Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning
Abstract
1. Introduction
2. Results
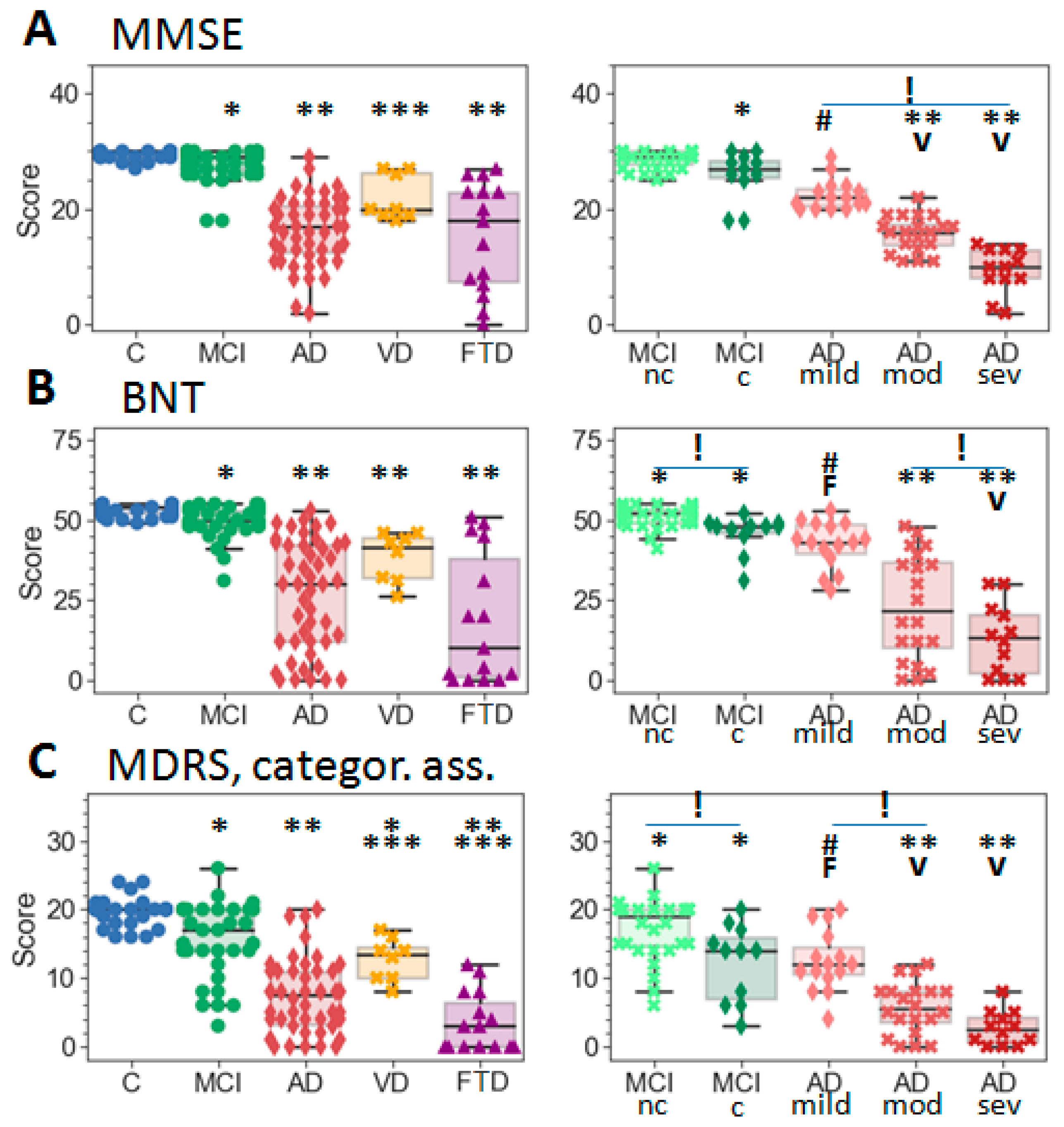
2.1. Subject Demographics
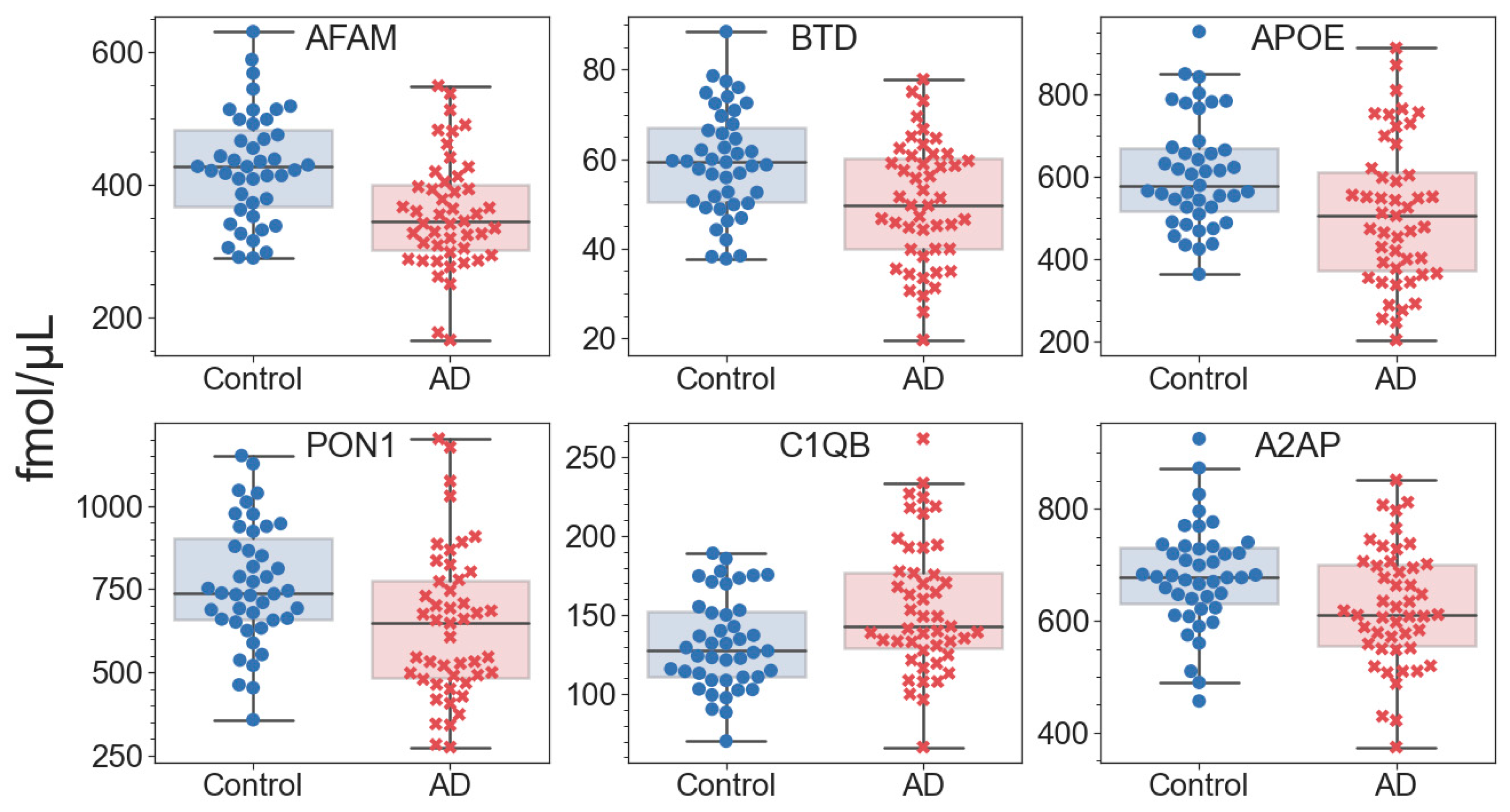
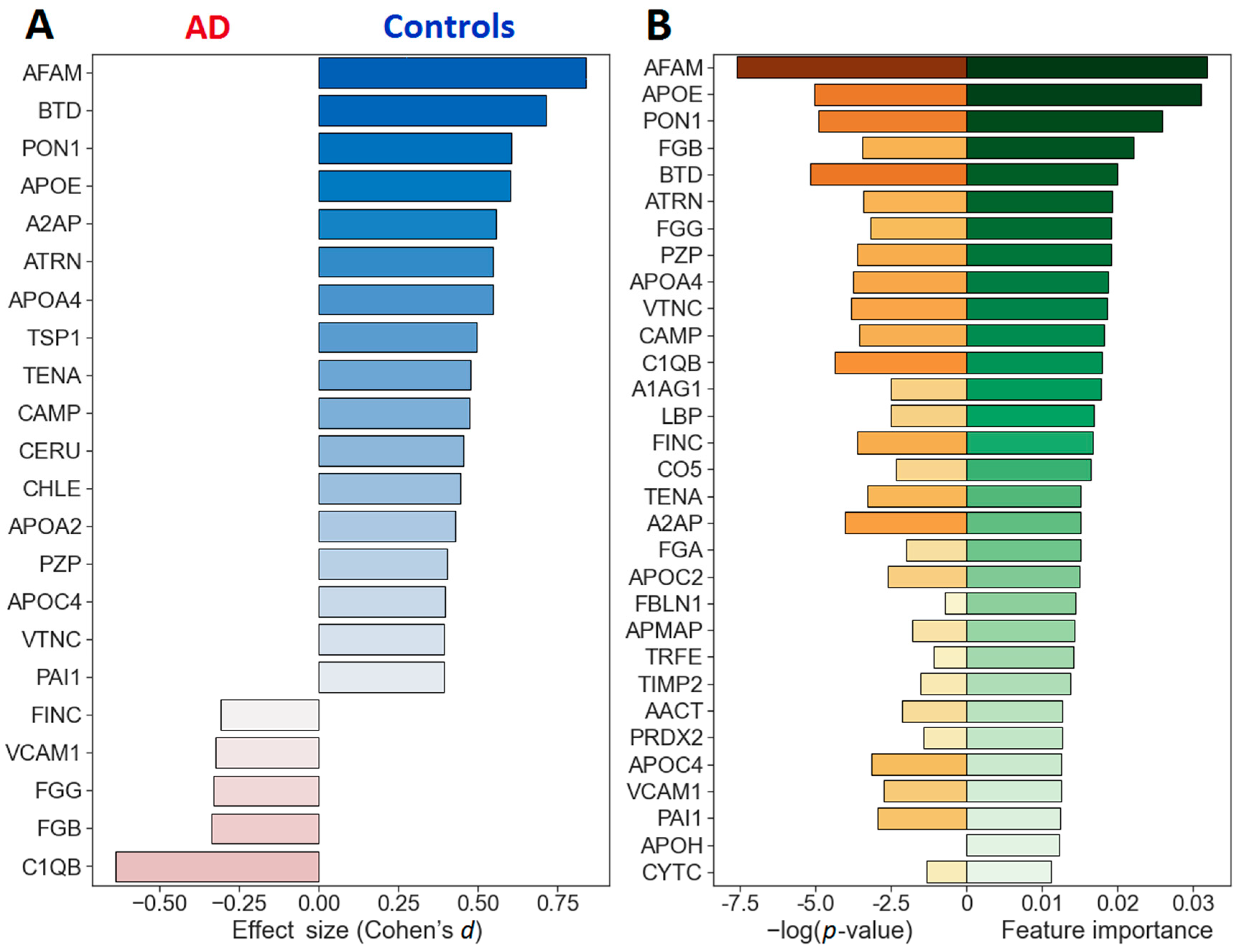
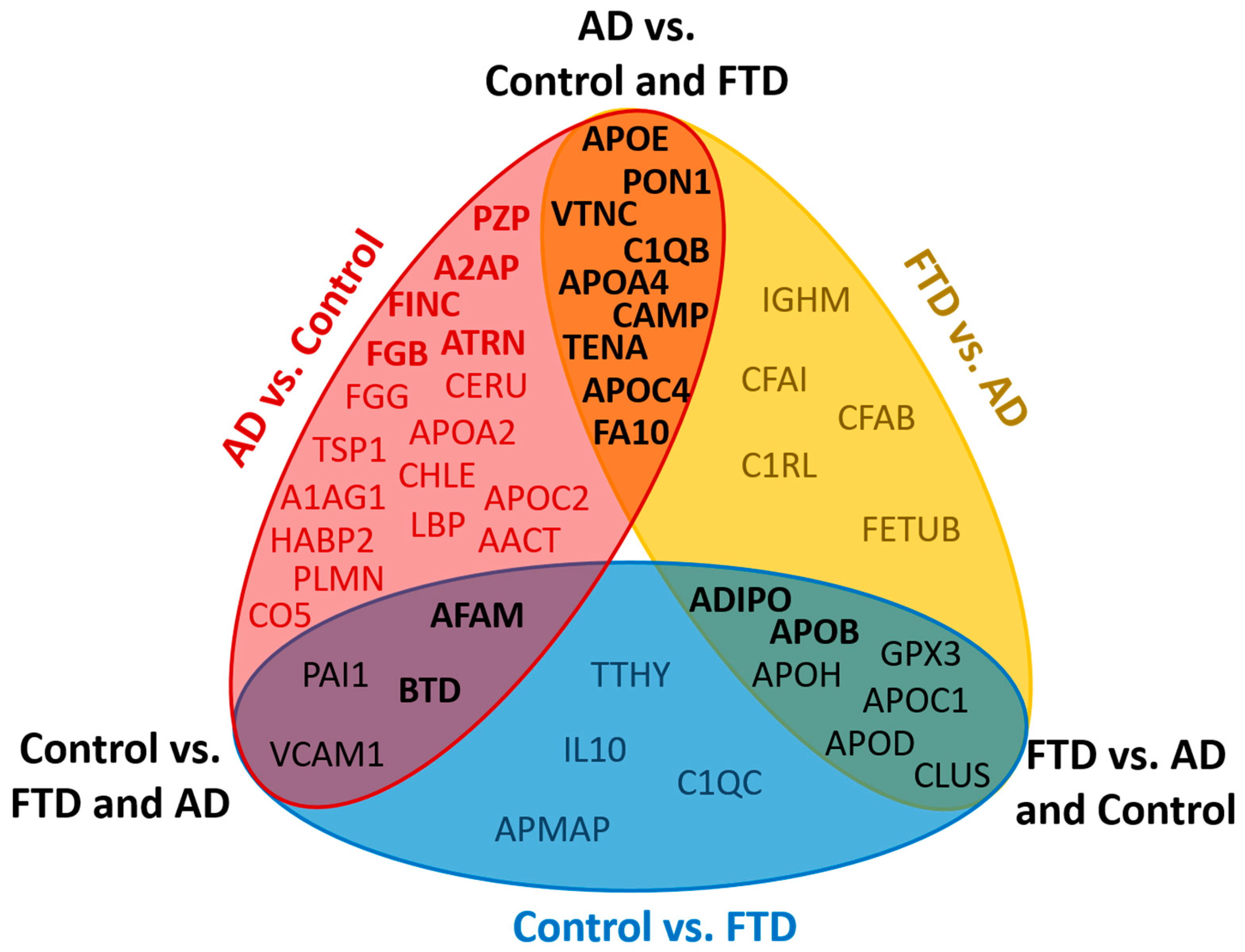
2.2. Quantitative Analysis of Blood Plasma Proteins
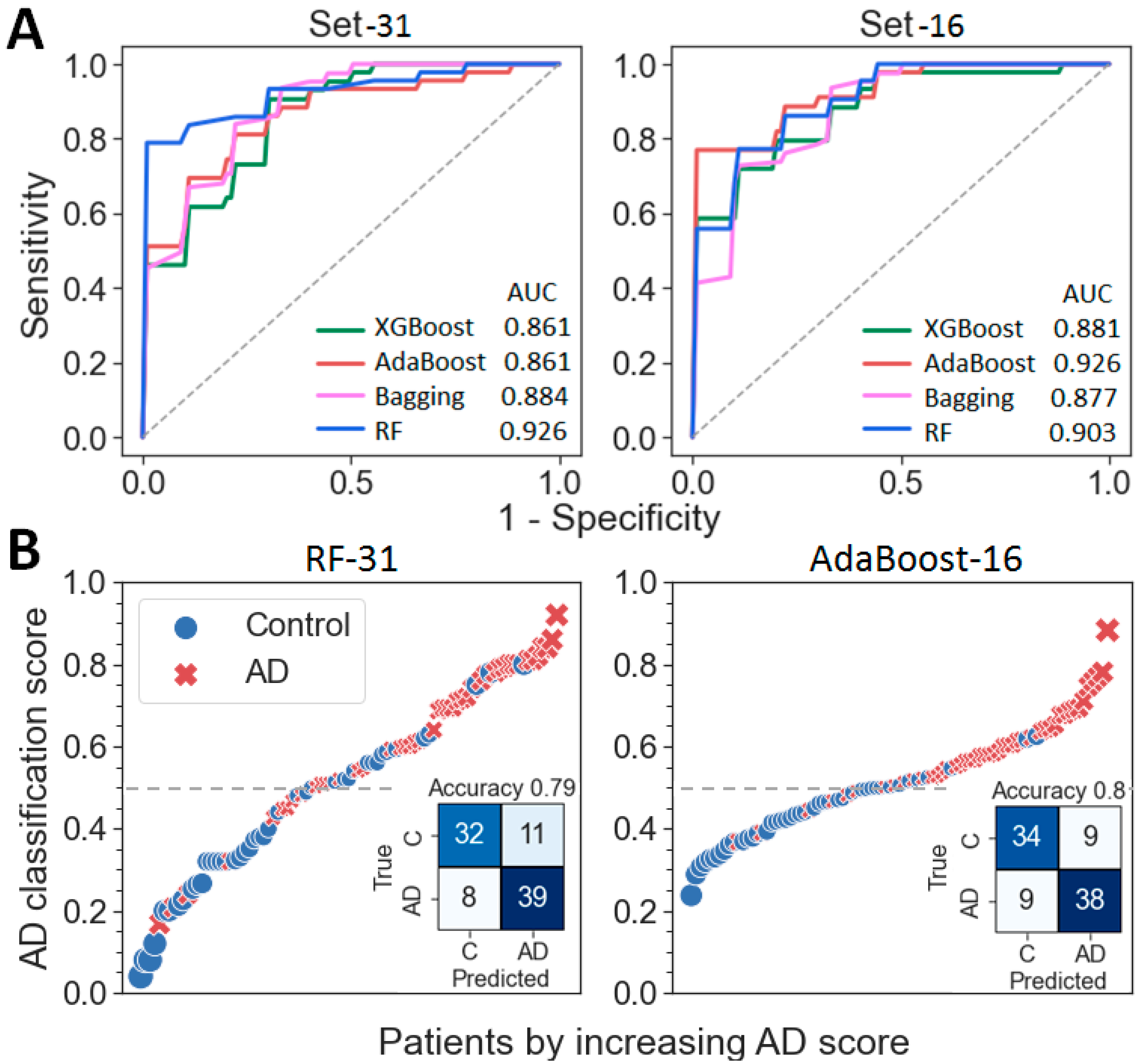
2.3. Building of a Binary Classifier (AD vs. Control)
| Protein Name | Abbr. | UniProt ID | Other Cohorts | p-Value | Effect Size | Relative Import. | |
|---|---|---|---|---|---|---|---|
| 1 | Afamin | AFAM | P43652 | 3 [26] | 1.7 × 10−4 | 0.840 | 0.0318 |
| 2 | Apolipoprotein E | APOE | P02649 | 2–5 [25,26] | 3.1 × 10−3 | 0.601 | 0.0310 |
| 3 | Serum paraoxonase/arylesterase 1 | PON1 | P27169 | 1 [25,26] | 3.6 × 10−3 | 0.605 | 0.0257 |
| 4 | Fibrinogen beta chain | FGB | P02675 | 1–2 [25,26] | 0.0192 | −0.339 | 0.0220 |
| 5 | Biotinidase | BTD | P43251 | 1 [45] | 2.7 × 10−3 | 0.714 | 0.0201 |
| 6 | Pregnancy zone protein | PZP | P20742 | 1 [26] | 0.0161 | 0.403 | 0.0192 |
| 7 | Attractin | ATRN | O75882 | 1 [25,26] | 0.0200 | 0.548 | 0.0191 |
| 8 | Fibrinogen gamma chain | FGG | P02679 | 3–4 [25,26] | 0.0263 | −0.331 | 0.0191 |
| 9 | Apolipoprotein A-IV | APOA4 | P06727 | 3 [26] | 0.0134 | 0.548 | 0.0187 |
| 10 | Vitronectin | VTNC | P04004 | 1–3 [25,26] | 0.0128 | 0.395 | 0.0185 |
| 11 | Cathelicidin antimicrobial peptide | CAMP | P49913 | - | 0.0176 | 0.473 | 0.0181 |
| 12 | Complement C1q subcomponent subunit B | C1QB | P02746 | - | 6.7 × 10−3 | −0.638 | 0.0180 |
| 13 | Alpha-1-acid glycoprotein 1 | A1AG1 | P02763 | 1 [25] | 0.0566 | −0.447 | 0.0178 |
| 14 | Lipopolysaccharide-binding protein | LBP | P18428 | - | 0.0577 | 0.332 | 0.0169 |
| 15 | Fibronectin | FINC | P02751 | 1–3 [25,26] | 0.0157 | −0.308 | 0.0167 |
| 16 | Complement C5 | CO5 | P01031 | 1 [25,26] | 0.0679 | −0.382 | 0.0165 |
| 17 | Tenascin | TENA | P24821 | 1–3 [25,26,37] | 0.0232 | 0.476 | 0.0152 |
| 18 | Alpha-2-antiplasmin | A2AP | P08697 | - | 0.0102 | 0.557 | 0.0151 |
| 19 | Fibrinogen alpha chain | FGA | P02671 | 2–3 [25,26] | 0.103 | −0.250 | 0.0150 |
| 20 | Apolipoprotein C-II | APOC2 | P02655 | 1 [45] | 0.0516 | 0.530 | 0.0149 |
| 21 | Fibulin-1 | FBLN1 | P23142 | - | 0.433 | 0.292 | 0.0144 |
| 22 | Adipocyte plasma membrane-associated protein | APMAP | Q9HDC0 | - | 0.127 | −0.351 | 0.0143 |
| 23 | Serotransferrin | TRFE | P02787 | 2 [25,26] | 0.286 | 0.243 | 0.0141 |
