Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses
Abstract
1. Introduction
2. Results
2.1. Identification of EgHsfs in Eucalyptus
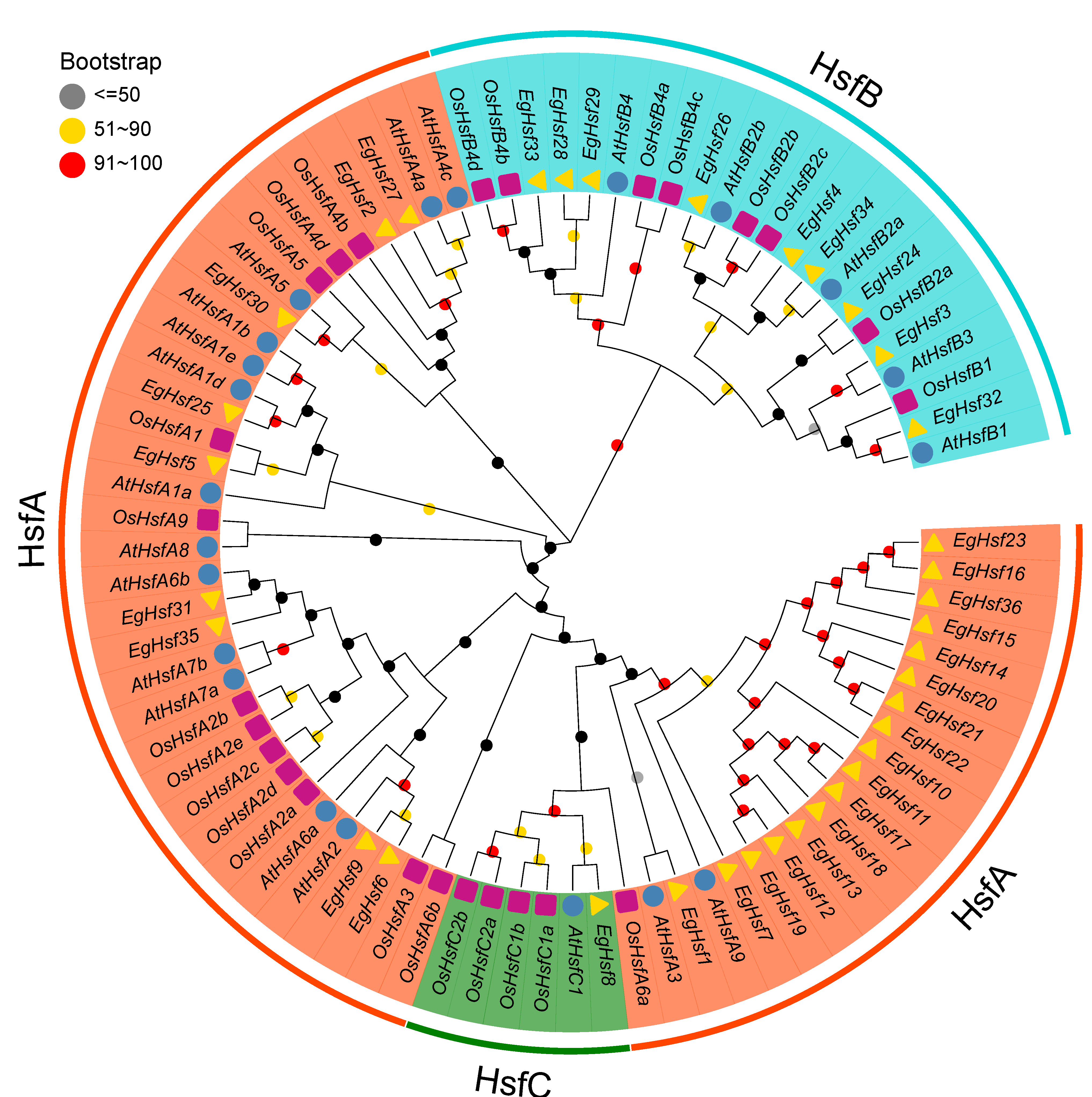
2.2. Phylogenetic Analysis and Classification of EgHSF Protein Members
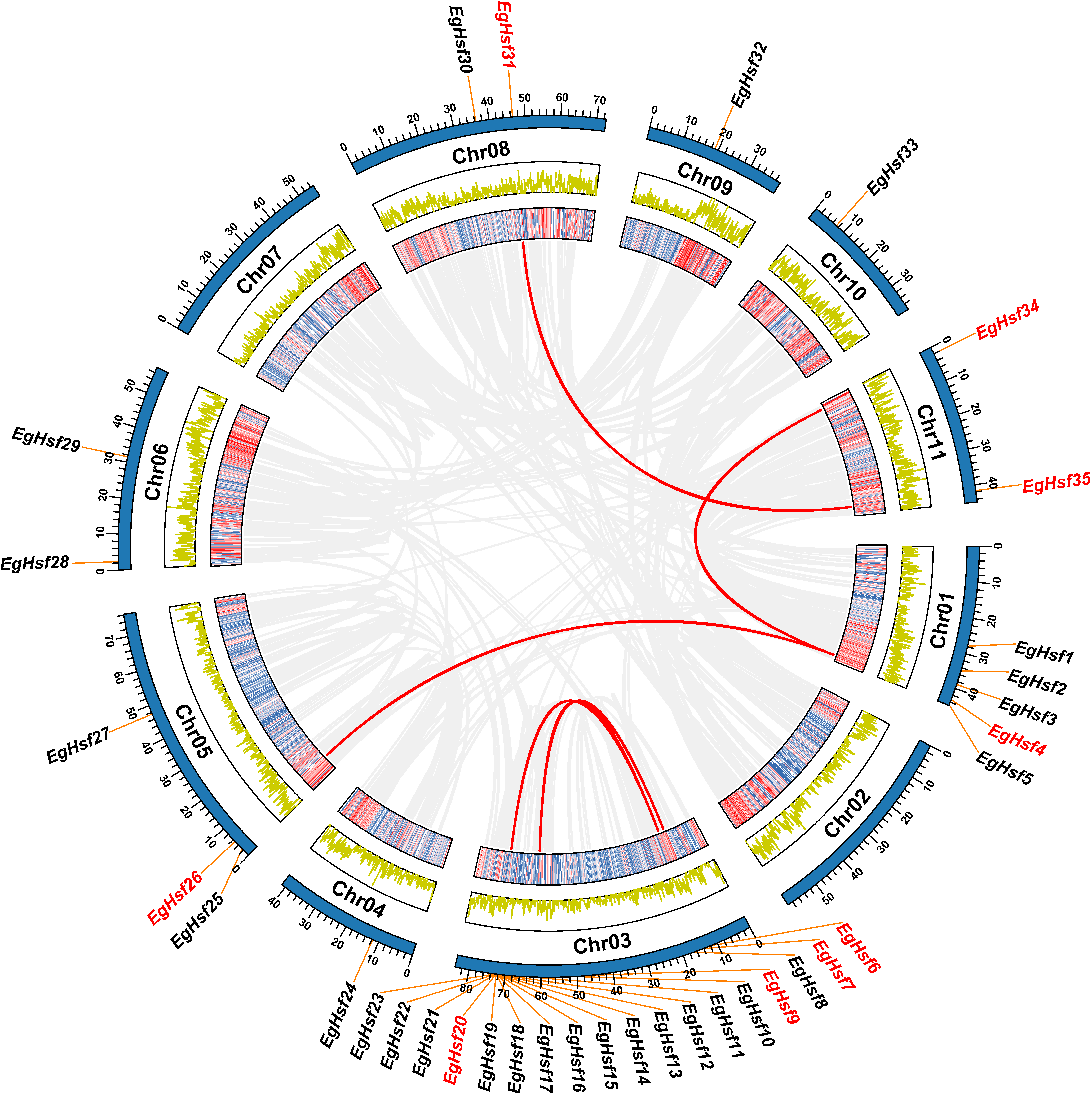
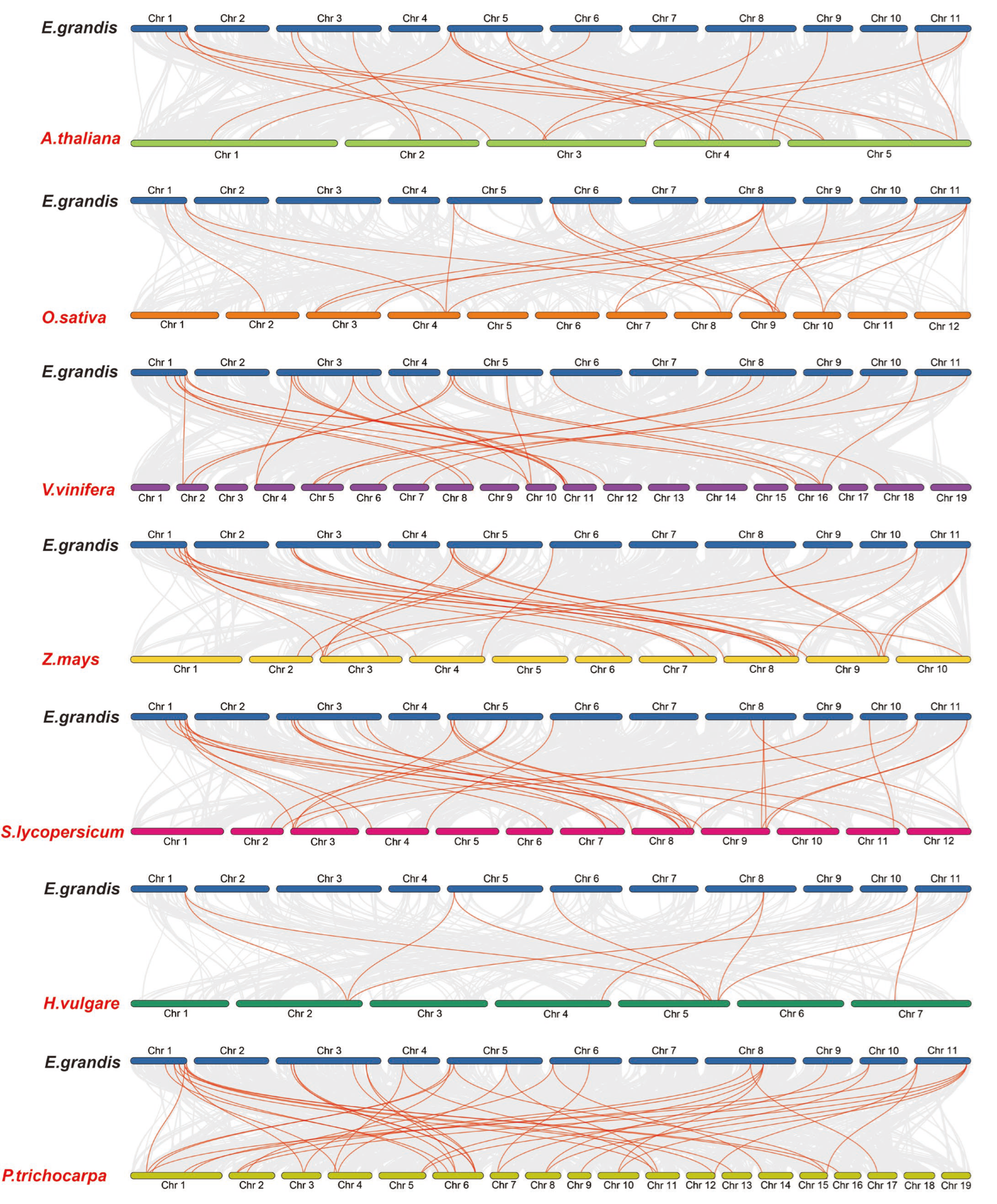
2.3. Chromosomal Distribution and Synteny Analysis of the EgHsf Genes
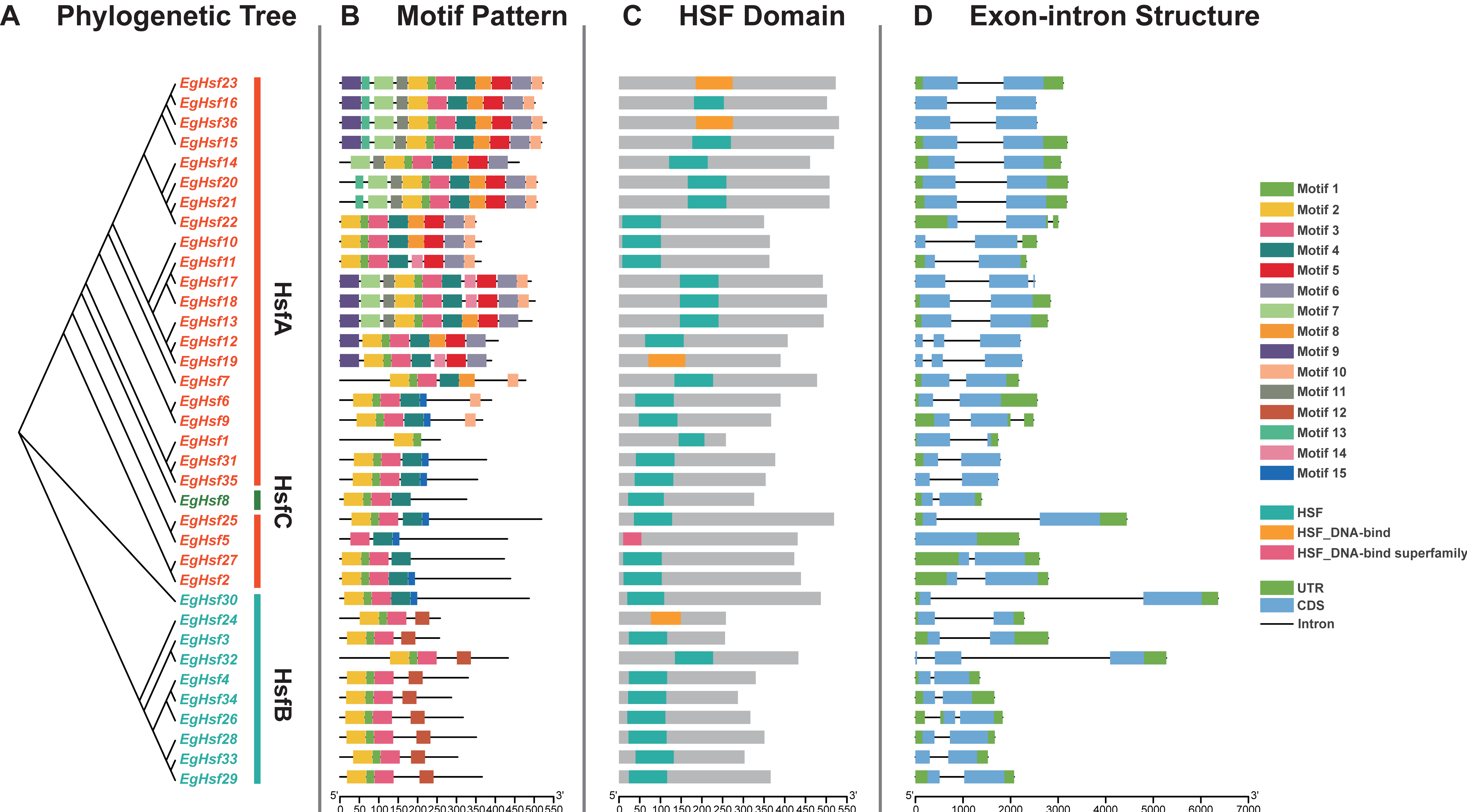
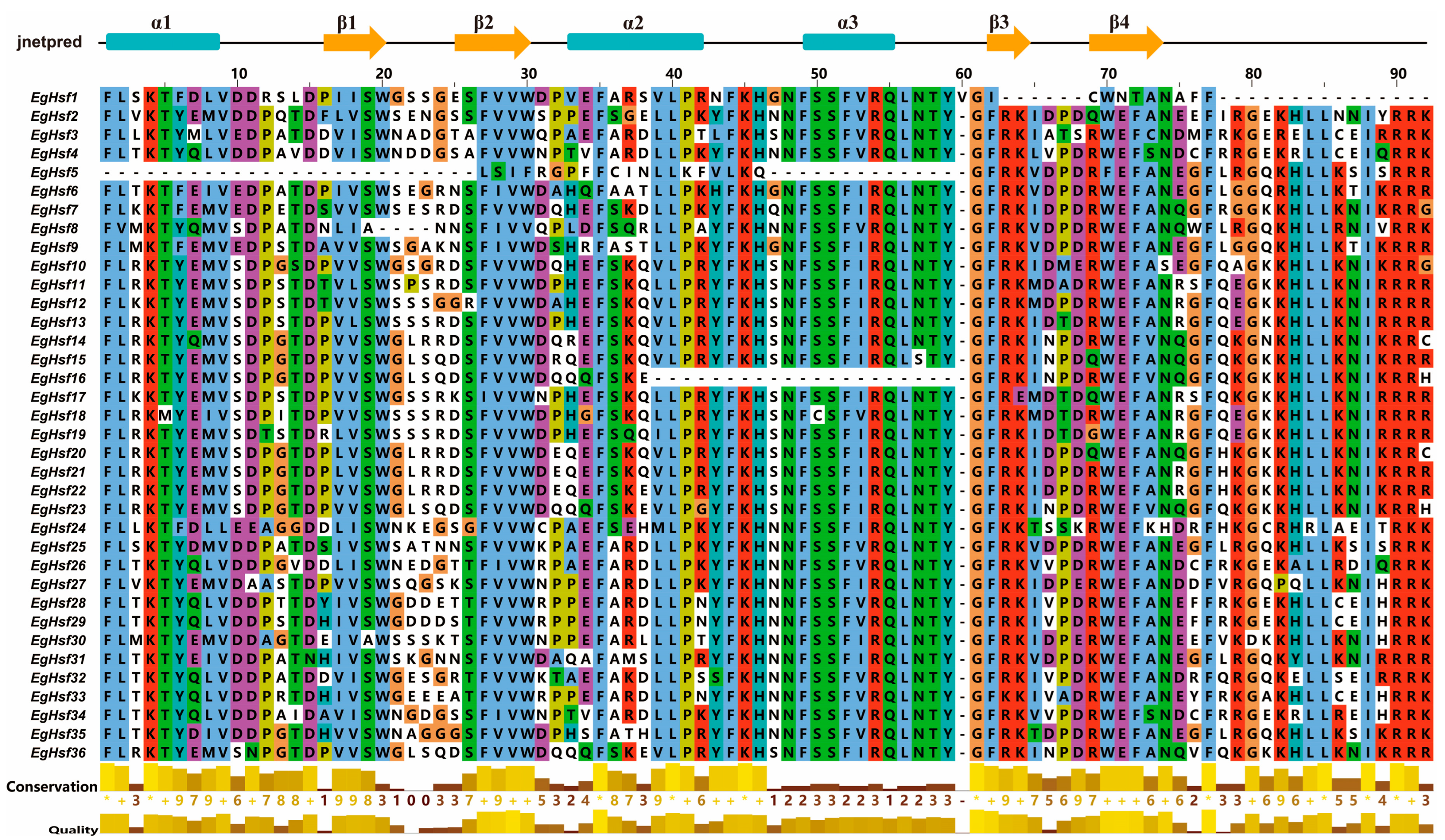
2.4. Structure and Motif Analysis of EgHsf Genes
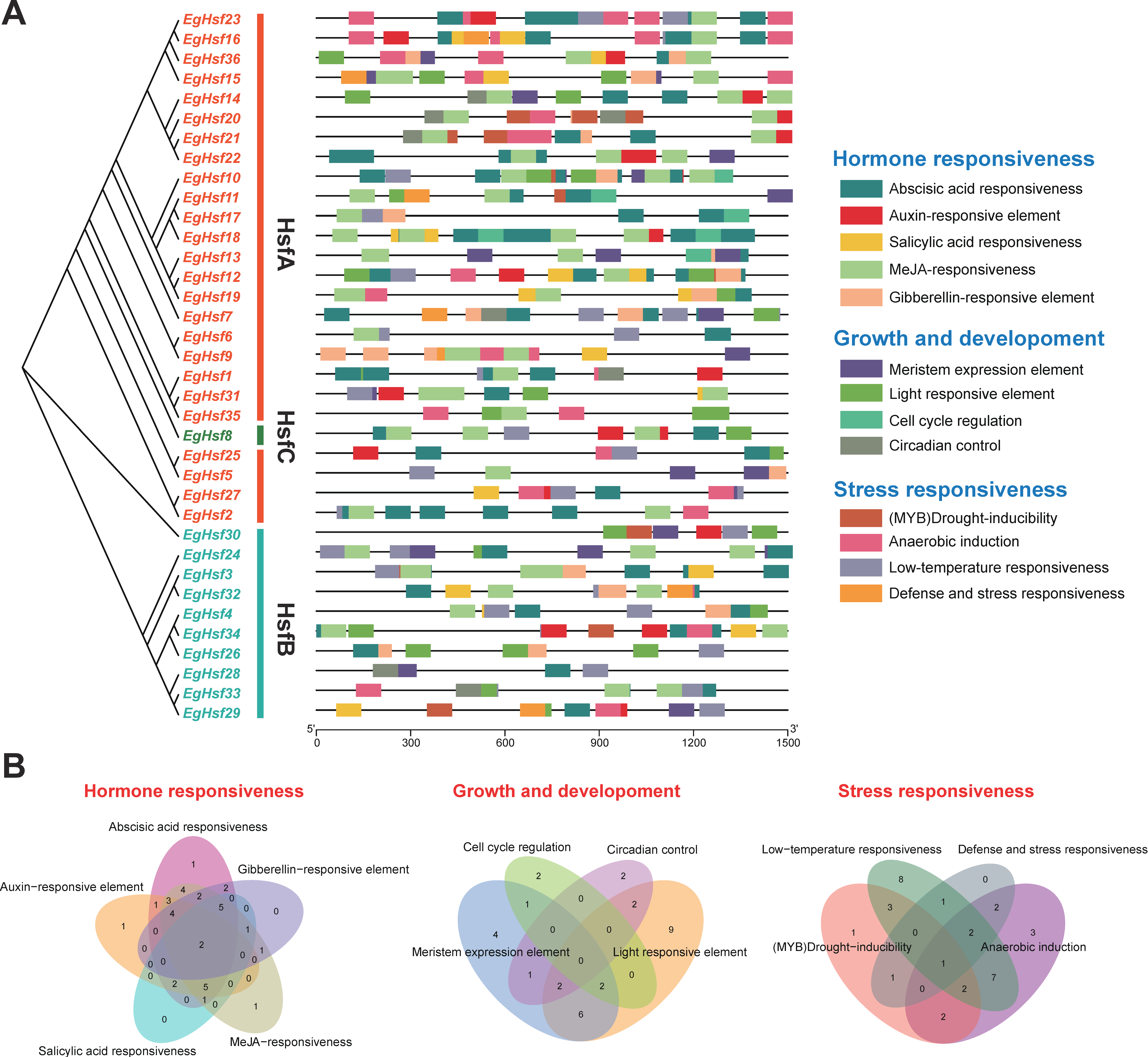
2.5. Promoter Cis-Element Analysis of EgHsf Genes
2.6. Expression of Hsf Genes in Eucalyptus under Abiotic Stress
2.7. Expression Map and Correlation Analysis of Hsf Genes in Different Tissues
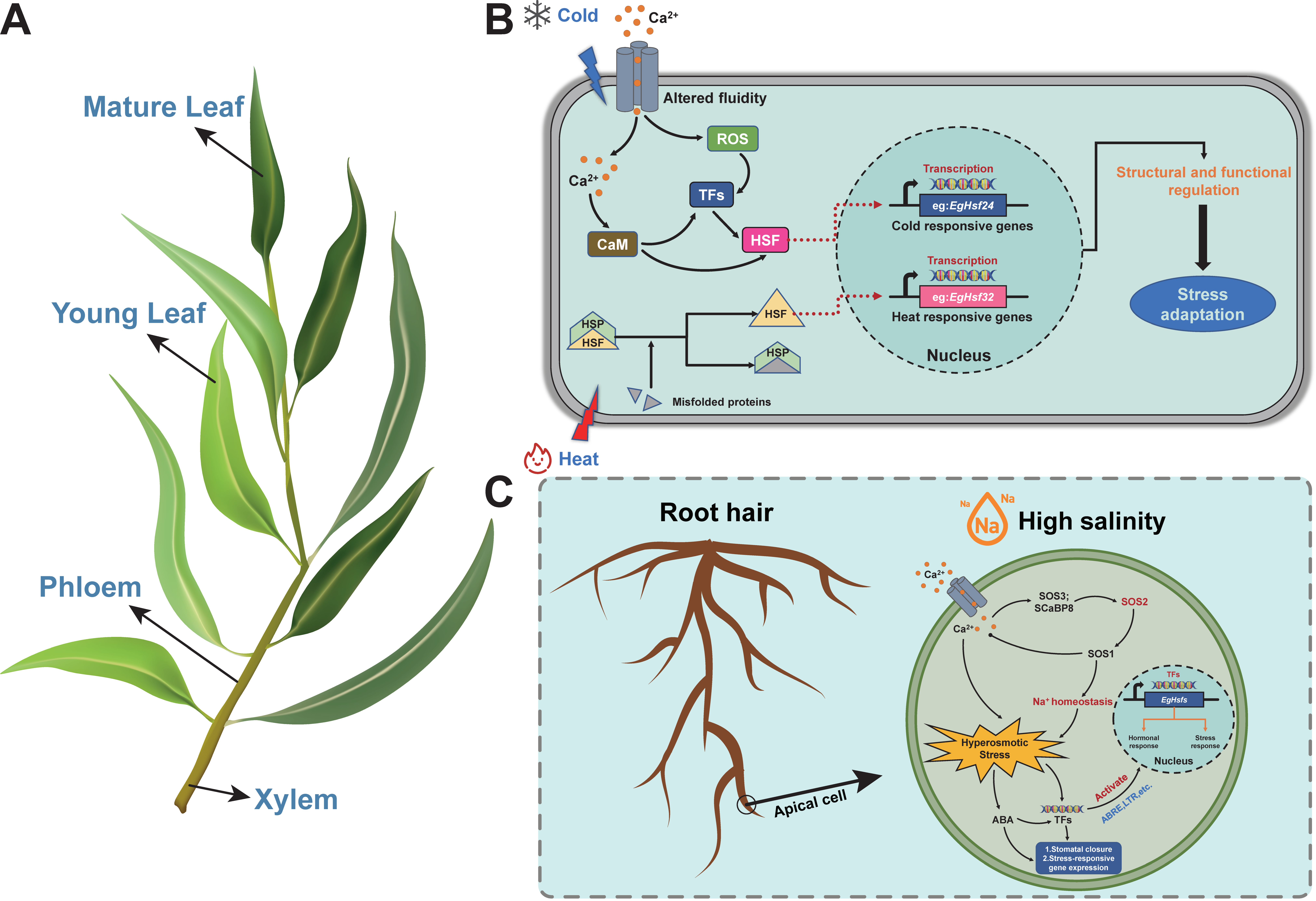
3. Discussion
4. Materials and Methods
4.1. Plant Material and Data Sources
4.2. Identification and Sequence Analysis of EgHSF Protein
4.3. Classification of EgHSF Protein Members and Construction of Phylogenetic Tree
4.4. Chromosomal Distribution and Synteny Analysis of the EgHsf Genes
4.5. Structure and Motif Analysis of EgHSFs
4.6. Promoter Cis-Element Analysis of EgHsf Genes
4.7. Heat Map Network Analysis of Hsf Gene Expression and Correlation Cluster Markers
4.8. RNA Extraction and qRT–PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, S.H.; Kim, S.; Chang, H.N.; Kim, H.J.; An, J.; Son, Y. Fine root biomass and production regarding root diameter in Pinusdensiflora and Quercusserrata forests: Soil depth effects and the relationship with net primary production. Turk. J. Agric. For. 2021, 45, 46–54. [Google Scholar] [CrossRef]
