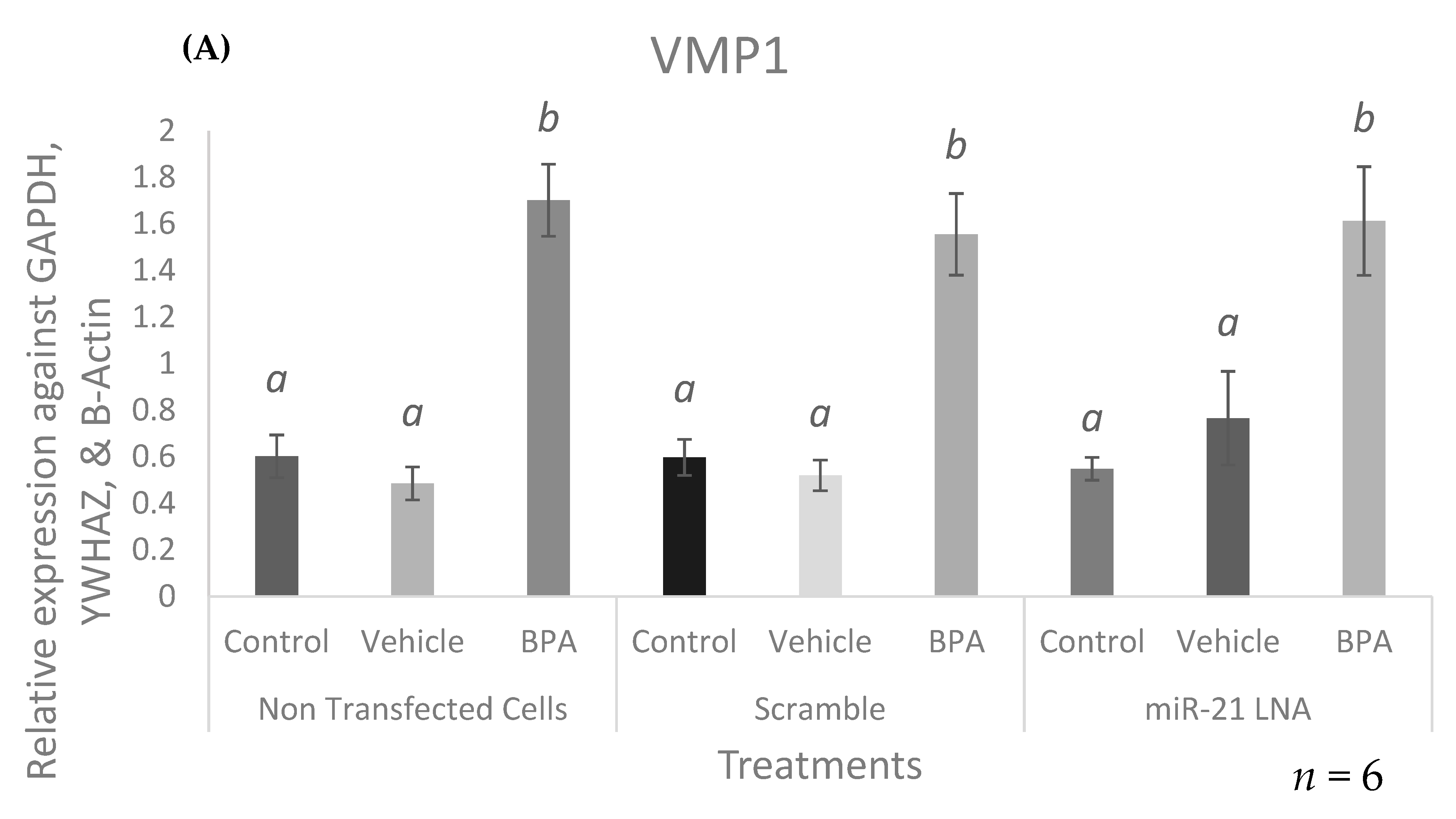
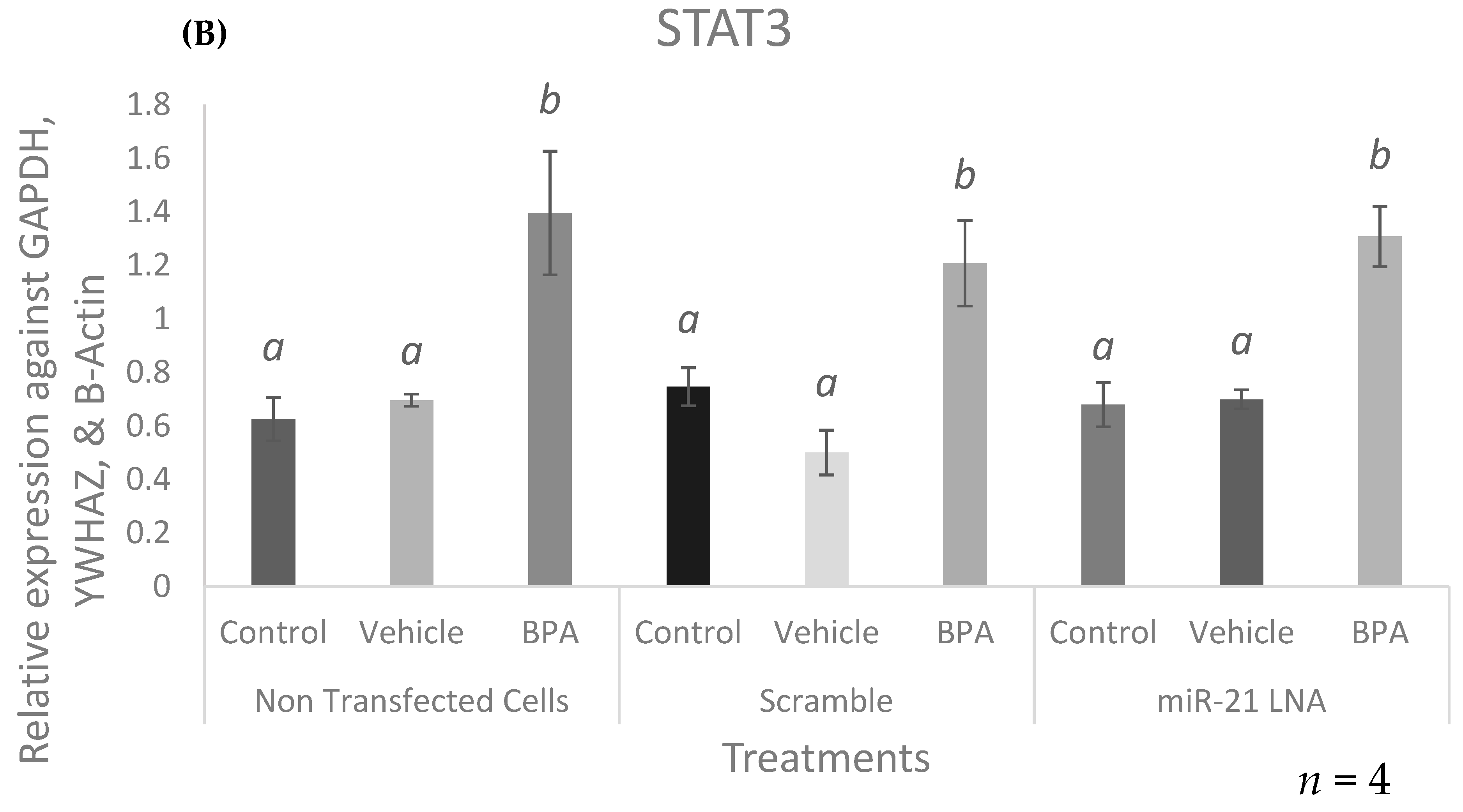
2.7. BPA Increases the Expression of the miR-21 Transcription Factor, STAT3, and the Primary Form of miR-21, VMP1-miR-21
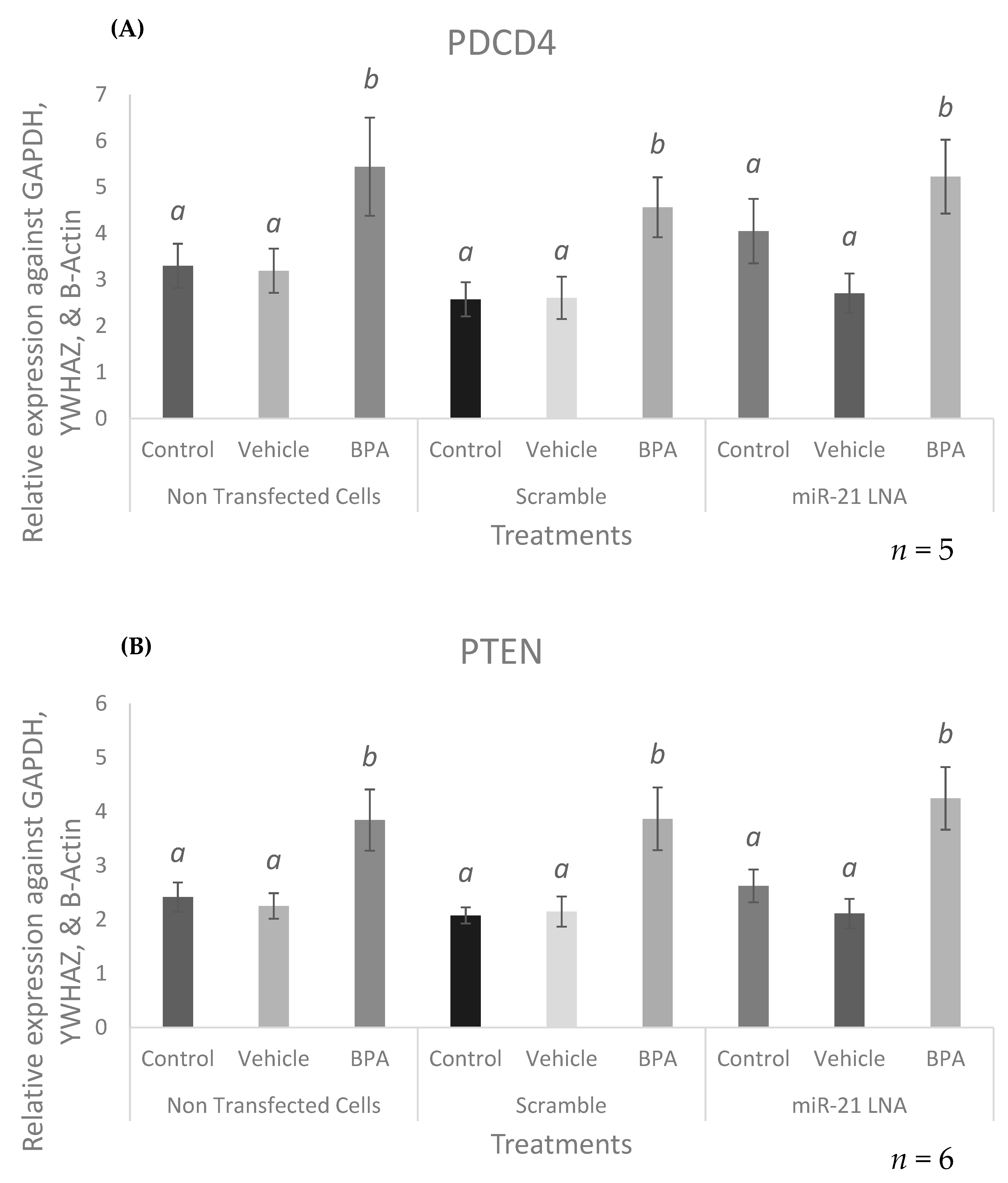
2.8. Known miR-21 Target Transcripts and Proteins Were Affected by BPA, but These Changes Were Not Reversed or Exaggerated by LNA-21 Knockdown
3. Discussion
4. Materials and Methods
4.1. Granulosa Cells Collection and In Vitro Granulosa Cell Culture
4.2. Cell Transfections with Locked Nucleic Acids (LNAs)
4.3. Confocal Microscopy
4.4. Flow Cytometry
4.5. RNA Isolation and cDNA Synthesis
4.6. Quantitative qPCR
4.7. Apoptosis Measurement Using Annexin Staining and Flow Cytometry
4.8. Protein Isolation and Western Blot Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y. Modern Epigenetics Methods in Biological Research. Methods 2021, 187, 104–113. [Google Scholar] [CrossRef]
- Vazquez, M.J.; Daza-Dueñas, S.; Tena-Sempere, M. Emerging Roles of Epigenetics in the Control of Reproductive Function: Focus on Central Neuroendocrine Mechanisms. J. Endocr. Soc. 2021, 5, bvab152. [Google Scholar] [CrossRef]
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging Role of Non-coding RNA in Health and Disease. Metab. Brain Dis. 2021, 36, 1119–1134. [Google Scholar] [CrossRef]
- Tu, J.; Cheung, A.H.H.; Chan, C.L.K.; Chan, W.Y. The Role of MicroRNAs in Ovarian Granulosa Cells in Health and Disease. Front. Endocrinol. 2019, 10, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front. Cell Dev. Biol. 2021, 9, 654028. [Google Scholar] [CrossRef]
- Breton, C.V.; Landon, R.; Kahn, L.G.; Enlow, M.B.; Peterson, A.K.; Bastain, T.; Braun, J.; Comstock, S.S.; Duarte, C.S.; Hipwell, A.; et al. Exploring the Evidence for Epigenetic Regulation of Environmental Influences on Child Health across Generations. Commun. Biol. 2021, 4, 769. [Google Scholar] [CrossRef]
- Hale, B.J.; Li, Y.; Adur, M.K.; Ross, J.W. Inhibition of Germinal Vesicle Breakdown Using IBMX Increases MicroRNA-21 in the Porcine Oocyte. Reprod. Biol. Endocrinol. 2020, 18, 39. [Google Scholar] [CrossRef]
- Khaghani, A.J.; Farrokh, P.; Zavareh, S. Epigenetic Effects of Bisphenol A on Granulosa Cells of Mouse Follicles during in vitro Culture: An Experimental Study. Int. J. Reprod. Biomed. 2021, 19, 129–136. [Google Scholar] [CrossRef]
- Turathum, B.; Gao, E.M.; Chian, R.C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 2021, 10, 2292. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, Z.; Mohammadi-Yeganeh, S.; Rezaee, D.; Salehi, M. MicroRNA-21 Is Involved in Oocyte Maturation, Blastocyst Formation, and Pre-Implantation Embryo Development. Dev. Biol. 2021, 480, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Uhde, K.; Van Tol, H.T.; Stout, T.A.; Roelen, B.A. MicroRNA Expression in Bovine Cumulus Cells in Relation to Oocyte Quality. Non-Coding RNA 2017, 3, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, E.C.; Hale, B.J.; Yang, C.X.; Njoka, J.G.; Ross, J.W. MicroRNA-21 and PDCD4 Expression during in vitro Oocyte Maturation in Pigs. Reprod. Biol. Endocrinol. RBE 2016, 14, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assou, S.; Al-Edani, T.; Haouzi, D.; Philippe, N.; Lecellier, C.H.; Piquemal, D.; Commes, T.; Aït-Ahmed, O.; Dechaud, H.; Hamamah, S. MicroRNAs: New Candidates for the Regulation of the Human Cumulus–Oocyte Complex. Hum. Reprod. 2013, 28, 3038–3049. [Google Scholar] [CrossRef] [Green Version]