| 24 | Metalloproteinase inhibitor 2 | TIMP2 | P16035 | - | 0.175 | 0.386 | 0.0138 |
| 25 | Alpha-1-antichymotrypsin | AACT | P01011 | 1 [25] | 0.0883 | −0.475 | 0.0128 |
| 26 | Peroxiredoxin-2 | PRDX2 | P32119 | 1 [25] | 0.199 | −0.009 | 0.0126 |
| 27 | Apolipoprotein C-IV | APOC4 | P55056 | - | 0.0274 | 0.396 | 0.0125 |
| 28 | Vascular cell adhesion protein 1 | VCAM1 | P19320 | 3 [25,26,37] | 0.0443 | −0.323 | 0.0125 |
| 29 | Plasminogen activator inhibitor 1 | PAI1 | P05121 | 1 [37] | 0.0343 | 0.392 | 0.0123 |
| 30 | Beta-2-glycoprotein 1 | APOH | P02749 | 2–3 [25,26] | 1.0 | 0.007 | 0.0122 |
| 31 | Cystatin-C | CYTC | P01034 | 1 [25] | 0.217 | −0.420 | 0.0113 |
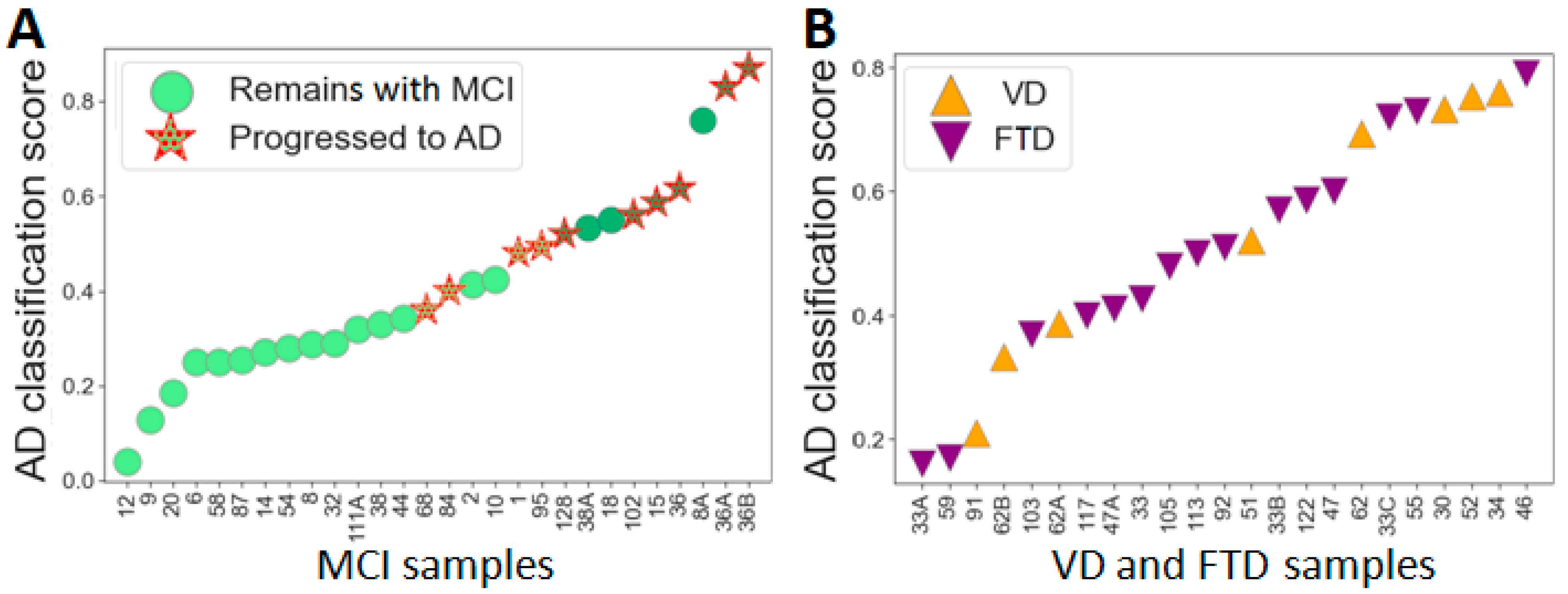
2.4. The Differentiation of MCI Subgroups with the Developed Classifiers
2.5. Proteomic Differences between AD, FTD and VD Samples
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Plasma Samples Collection and Preparation for MS
4.3. LC-MS/MS Analysis and MS Data Processing
4.4. Statistical Analysis
4.5. Machine Learning for Diagnosis Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimer’s Dement. 2013, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2016 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2016, 12, 459–509. [Google Scholar] [CrossRef] [PubMed]
- Perrin, J.; Fagan, A.M.; Holtzman, D.M. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature 2009, 461, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Mollenhauer, B.; Bibl, M.; Wiltfang, J.; Steinacker, P.; Ciesielczyk, B.; Trenkwalder, C.; Otto, M. Total tau protein, phosphorylated tau (181p) protein, β-amyloid1-42, and β-amyloid1-40 in cerebrospinal fluid of patients with dementia with Lewy bodies. Clin. Chem. Lab. Med. 2006, 44, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, C.; Smailagic, N.; Ladds, E.C.; Noel-Storr, A.H.; Ukoumunne, O.; Martin, S. CSF tau and the CSF tau/ABeta ratio for the diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst. Rev. 2013, 3, CD10803. [Google Scholar] [CrossRef]
- Rissman, R.A.; Trojanowski, J.O.; Shaw, L.M.; Aisen, P.S. Longitudinal plasma amyloid beta as a biomarker of Alzheimer’s disease. J. Neural. Transm. 2012, 119, 843–850. [Google Scholar] [CrossRef]
- Apostolova, L.G.; Hwang, K.S.; Andrawis, J.P.; Babakchanian, S.; Morra, J.H.; Cummings, J.L.; Toga, A.W.; Trojanowski, J.Q.; Shaw, L.M.; Jack, C.R.; et al. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol. Aging 2010, 31, 1284–1303. [Google Scholar] [CrossRef]