- Curovic, M.; Spalevic, V.; Sestras, P.; Motta, R.; Dan, C.; Garbarino, M.; Vitali, A.; Urbinati, C. Structural and ecological characteristics of mixed broadleaved old-growth forest (Biogradska Gora—Montenegro). Turk. J. Agric. For. 2020, 44, 428–438. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Forrester, D.I.; Smith, R.G.B. Faster growth of Eucalyptus grandis and Eucalyptus pilularis in mixed-species stands than monocultures. For. Ecol. Manag. 2012, 286, 81–86. [Google Scholar] [CrossRef]
- Bakshi, A.; Moin, M.; Kumar, M.U.; Reddy, A.B.; Ren, M.; Datla, R.; Siddiq, E.A.; Kirti, P.B. Ectopic expression of Arabidopsis Target of Rapamycin (AtTOR) improves water-use efficiency and yield potential in rice. Sci. Rep. 2017, 7, 42835. [Google Scholar] [CrossRef]
- Balazadeh, S. A ‘hot’ cocktail: The multiple layers of thermomemory in plants. Curr. Opin. Plant Biol. 2021, 65, 102147. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Young, J.C.; Barral, J.M.; Ulrich Hartl, F. More than folding: Localized functions of cytosolic chaperones. Trends Biochem. Sci. 2003, 28, 541–547. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K. The HSP90 complex of plants. Biochim. Biophys. Acta 2012, 1823, 689–697. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Valle, E.M. Investigating the role of plant heat shock proteins during oxidative stress. Plant Signal Behav. 2008, 3, 856–857. [Google Scholar] [CrossRef]