- McBride, D.; Carré, W.; Sontakke, S.D.; Hogg, C.O.; Law, A.; Donadeu, F.X.; Clinton, M. Identification of MiRNAs Associated with the Follicular-Luteal Transition in the Ruminant Ovary. Reproduction 2012, 144, 221–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tscherner, A.; Brown, A.C.; Stalker, L.; Kao, J.; Dufort, I.; Sirard, M.A.; LaMarre, J. STAT3 Signaling Stimulates MiR-21 Expression in Bovine Cumulus Cells during in vitro Oocyte Maturation. Sci. Rep. 2018, 8, 11527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartolucci, A.F.; Uliasz, T.; Peluso, J.J. MicroRNA-21 as a Regulator of Human Cumulus Cell Viability and Its Potential Influence on the Developmental Potential of the Oocyte. Biol. Reprod. 2020, 103, 94–103. [Google Scholar] [CrossRef]
- Rasdi, Z.; Kamaludin, R.; Rahim, S.A.; Syed Ahmad Fuad, S.B.; Othman, M.H.D.; Siran, R.; Mohd Nor, N.S.; Abdul Hamid Hasani, N.; Sheikh Abdul Kadir, S.H. The Impacts of Intrauterine Bisphenol a Exposure on Pregnancy and Expression of MiRNAs Related to Heart Development and Diseases in Animal Model. Sci. Rep. 2020, 10, 5882. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; He, Y.; Wang, X.; Peng, D.; Chen, X.; Li, X.; Wang, Q. Overexpression of MiR-21 in Stem Cells Improves Ovarian Structure and Function in Rats with Chemotherapy-Induced Ovarian Damage by Targeting PDCD4 and PTEN to Inhibit Granulosa Cell Apoptosis. Stem Cell Res. Ther. 2017, 8, 187. [Google Scholar] [CrossRef] [Green Version]
- Ji, K.; Wang, X.; Zhang, A.; Wen, H. Prognostic Value of MicroRNA-21 in Epithelial Ovarian Carcinoma: A Protocol for Systematic Review and Meta Analysis. Medicine 2020, 99, e23849. [Google Scholar] [CrossRef]
- Karakaya, C.; Guzeloglu-Kayisli, O.; Uyar, A.; Kallen, A.N.; Babayev, E.; Bozkurt, N.; Unsal, E.; Karabacak, O.; Seli, E. Poor Ovarian Response in Women Undergoing in vitro Fertilization Is Associated with Altered MicroRNA Expression in Cumulus Cells. Fertil. Steril. 2015, 103, 1469–1476.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimmino, I.; Fiory, F.; Perruolo, G.; Miele, C.; Beguinot, F.; Formisano, P.; Oriente, F. Potential Mechanisms of Bisphenol A (BPA) Contributing to Human Disease. Int. J. Mol. Sci. 2020, 21, 5761. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Das, M.K.; Duttaroy, A.K. Plastics Derived Endocrine-Disrupting Compounds and Their Effects on Early Development. Birth Defects Res. 2020, 112, 1308–1325. [Google Scholar] [CrossRef] [PubMed]
- Eichenlaub-Ritter, U.; Pacchierotti, F. Bisphenol a Effects on Mammalian Oogenesis and Epigenetic Integrity of Oocytes: A Case Study Exploring Risks of Endocrine Disrupting Chemicals. BioMed Res. Int. 2015, 2015, 698795. [Google Scholar] [CrossRef] [Green Version]
- Sabry, R.; Nguyen, M.; Younes, S.; Favetta, L.A. BPA and Its Analogs Increase Oxidative Stress Levels in in vitro Cultured Granulosa Cells by Altering Anti-Oxidant Enzymes Expression. Mol. Cell. Endocrinol. 2022, 545, 111574. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Sabry, R.; Davis, O.S.; Favetta, L.A. Effects of BPA, BPS, and BPF on Oxidative Stress and Antioxidant Enzyme Expression in Bovine Oocytes and Spermatozoa. Genes 2022, 13, 142. [Google Scholar] [CrossRef]
- Saleh, A.C.; Sabry, R.; Mastromonaco, G.F.; Favetta, L.A. BPA and BPS Affect the Expression of Anti-Mullerian Hormone (AMH) and Its Receptor during Bovine Oocyte Maturation and Early Embryo Development. Reprod. Biol. Endocrinol. 2021, 19, 119. [Google Scholar] [CrossRef]