- Drzezga, A. Amyloid-plaque imaging in early and differential diagnosis of dementia. Ann. Nucl. Med. 2010, 24, 55–66. [Google Scholar] [CrossRef]
- Norberg, A.; Langstrom, B.; Scheinin, N.; Karrasch, M.; Grimmer, T.; Miederer, I.; Edison, P.; Okello, A.; Van Laere, K.; Nelissen, N.; et al. A European multicentre PET study of fibrillar amyloid in Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 104–114. [Google Scholar] [CrossRef]
- Dickerson, B.C.; Wolk, D.A. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology 2012, 78, 84–90. [Google Scholar] [CrossRef]
- Henriksen, K.; Bryant, S.E.; Hampel, H.; Trojanowski, J.Q.; Montine, T.J.; Jeromin, A.; Blennow, K.; Lonneborg, A.; Wyss-Coray, T.; Soares, H.; et al. The future of blood-based biomarkers for Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Grimmer, T.; Riemenschneider, M.; Fors, H.; Henriksen, G.; Klunk, W.E.; Mathis, C.A.; Shiga, T.; Wester, H.-J.; Kurz, A.; Drzezga, A. Beta amyloid in Alzheimer’s disease: Increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol. Psychiatry 2009, 65, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Galozzi, S.; Marcus, K.; Barkovits, K. Amyloid-β as a biomarker for Alzheimer’s disease: Quantification methods in body fluids. Expert Rev. Proteom. 2015, 12, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Lehallier, B.; Essioux, L.; Gayan, J.; Alexandridis, R.; Nikolchaeva, T.; Wyss-Coray, T.; Britschgi, M. Combined plasma and cerebrospinal fluid signature for the prediction of midterm progression from mild cognitive impairment to Alzheimer disease. JAMA Neurol. 2016, 73, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Janelidze, S.; Stomrud, E.; Palmqvist, S.; Zetterberg, H.; Westen, D.; Jeromin, A.; Song, L.; Hanlon, D.; Tan Hehir, C.A.; Baker, D.; et al. Plasma beta-amyloid in Alzheimer’s disease and vascular disease. Sci. Rep. 2016, 6, 26801. [Google Scholar] [CrossRef] [PubMed]
- Killiany, R.J.; Gomez-Isla, T.; Moss, M.; Kikins, R.; Sandor, T.; Tanzi, R.; Jones, K.; Hyman, B.T.; Albert, M.S. Use of structural magnetic resonance imaging to predict who will get Alzheimer’s disease. Ann. Neurol. 2000, 47, 430–439. [Google Scholar] [CrossRef]
- Jack, C.R.; Petersen, R.C.; Xu, Y.C.; O’Brien, P.C.; Smith, G.E.; Ivnik, R.J.; Boeve, B.F.; Warning, S.C.; Tangalos, E.G.; Kokmen, E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 1999, 52, 1397. [Google Scholar] [CrossRef]