- Arrigo, A.P. Human small heat shock proteins: Protein interactomes of homo- and hetero-oligomeric complexes: An update. FEBS Lett. 2013, 587, 1959–1969. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Colinet, H.; Lee, S.F.; Hoffmann, A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 2010, 277, 174–185. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2012, 1819, 104–119, Erratum in Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2018, 1861, 60. [Google Scholar] [CrossRef]
- Zhang, Q.; Geng, J.; Du, Y.; Zhao, Q.; Zhang, W.; Fang, Q.; Yin, Z.; Li, J.; Yuan, X.; Fan, Y.; et al. Heat shock transcription factor (Hsf) gene family in common bean (Phaseolus vulgaris): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses at the sprout stage under abiotic stress. BMC Plant Biol. 2022, 22, 33. [Google Scholar] [CrossRef]
- Waters, E.R.; Lee, G.J.; Vierling, E. Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 1996, 47, 325–338. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Giesguth, M.; Sahm, A.; Simon, S.; Dietz, K.J. Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 2015, 589, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Baruah, P.M.; Krishnatreya, D.B.; Bordoloi, K.S.; Gill, S.S.; Agarwala, N. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol. Biochem. 2021, 162, 221–236. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, H.; Liu, J.; Du, Q.; Lu, S.; Liu, C. Genome-wide identification and functional characterization of natural antisense transcripts in Salvia miltiorrhiza. Sci. Rep. 2021, 11, 4769. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.M.; Kumar, A.; et al. Genome-wide analysis and characterization of the proline-rich extensin-like receptor kinases (PERKs) gene family reveals their role in different developmental stages and stress conditions in wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.-x.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2020, 93, 89–103. [Google Scholar] [CrossRef]