- Sabry, R.; Saleh, A.C.; Stalker, L.; LaMarre, J.; Favetta, L.A. Effects of Bisphenol A and Bisphenol S on MicroRNA Expression during Bovine (Bos Taurus) Oocyte Maturation and Early Embryo Development. Reprod. Toxicol. 2021, 99, 96–108. [Google Scholar] [CrossRef]
- Ye, Y.; Tang, Y.; Xiong, Y.; Feng, L.; Li, X. Bisphenol a Exposure Alters Placentation and Causes Preeclampsia-like Features in Pregnant Mice Involved in Reprogramming of DNA Methylation of WNT2. FASEB J. 2019, 33, 2732. [Google Scholar] [CrossRef] [Green Version]
- Deb, P.; Bhan, A.; Hussain, I.; Ansari, K.I.; Bobzean, S.A.; Pandita, T.K.; Perrotti, L.I.; Mandal, S.S. Endocrine Disrupting Chemical, Bisphenol-A, Induces Breast Cancer Associated Gene HOXB9 Expression in vitro and in vivo. Gene 2016, 590, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Oldenburg, J.; Fürhacker, M.; Hartmann, C.; Steinbichl, P.; Banaderakhshan, R.; Haslberger, A. Different Bisphenols Induce Non-Monotonous Changes in MiRNA Expression and LINE-1 Methylation in Two Cell Lines. Environ. Epigenet. 2021, 7, dvab011. [Google Scholar] [CrossRef]
- Rodosthenous, R.S.; Baccarelli, A.A.; Mansour, A.; Adir, M.; Israel, A.; Racowsky, C.; Hauser, R.; Bollati, V.; Machtinger, R. Supraphysiological Concentrations of Bisphenol a Alter the Expression of Extracellular Vesicle-Enriched MiRNAs from Human Primary Granulosa Cells. Toxicol. Sci. 2019, 169, 5–13. [Google Scholar] [CrossRef]
- Lite, C.; Ahmed, S.S.S.J.; Santosh, W.; Seetharaman, B. Prenatal Exposure to Bisphenol-A Altered MiRNA-224 and Protein Expression of Aromatase in Ovarian Granulosa Cells Concomitant with Elevated Serum Estradiol Levels in F1 Adult Offspring. J. Biochem. Mol. Toxicol. 2019, 33, e22317. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ibarra, A.; Martínez-Razo, L.D.; Vázquez-Martínez, E.R.; Martínez-Cruz, N.; Flores-Ramírez, R.; García-Gómez, E.; López-López, M.; Ortega-González, C.; Camacho-Arroyo, I.; Cerbón, M. Unhealthy Levels of Phthalates and Bisphenol A in Mexican Pregnant Women with Gestational Diabetes and Its Association to Altered Expression of MiRNAs Involved with Metabolic Disease. Int. J. Mol. Sci. 2019, 20, 3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, B.G.; Babayev, S.N.; Chen, L.X.; Carr, B.R.; Ann Word, R.; Jimenez, P.T. Estrogen-Regulated MiRNA-27b Is Altered by Bisphenol A in Human Endometrial Stromal Cells. Reproduction 2018, 156, 559–567. [Google Scholar] [CrossRef] [Green Version]
- Tilghman, S.L.; Bratton, M.R.; Segar, H.C.; Martin, E.C.; Rhodes, L.V.; Li, M.; McLachlan, J.A.; Wiese, T.E.; Nephew, K.P.; Burow, M.E. Endocrine Disruptor Regulation of MicroRNA Expression in Breast Carcinoma Cells. PLoS ONE 2012, 7, e32754. [Google Scholar] [CrossRef]
- Hui, L.; Li, H.; Lu, G.; Chen, Z.; Sun, W.; Shi, Y.; Fu, Z.; Huang, B.; Zhu, X.; Lu, W.; et al. Low Dose of Bisphenol a Modulates Ovarian Cancer Gene Expression Profile and Promotes Epithelial to Mesenchymal Transition via Canonical Wnt Pathway. Toxicol. Sci. 2018, 164, 527–538. [Google Scholar] [CrossRef]
- Avissar-Whiting, M.; Veiga, K.R.; Uhl, K.M.; Maccani, M.A.; Gagne, L.A.; Moen, E.L.; Marsit, C.J. Bisphenol a Exposure Leads to Specific MicroRNA Alterations in Placental Cells. Reprod. Toxicol. 2010, 29, 401–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Felice, B.; Manfellotto, F.; Palumbo, A.; Troisi, J.; Zullo, F.; di Carlo, C.; Sardo, A.D.S.; de Stefano, N.; Ferbo, U.; Guida, M.; et al. Genome-Wide MicroRNA Expression Profiling in Placentas from Pregnant Women Exposed to BPA. BMC Med. Genom. 2015, 8, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Ding, D.; Wang, T.; Liu, Q.; Lin, Y. MiR-338 Controls BPA-Triggered Pancreatic Islet Insulin Secretory Dysfunction from Compensation to Decompensation by Targeting Pdx-1. FASEB J. 2017, 31, 5184–5195. [Google Scholar] [CrossRef] [Green Version]