- Zhang, S.; Han, D.; Tan, X.; Feng, J.; Guo, Y.; Ding, Y. Diagnostic accuracy of 18 F-FDG and 11 C-PIB–PET for prediction of short-term conversion to Alzheimer’s disease in subjects with mild cognitive impairment. Int. J. Clin. Pract. 2012, 66, 185–198. [Google Scholar] [CrossRef]
- Shaw, L.M. Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal fluid biomarker signature in Alzheimer’s Disease Neuroimaging Initiative subjects. Ann. Neurol. 2009, 65, 403–413. [Google Scholar] [CrossRef]
- Hampel, H.; O’Bryant, S.E.; Molinuevo, J.L.; Zetterberg, H.; Masters, C.; Lista, S.; Kiddle, S.J.; Batrla, R.; Blennow, K. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat. Rev. Neurol. 2018, 14, 639–652. [Google Scholar] [CrossRef]
- Ovod, V.; Ramsey, K.N.; Mawuenyega, K.G.; Bollinger, J.G.; Hicks, T.; Schneider, T.; Sullivan, M.; Paumier, K.; Holtzman, D.M.; Morris, J.C.; et al. Amyloid β concentrations and stable isotope labeling kinetics of human plasma specific to central nervous system amyloidosis. Alzheimer’s Dement. 2017, 13, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kaneko, N.; Villemagne, V.L.; Kato, T.; Doecke, J.; Dore, V.; Fowler, C.; Li, Q.X.; Martins, R.; Rowe, C.; et al. High performance plasma amyloid-beta biomarkers for Alzheimer’s disease. Nature 2018, 554, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Zetterberg, H.; Blennow, K. Moving fluid biomarkers for Alzheimer’s disease from research tools to routine clinical diagnostics. Mol. Neurodegener. 2021, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, N.R.; Horie, K.; Sato, C.; Bateman, R.J. Blood plasma phosphorylated-tau isoforms track CNS change in Alzheimer’s disease. J. Exp. Med. 2020, 217, e20200861. [Google Scholar] [CrossRef] [PubMed]
- Kiddle, S.J.; Sattlrcker, M.; Proitsi, P.; Simmons, A.; Westman, E.; Bazenet, C.; Nelson, S.K.; Williams, S.; Hodges, A.; Johnston, C.; et al. Candidate blood proteome markers of Alzheimer’s disease onset and progression: A systematic review and replication study. J. Alzheimer’s Dis. 2014, 38, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Rehiman, S.H.; Lim, S.M.; Neoh, C.F.; Majeed, A.B.A.; Chin, A.-V.; Tan, M.P.; Kamaruzzaman, S.B.; Ramasamy, K. Proteomics as a reliable approach for discovery of blood-based Alzheimer’s disease biomarkers: A systematic review and meta-analysis. Ageing Res. Rev. 2020, 60, 101066. [Google Scholar] [CrossRef]