- Luo, J.; Abid, M.; Tu, J.; Gao, P.; Wang, Z.; Huang, H. Genome-wide identification of the LHC gene family in kiwifruit and regulatory role of AcLhcb3.1/3.2 for chlorophyll a content. Int. J. Mol. Sci. 2022, 23, 6528. [Google Scholar] [CrossRef]
- Duan, S.; Liu, B.; Zhang, Y.; Li, G.; Guo, X. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 2019, 20, 257. [Google Scholar] [CrossRef]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide identification and classification of HSF family in grape, and their transcriptional analysis under heat acclimation and heat stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.P.; Zhai, Y.F.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef]
- Huang, B.; Huang, Z.; Ma, R.; Chen, J.; Zhang, Z.; Yrjala, K. Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2021, 11, 16492. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Z.; Tang, R.; Wang, L.; Chen, C.; Ren, Z. Genome-wide identification and expression analysis of Hsf and Hsp gene families in cucumber (Cucumis sativus L.). Plant Growth Regul. 2021, 95, 223–239. [Google Scholar] [CrossRef]
- Cao, J.; Schneeberger, K.; Ossowski, S.; Gunther, T.; Bender, S.; Fitz, J.; Koenig, D.; Lanz, C.; Stegle, O.; Lippert, C.; et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011, 43, 956–963. [Google Scholar] [CrossRef]
- Sasaki, T.; Matsumoto, T.; Baba, T.; Yamamoto, K.; Wu, J.; Katayose, Y.; Sakata, K. The international rice genome sequencing project: Progress and prospects. In Rice Genetics IV; International Rice Research Institute (IRRI): Los Baños, Philippines, 2008; pp. 189–196. [Google Scholar]