- Kasuya, T.; Hori, S.I.; Watanabe, A.; Nakajima, M.; Gahara, Y.; Rokushima, M.; Yanagimoto, T.; Kugimiya, A. Ribonuclease H1-Dependent Hepatotoxicity Caused by Locked Nucleic Acid-Modified Gapmer Antisense Oligonucleotides. Sci. Rep. 2016, 6, 30377. [Google Scholar] [CrossRef]
- Sabry, R.; Yamate, J.; Favetta, L.; LaMarre, J. MicroRNAs: Potential Targets and Agents of Endocrine Disruption in Female Reproduction. J. Toxicol. Pathol. 2019, 32, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; Yu, S.Y.; Yoon, H.J.; Lee, S.Y.; Youn, J.P.; Hwang, S.Y. Epigenetic Regulation of MiR-22 in a BPA-Exposed Human Hepatoma Cell. BioChip J. 2015, 9, 76–84. [Google Scholar] [CrossRef]
- Zhang, W.; Fang, Y.; Shi, X.; Zhang, M.; Wang, X.; Tan, Y. Effect of Bisphenol A on the EGFR-STAT3 Pathway in MCF-7 Breast Cancer Cells. Mol. Med. Rep. 2012, 5, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Zhou, J.; Sun, X.; He, C.; Zhang, K.; Hu, K. Effects of Bisphenol A on Apoptosis of Ovarian Preantral Follicular Granulosa Cells and Ovarian Development in Mice. Nan Fang Yi Ke Da Xue Xue Bao 2021, 41, 93–99. [Google Scholar] [CrossRef]
- Téteau, O.; Jaubert, M.; Desmarchais, A.; Papillier, P.; Binet, A.; Maillard, V.; Elis, S. Bisphenol A and S Impaired Ovine Granulosa Cell Steroidogenesis. Reproduction 2020, 159, 571–583. [Google Scholar] [CrossRef]
- Mansur, A.; Israel, A.; Combelles, C.M.H.; Adir, M.; Racowsky, C.; Hauser, R.; Baccarelli, A.A.; Machtinger, R. Bisphenol-A Exposure and Gene Expression in Human Luteinized Membrana Granulosa Cells in vitro. Hum. Reprod. 2017, 32, 409–417. [Google Scholar] [CrossRef]
- Naji, M.; Aleyasin, A.; Nekoonam, S.; Arefian, E.; Mahdian, R.; Amidi, F. Differential Expression of MiR-93 and MiR-21 in Granulosa Cells and Follicular Fluid of Polycystic Ovary Syndrome Associating with Different Phenotypes. Sci. Rep. 2017, 7, 14671. [Google Scholar] [CrossRef]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. MicroRNA-21 Promotes Breast Cancer Proliferation and Metastasis by Targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef] [Green Version]
- Yenuganti, V.R.; Vanselow, J. Cultured Bovine Granulosa Cells Rapidly Lose Important Features of Their Identity and Functionality but Partially Recover under Long-Term Culture Conditions. Cell Tissue Res. 2017, 368, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-MiRNA Oligonucleotides: A Comprehensive Guide for Design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Larcher, L.M.; Ma, L.; Veedu, R.N. Systematic Screening of Commonly Used Commercial Transfection Reagents towards Efficient Transfection of Single-Stranded Oligonucleotides. Molecules 2018, 23, 2564. [Google Scholar] [CrossRef] [Green Version]
- Carletti, M.Z.; Fiedler, S.D.; Christenson, L.K. MicroRNA 21 Blocks Apoptosis in Mouse Periovulatory Granulosa Cells. Biol. Reprod. 2010, 83, 286–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.; Tran, N. Global MiRNA to MiRNA Interactions: Impacts for MiR-21. Trends Cell Biol. 2021, 31, 3–5. [Google Scholar] [CrossRef]