- Morgan, A.R.; Touchard, S.; Leckey, C.; O’Hagan, C.; Nevado-Holgado, A.J.; Consortium, N.; Barkhof, F.; Bertram, L.; Blin, O.; Bos, I.; et al. Inflammatory biomarkers in Alzheimer’s disease plasma. Alzheimer’s Dement. 2019, 15, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Doecke, J.D.; Laws, S.M.; Faux, N.G.; Wilson, W.; Burnham, S.C.; Lam, C.P.; Mondal, A.; Bedo, J.; Bush, A.I.; Brown, B.; et al. Alzheimer’s Disease Neuroimaging Initiative; Australian Imaging Biomarker and Lifestyle Research Group. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch. Neurol. 2012, 69, 1318–1325. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Xiao, G.; Barber, R.; Reisch, J.; Doody, R.; Fairchild, T.; Adams, P.; Waring, S.; Diaz-Arrastia, R.; Texas Alzheimer’s Research Consortium. A serum protein-based algorithm for the detection of Alzheimer disease. Arch. Neurol. 2010, 67, 1077–1081. [Google Scholar] [CrossRef]
- Ray, S.; Britschgi, M.; Herbert, C.; Takeda-Uchimura, Y.; Boxer, A.; Blennow, K.; Friedman, L.F.; Galasko, D.R.; Jutel, M.; Karydas, A.; et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat. Med. 2007, 13, 1359–1362. [Google Scholar] [CrossRef]
- Hye, A.; Riddoch-Contreras, J.; Baird, A.L.; Ashton, N.J.; Bazenet, C.; Leung, R.; Westman, E.; Simmons, A.; Dobson, R.; Sattlecker, M.; et al. Plasma proteins predict conversion to dementia from prodromal disease. Alzheimer’s Demen. 2014, 10, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liu, Y.P.; Liu, H.L.; Li, J.; Xiang, Y.; Liu, Y.H.; Sheng, S.; Liu, L.; Wang, Y.; Fu, W. Serum protein-based profiles as novel biomarkers for the diagnosis of Alzheimer’s disease. Mol. Neurobiol. 2018, 55, 3999–4008. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Buckley, N.J.; Bos, I.; Engelborghs, S.; Sleegers, K.; Frisoni, G.B.; Wallin, A.; Lléo, A.; Popp, J.; Martinez-Lage, P.; et al. Plasma Proteomic Biomarkers Relating to Alzheimer’s Disease: A Meta-Analysis Based on Our Own Studies. Front. Aging Neurosci. 2021, 13, 712545. [Google Scholar] [CrossRef] [PubMed]
- Henkel, A.W.; Muller, K.; Lewczuk, P.; Muller, T.; Marcus, K.; Kornhuber, J.; Wiltfang, J. Multidimensional plasma protein separation technique for identification of potential Alzheimer’s disease plasma biomarkers: A pilot study. J. Neural. Transm. 2012, 119, 779–788. [Google Scholar] [CrossRef]
- Walker, K.A.; Chen, J.; Zhang, J.; Fornage, M.; Yang, Y.; Zhou, L.; Grams, M.E.; Tin, A.; Daya, N.; Hoogeveen, R.C.; et al. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat. Aging 2021, 1, 473–489. [Google Scholar] [CrossRef]