- Asamizu, E. Tomato genome sequencing: Deciphering the euchromatin region of the chromosome 8. Plant Biotechnol. 2007, 24, 5–9. [Google Scholar] [CrossRef][Green Version]
- Hu, Y.; Colantonio, V.; Muller, B.S.F.; Leach, K.A.; Nanni, A.; Finegan, C.; Wang, B.; Baseggio, M.; Newton, C.J.; Juhl, E.M.; et al. Genome assembly and population genomic analysis provide insights into the evolution of modern sweet corn. Nat. Commun. 2021, 12, 1227. [Google Scholar] [CrossRef]
- Douglas, C.J.; DiFazio, S.P. The populus genome and comparative genomics. In Genetics and Genomics of Populus; Jansson, S., Bhalerao, R., Groover, A., Eds.; Springer: New York, NY, USA, 2010; pp. 67–90. [Google Scholar]
- Li, P.-S.; Yu, T.-F.; He, G.-H.; Chen, M.; Zhou, Y.-B.; Chai, S.-C.; Xu, Z.-S.; Ma, Y.-Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef]
- Chung, E.; Kim, K.M.; Lee, J.H. Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genom. 2013, 40, 127–135. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Doring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Zhang, D.; Li, D. Expression analysis of segmentally duplicated ZmMPK3-1 and ZmMPK3-2 genes in Maize. Plant Mol. Biol. Rep. 2012, 31, 457–463. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Long, Y.; Si, W.; Cheng, B.; Jiang, H. A novel heat shock transcription factor (ZmHsf08) negatively regulates salt and drought stress responses in Maize. Int. J. Mol. Sci. 2021, 22, 11922. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Briere, C.; Owens, G.L.; Carrere, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, Y.; Yang, X.; Zhang, B.; Xu, F.; Wang, Y.; Song, C.; Yi, M.; Ma, N.; Zhou, X.; et al. The heat stress transcription factor LlHsfA4 enhanced basic thermotolerance through regulating ROS metabolism in lilies (Lilium longiflorum). Int. J. Mol. Sci. 2022, 23, 572. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Procter, J.; Martin, D.A.; Barton, G.J. Jalview: Visualization and analysis of molecular sequences, alignments, and structures. BMC Bioinform. 2005, 6, P28. [Google Scholar] [CrossRef]
- Troshin, P.V.; Procter, J.B.; Sherstnev, A.; Barton, D.L.; Madeira, F.; Barton, G.J. JABAWS 2.2 distributed web services for bioinformatics: Protein disorder, conservation and RNA secondary structure. Bioinformatics 2018, 34, 1939–1940. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Best, D.J.; Roberts, D.E. Algorithm AS 89: The upper tail probabilities of Spearman’s Rho. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1975, 24, 377–379. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Number | Gene | Member | Chromosome Location | Gene Length(nt) | Protein Molecular Weight (MW, Da) | Protein Isoelectric Point (pI) | Predicted Subcellular Location |
|---|---|---|---|---|---|---|---|
| 1 | Eucgr.A01544 | EgHsf1 | Chr01 | 1737 | 27,565 | 4.75 | Nucleus |
| 2 | Eucgr.A01948 | EgHsf2 | Chr01 | 2791 | 49,802 | 5.19 | Nucleus |
| 3 | Eucgr.A02338 | EgHsf3 | Chr01 | 2793 | 29,215 | 8.25 | Nucleus |
| 4 | Eucgr.A02804 | EgHsf4 | Chr01 | 1349 | 35,566 | 4.78 | Nucleus |
| 5 | Eucgr.A02976 | EgHsf5 | Chr01 | 2179 | 47,615 | 5.67 | Nucleus |
| 6 | Eucgr.C00664 | EgHsf6 | Chr03 | 2559 | 43,514 | 4.94 | Nucleus |