- Emmerling, V.V.; Fischer, S.; Kleemann, M.; Handrick, R.; Kochanek, S.; Otte, K. MiR-483 Is a Self-Regulating MicroRNA and Can Activate Its Own Expression via USF1 in HeLa Cells. Int. J. Biochem. Cell Biol. 2016, 80, 81–86. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. MicroRNAs Regulating MicroRNAs in Cancer. Trends Cancer 2018, 4, 465–468. [Google Scholar] [CrossRef]
- Werry, N.; Russell, S.J.; Gillis, D.J.; Miller, S.; Hickey, K.; Larmer, S.; Lohuis, M.; Librach, C.; LaMarre, J. Characteristics of MiRNAs Present in Bovine Sperm and Associations with Differences in Fertility. Front. Endocrinol. 2022, 13, 912. [Google Scholar] [CrossRef]
- Hua, R.; Zhang, X.; Li, W.; Lian, W.; Liu, Q.; Gao, D.; Wang, Y.; Lei, M. Ssc-MiR-21-5p Regulates Endometrial Epithelial Cell Proliferation, Apoptosis and Migration via the PDCD4/AKT Pathway. J. Cell Sci. 2020, 133, jcs248898. [Google Scholar] [CrossRef]
- Liu, J.; Zhai, R.; Zhao, J.; Kong, F.; Wang, J.; Jiang, W.; Xin, Q.; Xue, X.; Luan, Y. Programmed Cell Death 4 Overexpression Enhances Sensitivity to Cisplatin via the JNK/c-Jun Signaling Pathway in Bladder Cancer. Int. J. Oncol. 2018, 52, 1633–1642. [Google Scholar] [CrossRef] [Green Version]
- Green, D.E.; Murphy, T.C.; Kang, B.Y.; Bedi, B.; Yuan, Z.; Sadikot, R.T.; Hart, C.M. Peroxisome Proliferator-Activated Receptor-γ Enhances Human Pulmonary Artery Smooth Muscle Cell Apoptosis through MicroRNA-21 and Programmed Cell Death 4. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2017, 313, L371–L383. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Yu, J.; Zhang, Y.; Bian, Y.; Ma, J.; Yan, J. Overexpression of PTEN Regulated by MiR-19b and MiR-494 in the Villous of Recurrent Spontaneous Abortion Patients. J. Reprod. Immunol. 2020, 140, 103133. [Google Scholar] [CrossRef]
- Li, X.; Xie, W.; Xie, C.; Huang, C.; Zhu, J.; Liang, Z.; Deng, F.; Zhu, M.; Zhu, W.; Wu, R.; et al. Curcumin Modulates MiR-19/PTEN/AKT/P53 Axis to Suppress Bisphenol A-Induced MCF-7 Breast Cancer Cell Proliferation. Phytother. Res. 2014, 28, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Zhang, J. MiRNA-21 Inhibition Suppresses the Human Epithelial Ovarian Cancer by Targeting PTEN Signal Pathway. Saudi J. Biol. Sci. 2019, 26, 2026–2029. [Google Scholar] [CrossRef]
- Yu, X.; Chen, Y.; Tian, R.; Li, J.; Li, H.; Lv, T.; Yao, Q. MiRNA-21 Enhances Chemoresistance to Cisplatin in Epithelial Ovarian Cancer by Negatively Regulating PTEN. Oncol. Lett. 2017, 14, 1807–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jiang, K.; Zhao, F. Icariin Regulates the Proliferation and Apoptosis of Human Ovarian Cancer Cells through MicroRNA-21 by Targeting PTEN, RECK and Bcl-2. Oncol. Rep. 2015, 33, 2829–2836. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Kinkade, J.A.; Bivens, N.J.; Rosenfeld, C.S. MiRNA Changes in the Mouse Placenta Due to Bisphenol a Exposure. Epigenomics 2021, 13, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Alm, H.; Choi, Y.H.; Love, L.; Heleil, B.; Torner, H.; Hinrichs, K. Holding Bovine Oocytes in the Absence of Maturation Inhibitors: Kinetics of in vitro Maturation and Effect on Blastocyst Development after in vitro Fertilization. Theriogenology 2008, 70, 1024–1029. [Google Scholar] [CrossRef]