- Whelan, C.D.; Mattsson, N.; Nagle, M.W.; Vijayaraghavan, S.; Hyde, C.; Janelidze, S.; Stomrud, E.; Lee, J.; Fitz, L.; Samad, T.A.; et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. Acta Neuropathol. Commun. 2019, 7, 169. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhou, X.; Ip, F.C.; Chan, P.; Chen, Y.; Lai, N.C.; Cheung, K.; Lo, R.M.N.; Tong, E.P.S.; Wong, B.W.Y.; et al. Large-scale proteomic profiling identifies a high-performance biomarker panel for Alzheimer’s disease screening and staging. Alzheimer’s Demen. 2022, 18, 88–102. [Google Scholar] [CrossRef]
- Song, F.; Poljak, A.; Kochan, N.A.; Raftery, M.; Brodaty, H.; Smythe, G.A.; Schdev, P.S. Plasma protein profiling of mild cognitive impairment and Alzheimer’s disease using iTRAQ quantitative proteomics. Proteome Sci. 2014, 12, 5. [Google Scholar] [CrossRef]
- Muenchhoff, J.; Poljak, A.; Song, F.; Raftery, M.; Brodaty, H.; Duncan, M.; McEvoy, M.; Attia, J.; Schofield, P.W.; Sachdev, P.S. Plasma protein profiling of mild cognitive impairment and Alzheimer’s disease across two independent cohorts. J. Alzheimer’s Dis. 2015, 43, 1355–1373. [Google Scholar] [CrossRef]
- Dayon, L.; Wojcik, J.; Galindo, N.; Corthesy, J.; Cominetti, O.; Oikonomidi, A.; Henry, H.; Migliavacca, E.; Bowman, G.L.; Popp, J. Plasma proteomic profiles of cerebrospinal fluid-defined Alzheimer’s disease pathology in older adults. J. Alzheimer’s Dis. 2017, 60, 1641–1652. [Google Scholar] [CrossRef]
- Dey, K.K.; Wang, H.; Niu, M.; Bai, B.; Wang, X.; Li, Y.; Cho, J.-H.; Tan, H.; Mishra, A.; High, A.A.; et al. Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer’s disease. Clin. Proteom. 2019, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Park, J.C.; Han, S.H.; Lee, H.; Jeong, H.; Byun, M.S.; Bae, J.; Kim, H.; Lee, D.Y.; Yi, D.; Shin, S.A.; et al. Prognostic plasma protein panel for Aβ deposition in the brain in Alzheimer’s disease. Prog. Neurobiol. 2019, 183, 101690. [Google Scholar] [CrossRef] [PubMed]
- Ashton, N.J.; Nevado-Holgado, A.J.; Barber, I.S.; Lynham, S.; Gupta, V.; Chatterjee, P.; Goozee, K.; Hone, E.; Pedrini, S.; Blennow, K.; et al. A plasma protein classifier for predicting amyloid burden for preclinical Alzheimer’s disease. Sci. Adv. 2019, 5, eaau7220. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, A.; Ashton, N.J.; Chatterjee, P.; Goozee, K.; Shen, K.; Fripp, J.; Ames, D.; Rowe, C.; Masters, C.; Villemagne, V.; et al. Plasma transferrin and hemopexin are associated with altered Aβ uptake and cognitive decline in Alzheimer’s disease pathology. Alzheimer’s Res. Ther. 2020, 12, 72. [Google Scholar] [CrossRef]