| 7 | Eucgr.C00873 | EgHsf7 | Chr03 | 2170 | 52,813 | 4.61 | Nucleus |
| 8 | Eucgr.C01043 | EgHsf8 | Chr03 | 1389 | 35,714 | 5.31 | Nucleus |
| 9 | Eucgr.C03056 | EgHsf9 | Chr03 | 2482 | 41,266 | 4.56 | Nucleus |
| 10 | Eucgr.C03424 | EgHsf10 | Chr03 | 2550 | 41,407 | 5.78 | Nucleus |
| 11 | Eucgr.C03431 | EgHsf11 | Chr03 | 2333 | 41,444 | 5.99 | Nucleus |
| 12 | Eucgr.C03433 | EgHsf12 | Chr03 | 2205 | 45,858 | 6.11 | Nucleus |
| 13 | Eucgr.C03434 | EgHsf13 | Chr03 | 2780 | 54,933 | 5.21 | Nucleus |
| 14 | Eucgr.C03435 | EgHsf14 | Chr03 | 3056 | 51,533 | 5.92 | Nucleus |
| 15 | Eucgr.C03440 | EgHsf15 | Chr03 | 3191 | 57,928 | 6.06 | Nucleus |
| 16 | Eucgr.C03441 | EgHsf16 | Chr03 | 2534 | 55,594 | 5.76 | Nucleus |
| 17 | Eucgr.C03447 | EgHsf17 | Chr03 | 2508 | 54,914 | 5.24 | Nucleus |
| 18 | Eucgr.C03449 | EgHsf18 | Chr03 | 2838 | 55,702 | 5.05 | Nucleus |
| 19 | Eucgr.C03452 | EgHsf19 | Chr03 | 2243 | 44,186 | 5.84 | Nucleus |
| 20 | Eucgr.C03454 | EgHsf20 | Chr03 | 3203 | 57,199 | 5.91 | Nucleus |
| 21 | Eucgr.C03456 | EgHsf21 | Chr03 | 3183 | 57,348 | 6.07 | Nucleus |
| 22 | Eucgr.C03457 | EgHsf22 | Chr03 | 3004 | 40,621 | 5.72 | Nucleus |
| 23 | Eucgr.C03465 | EgHsf23 | Chr03 | 3107 | 58,095 | 5.98 | Nucleus |
| 24 | Eucgr.D00627 | EgHsf24 | Chr04 | 2286 | 29,766 | 6.33 | Nucleus |
| 25 | Eucgr.E00253 | EgHsf25 | Chr05 | 4442 | 56,475 | 4.72 | Nucleus |
| 26 | Eucgr.E00555 | EgHsf26 | Chr05 | 1834 | 33,858 | 9.41 | Nucleus |
| 27 | Eucgr.E02813 | EgHsf27 | Chr05 | 2604 | 47,555 | 5.09 | Nucleus |
| 28 | Eucgr.F00215 | EgHsf28 | Chr06 | 1668 | 39,371 | 7.11 | Nucleus |
| 29 | Eucgr.F02327 | EgHsf29 | Chr06 | 2075 | 40,710 | 8.44 | Nucleus |