- Klinge, C. Estrogen Regulation of MicroRNA Expression. Curr. Genom. 2009, 10, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Shanle, E.K.; Xu, W. Endocrine Disrupting Chemicals Targeting Estrogen Receptor Signaling: Identification and Mechanisms of Action. Chem. Res. Toxicol. 2011, 24, 6–19. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.J.; Ren, G.; Liu, J.L.; Zhao, Z.A.; Yu, Y.S.; Su, R.W.; Ma, X.H.; Ni, H.; Lei, W.; Yang, Z.M. MicroRNA Expression and Regulation in Mouse Uterus during Embryo Implantation. J. Biol. Chem. 2008, 283, 23473–23484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Peng, R.; Zeng, M.; Zhang, Z.; Liu, S.; Jiang, D.; Lu, Y.; Zou, F. An Autoregulatory Feedback Loop of MiR-21/VMP1 Is Responsible for the Abnormal Expression of MiR-21 in Colorectal Cancer Cells. Cell Death Dis. 2020, 11, 1067. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Garrido, J.; Carilla-Latorre, S.; Lázaro-Diéguez, F.; Egea, G.; Escalante, R. Vacuole Membrane Protein 1 Is an Endoplasmic Reticulum Protein Required for Organelle Biogenesis, Protein Secretion, and Development. Mol. Biol. Cell 2008, 19, 3442–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Kou, D.; Le, W. Roles of VMP1 in Autophagy and ER–Membrane Contact: Potential Implications in Neurodegenerative Disorders. Front. Mol. Neurosci. 2020, 13, 42. [Google Scholar] [CrossRef]
- Tošić, I.; Frank, D.A. STAT3 as a Mediator of Oncogenic Cellular Metabolism: Pathogenic and Therapeutic Implications. Neoplasia 2021, 23, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.M.; Wang, C.G.; Zhu, M.; Xing, R.; Cui, J.T.; Li, W.M.; Yu, D.D.; Wang, S.B.; Zhu, W.; Ye, Y.J.; et al. STAT3 Signaling Drives EZH2 Transcriptional Activation and Mediates Poor Prognosis in Gastric Cancer. Mol. Cancer 2016, 15, 79. [Google Scholar] [CrossRef] [Green Version]
- Brion, C.; Lutz, S.M.; Albert, F.W. Simultaneous Quantification of Mrna and Protein in Single Cells Reveals Post-Transcriptional Effects of Genetic Variation. Elife 2020, 9, e60645. [Google Scholar] [CrossRef]
- Ajuyah, P.; Hill, M.; Ahadi, A.; Lu, J.; Hutvagner, G.; Tran, N. MicroRNA (MiRNA)-to-MiRNA Regulation of Programmed Cell Death 4 (PDCD4). Mol. Cell. Biol. 2019, 39, e00086-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Gee, H.; Rose, B.; Lee, C.S.; Clark, J.; Elliott, M.; Gamble, J.R.; Cairns, M.J.; Harris, A.; Khoury, S.; et al. Regulation of the Tumour Suppressor PDCD4 by MiR-499 and MiR-21 in Oropharyngeal Cancers. BMC Cancer 2016, 16, 86. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting Apoptosis in Cancer Therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Surova, O.; Zhivotovsky, B. Various Modes of Cell Death Induced by DNA Damage. Oncogene 2012, 32, 3789–3797. [Google Scholar] [CrossRef] [Green Version]
- Dumitrascu, M.C.; Mares, C.; Petca, R.C.; Sandru, F.; Popescu, R.I.; Mehedintu, C.; Petca, A. Carcinogenic Effects of Bisphenol A in Breast and Ovarian Cancers (Review). Oncol. Lett. 2020, 20, 282. [Google Scholar] [CrossRef]
- Teng, Y.; Manavalan, T.T.; Hu, C.; Medjakovic, S.; Jungbauer, A.; Klinge, C.M. Endocrine Disruptors Fludioxonil and Fenhexamid Stimulate MiR-21 Expression in Breast Cancer Cells. Toxicol. Sci. 2013, 131, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Fu, W.; Quan, C.; Yan, M.; Liu, C.; Qi, S.; Yang, K. The Role of Pten/Akt Signaling Pathway Involved in BPA-Induced Apoptosis of Rat Sertoli Cells. Environ. Toxicol. 2015, 30, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jenkins, S.; Lamartiniere, C.A. Cell Proliferation and Apoptosis in Rat Mammary Glands Following Combinational Exposure to Bisphenol A and Genistein. BMC Cancer 2014, 14, 379. [Google Scholar] [CrossRef] [Green Version]