- Chen, M.; Xia, W. Proteomic profiling of plasma and brain tissue from Alzheimer’s disease patients reveals candidate network of plasma biomarkers. J. Alzheimer’s Dis. 2020, 76, 349–368. [Google Scholar] [CrossRef]
- Khan, M.J.; Desaire, H.; Lopez, O.L.; Kamboh, M.I.; Robinson, R.A.S. Why Inclusion Matters for Alzheimer’s Disease Biomarker Discovery in Plasma. J. Alzheimer’s Dis. 2021, 79, 1327–1344. [Google Scholar] [CrossRef]
- Kitamura, Y.; Usami, R.; Ichichara, S.; Kida, H.; Satoh, M.; Tomimoto, H.; Murata, M.; Oikawa, S. Plasma protein profiling for potential biomarkers in the early diagnosis of Alzheimer’s disease. Neurol. Res. 2017, 39, 231–238. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Verma, A.; Mishra, V.N. Proteomics based identification of differential plasma proteins and changes in white matter integrity as markers in early detection of mild cognitive impaired subjects at high risk of Alzheimer’s disease. Neurosci. Lett. 2018, 676, 71–77. [Google Scholar] [CrossRef]
- Soares, H.D.; Potter, W.Z.; Pickering, E.; Kuhn, M.; Immermann, F.W.; Shera, D.M.; Ferm, M.; Dean, R.A.; Simon, A.J.; Swenson, F.; et al. Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics Project. Plasma biomarkers associated with the apolipoprotein E genotype and Alzheimer. Arch. Neurol. 2012, 69, 1310–1317. [Google Scholar] [CrossRef]
- Thambisetty, M.; Tripaldi, R.; Riddoch-Contreras, J.; Hye, A.; An, Y.; Campbell, J.; Sojkova, J.; Kinsey, A.; Lynham, S.; Zhou, Y.; et al. Proteome-based plasma markers of brain amyloid-β deposition in non-demented older individuals. J. Alzheimer’s Dis. 2010, 22, 1099–1109. [Google Scholar] [CrossRef]
- Zhao, X.; Lejnine, S.; Spond, J.; Zhang, C.; Ramaraj, T.C.; Holder, D.J.; Dai, H.; Weiner, R.; Lacterza, O.F. A candidate plasma protein classifier to identify Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 43, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.R.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H.; et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotech. 2009, 27, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Gaither, C.; Popp, R.; Mohammed, Y.; Bochers, C.H. Determination of the concentration range for 267 proteins from 21 lots of commercial human plasma using highly multiplexed multiple reaction monitoring mass spectrometry. Analyst 2020, 145, 3634–3644. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.M.; Geyer, P.E.; Müller, J.B.; Strauss, M.T.; Koch, M.; Leypoldt, F.; Koertvelyessy, P.; Bittner, D.; Schipke, C.G.; Incesoy, E.I.; et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol. Syst. Biol. 2020, 16, e9356. [Google Scholar] [CrossRef]