| 30 | Eucgr.H02675 | EgHsf30 | Chr08 | 6363 | 54,320 | 5.93 | Nucleus |
| 31 | Eucgr.H03412 | EgHsf31 | Chr08 | 1784 | 42,895 | 5.35 | Nucleus |
| 32 | Eucgr.I00929 | EgHsf32 | Chr09 | 5275 | 46,303 | 6.68 | Nucleus |
| 33 | Eucgr.J00680 | EgHsf33 | Chr10 | 1523 | 34,909 | 7.11 | Nucleus |
| 34 | Eucgr.K00238 | EgHsf34 | Chr11 | 1658 | 31,565 | 9.33 | Nucleus |
| 35 | Eucgr.K03325 | EgHsf35 | Chr11 | 1740 | 39,837 | 5.33 | Nucleus |
| 36 | Eucgr.L01331 | EgHsf36 | scaffold_170 | 2553 | 59,378 | 6.31 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, T.; Liang, J.; Dai, J.; Zhou, X.-R.; Liao, W.; Guo, M.; Aslam, M.; Li, S.; Cao, G.; Cao, S. Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses. Int. J. Mol. Sci. 2022, 23, 8044. https://doi.org/10.3390/ijms23148044
Yuan T, Liang J, Dai J, Zhou X-R, Liao W, Guo M, Aslam M, Li S, Cao G, Cao S. Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses. International Journal of Molecular Sciences. 2022; 23(14):8044. https://doi.org/10.3390/ijms23148044
Chicago/Turabian StyleYuan, Tan, Jianxiang Liang, Jiahao Dai, Xue-Rong Zhou, Wenhai Liao, Mingliang Guo, Mohammad Aslam, Shubin Li, Guangqiu Cao, and Shijiang Cao. 2022. "Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses" International Journal of Molecular Sciences 23, no. 14: 8044. https://doi.org/10.3390/ijms23148044
APA StyleYuan, T., Liang, J., Dai, J., Zhou, X.-R., Liao, W., Guo, M., Aslam, M., Li, S., Cao, G., & Cao, S. (2022). Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses. International Journal of Molecular Sciences, 23(14), 8044. https://doi.org/10.3390/ijms23148044