- Rashmi; Sah, S.; Shyam, S.; Singh, M.K.; Palta, P. Treatment of Buffalo (Bubalus Bubalis) SCNT Embryos with MicroRNA-21 Mimic Improves Their Quality and Alters Gene Expression but Does not Affect Their Developmental Competence. Theriogenology 2019, 126, 8–16. [Google Scholar] [CrossRef]
- Hilker, R.E.; Pan, B.; Zhan, X.; Li, J. MicroRNA-21 Enhances Estradiol Production by Inhibiting WT1 Expression in Granulosa Cells. J. Mol. Endocrinol. 2022, 68, 11–22. [Google Scholar] [CrossRef]
- Jakic, B.; Buszko, M.; Cappellano, G.; Wick, G. Elevated Sodium Leads to the Increased Expression of HSP60 and Induces Apoptosis in HUVECs. PLoS ONE 2017, 12, e0179383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurashina, Y.; Imashiro, C.; Hirano, M.; Kuribara, T.; Totani, K.; Ohnuma, K.; Friend, J.; Takemura, K. Enzyme-Free Release of Adhered Cells from Standard Culture Dishes Using Intermittent Ultrasonic Traveling Waves. Commun. Biol. 2019, 2, 393. [Google Scholar] [CrossRef] [Green Version]
- Hooper, A.L. MicroRNA Regulation of Ovarian Angiogenesis and Folliculogenesis in Bovine Cystic Ovarian Disease. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2018. [Google Scholar]
- Gu, Z.; Eleswarapu, S.; Jiang, H. Identification and Characterization of MicroRNAs from the Bovine Adipose Tissue and Mammary Gland. FEBS Lett. 2007, 581, 981–988. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Long, K.; Ma, J.; Zhang, J.; He, D.; Jin, L.; Tang, Q.; Jiang, A.; Wang, X.; Hu, Y.; et al. Comparative Analysis of the MicroRNA Transcriptome between Yak and Cattle Provides Insight into High-Altitude Adaptation. PeerJ 2017, 2017, e3959. [Google Scholar] [CrossRef] [Green Version]
- Lawless, N.; Vegh, P.; O’Farrelly, C.; Lynn, D.J. The Role of MicroRNAs in Bovine Infection and Immunity. Front. Immunol. 2014, 5, 611. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.P.; Luoreng, Z.M.; Zan, L.S.; Li, F.; Li, N. Bovine MiR-146a Regulates Inflammatory Cytokines of Bovine Mammary Epithelial Cells via Targeting the TRAF6 Gene. J. Dairy Sci. 2017, 100, 7648–7658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian MicroRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Sharma, J. Role of Hippo Signaling Pathway in Bovine Preimplantation Embryo Development. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2016. [Google Scholar]
- Ferris, J.; Mahboubi, K.; MacLusky, N.; King, W.A.; Favetta, L.A. BPA Exposure during in vitro Oocyte Maturation Results in Dose-Dependent Alterations to Embryo Development Rates, Apoptosis Rate, Sex Ratio and Gene Expression. Reprod. Toxicol. 2016, 59, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cong Kong, L.; Jia, B.Y.; Chen, J.R.; Dong, Y.; Jiang, X.Y.; Ma, H.X. Analysis of the MicroRNA Expression Profile of Bovine Monocyte-Derived Macrophages Infected with Mycobacterium Avium Subsp. Paratuberculosis Reveals That MiR-150 Suppresses Cell Apoptosis by Targeting PDCD4. Int. J. Mol. Sci. 2019, 20, 2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.S. TGF Beta1 Dependent Regulation of MiR-21 in Bovine Fetal Fibroblasts. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2016. [Google Scholar]
- Meng, F.; Forrester-Gauntlett, B.; Turner, P.; Henderson, H.; Oback, B. Signal Inhibition Reveals JAK/STAT3 Pathway as Critical for Bovine Inner Cell Mass Development. Biol. Reprod. 2015, 93, 132. [Google Scholar] [CrossRef]
- Lakshmanan, I.; Batra, S.K. Protocol for Apoptosis Assay by Flow Cytometry Using Annexin V Staining Method. Bio-Protocol 2013, 3, e374. [Google Scholar] [CrossRef] [Green Version]
- Farahani, M.; Rezaei-Tavirani, M.; Arjmand, B. A Systematic Review of MicroRNA Expression Studies with Exposure to Bisphenol A. J. Appl. Toxicol. 2021, 41, 4–19. [Google Scholar] [CrossRef]
- Huang, M.; Huang, M.; Li, X.; Liu, S.; Fu, L.; Jiang, X.; Yang, M. Bisphenol A Induces Apoptosis through GPER-Dependent Activation of the ROS/Ca 2+-ASK1-JNK Pathway in Human Granulosa Cell Line KGN. Ecotoxicol. Environ. Saf. 2021, 208, 111429. [Google Scholar] [CrossRef]
- Harnett, K.G.; Chin, A.; Schuh, S.M. BPA and BPA Alternatives BPS, BPAF, and TMBPF, Induce Cytotoxicity and Apoptosis in Rat and Human Stem Cells. Ecotoxicol. Environ. Saf. 2021, 216, 112210. [Google Scholar] [CrossRef]



| MicroRNA | Primer ID | Accession Number | Sequence (5′-3′) | E (%) | Source |
|---|---|---|---|---|---|
| miR-191 | hsa-miR-191-5p | MIMAT0000440 | AACGGAATCCCAAAAGCAG | 99.7 | [88] |
| miR-106a | hsa-miR-106a-5p | MIMAT0000103 | CGCCAAAAGTGCTTACAGTGC | 92.4 | |
| miR-21 | bta-miR-21-5p | MIMAT0003528 | TAGCTTATCAGACTGATGTTGACT | 96.7 | [89] |
| miR-34c | bta-miR-34c | MIMAT0003854 | AGGCAGTGTAGTTAGCTGATTGC | 99.6 | [90] |
| miR-155 | hsa-miR-155-5p | MIMAT0000646 | TGCTAATCGTGATAGGGGTAAA | 100 | [91] |
| miR-146a | bta-miR-146a | MIMAT0009236 | TGAGAACTGAATTCCATAGGTTG | 100.2 | [92] |
| miR-10b | hsa-miR-10b-3p | MIMAT0000267 | GACAGATTCGATTCTAGGGGAAT | 101.5 | [93] |
| Gene Symbol | Gene Name | Product Size (bp) | Accession Number | Primer Sequence Sets (5′-3′) | E (%) | Source |
|---|---|---|---|---|---|---|
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 120 | NM_174814.2 | F: GCATCCCACAGACTATTTCC R: GCAAAGACAATGACAGACCA | 100.3 | [94] |
| ACTB | Beta-actin | 186 | NM_173979.3 | F: CCTTCCTGGGCATGGAATCCT R: TCTTCATTGTGCTGGGTGCC | 97 | [15] |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 153 | NM_001034034.2 | F: TTCCTGGTACGACAATGAATTTG R: GGAGATGGGGCAGGACTC | 99.8 | [95] |
| PDCD4 | Programmed cell death 4 | 108 | NM_001083647.1 | F: AAAGACTCTGACACCGATTA R: CAAGGACACTGCCAACAC | 99.4 | [96] |
| PTEN | Phosphatase and tensin homolog | 151 | NM_001319898.1 | F: TGCAGAGTTGCACAGTATCCC R: CACCAGTTCGTCCCTTTCCA | 100 | [97] |
| VMP1 | Vacuole membrane protein 1 | 266 | NM_001075368.2 | F: GACCAGAGACGTGTAGCAATG R: ACAATGCTTTGACGATGCCATA | 99.4 | [72] |
| STAT3 | Signal transducer and activator of transcription 3 | 200 | NM_001012671.2 | F: GTGCATTGACAAAGACTCCG R: AATCAGGGAGGCATCACAAT | 100.1 | [98] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabry, R.; Williams, M.; Werry, N.; LaMarre, J.; Favetta, L.A. BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition. Int. J. Mol. Sci. 2022, 23, 8276. https://doi.org/10.3390/ijms23158276
Sabry R, Williams M, Werry N, LaMarre J, Favetta LA. BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition. International Journal of Molecular Sciences. 2022; 23(15):8276. https://doi.org/10.3390/ijms23158276
Chicago/Turabian StyleSabry, Reem, Makenna Williams, Nicholas Werry, Jonathan LaMarre, and Laura A. Favetta. 2022. "BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition" International Journal of Molecular Sciences 23, no. 15: 8276. https://doi.org/10.3390/ijms23158276
APA StyleSabry, R., Williams, M., Werry, N., LaMarre, J., & Favetta, L. A. (2022). BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition. International Journal of Molecular Sciences, 23(15), 8276. https://doi.org/10.3390/ijms23158276