- Xie, C.; Zhuang, X.X.; Niu, Z.; Ai, R.; Lautrup, S.; Zheng, S.; Jiang, Y.; Han, R.; Gupta, T.S.; Cao, S.; et al. Amelioration of Alzheimer’s disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat. Biomed. Eng. 2022, 6, 76–93. [Google Scholar] [CrossRef]
- Gaither, C.; Popp, R.; Bochers, S.P.; Skarphedinsson, K.; Eiriksson, F.F.; Thorsteinsdottir, M.; Mohammed, Y.; Borchers, C.H. Performance assessment of a 125 human plasma peptide mixture stored at room temperature for multiple reaction monitoring-mass spectrometry. J. Proteome Res. 2021, 20, 4292–4302. [Google Scholar] [CrossRef]
- Trollor, J.N.; Smith, E.; Baune, B.T.; Kochan, N.A.; Campbell, L.; Samaras, K.; Crawford, J.; Brodaty, H.; Sachdev, P. Systemic inflammation is associated with MCI and its subtypes: The Sydney Memory and Aging Study. Dement. Geriatr. Cogn. Disord. 2010, 30, 569–578. [Google Scholar] [CrossRef]
- Shen, X.N.; Lu, Y.; Tan, C.T.Y.; Liu, L.Y.; Yu, J.T.; Feng, L.; Larbi, A. Identification of inflammatory and vascular markers associated with mild cognitive impairment. Aging 2019, 11, 2403–2419. [Google Scholar] [CrossRef]
- Yi, L.; Wu, T.; Luo, W.; Zhou, W.; Wu, J. A non-invasive, rapid method to genotype late-onset Alzheimer’s disease-related apolipoprotein E gene polymorphisms. Neural Regen. Res. 2014, 9, 69–75. [Google Scholar] [CrossRef]
- Folstein, M.E. A practical method for grading the cognitive state of patients for the children. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Rouleau, I.; Salmon, D.P.; Butters, N.; Kennedy, C.; McGuire, K. Quantitative and qualitative analyses of clock drawings in Alzheimer’s and Huntington’s disease. Brain Cogn. 1992, 18, 70–87. [Google Scholar] [CrossRef]
- Kaplan, E.; Goodglass, H.; Weintraub, S. Boston Naming Test; Pro-Ed: Austin, TX, USA, 2001. [Google Scholar]
- Luria, A.R. Neuropsychological studies in the USSR. A review (part II). Proc. Natl Acad. Sci. USA 1973, 70, 1278. [Google Scholar] [CrossRef] [PubMed]
- Altepeter, T.S.; Adams, R.L.; Buchanan, W.L.; Buck, P. Luria Memory Words Test and Wechsler Memory Scale: Comparison of utility in discriminating neurologically impaired from controls. J. Clin. Psychol. 1990, 46, 190–193. [Google Scholar] [CrossRef]
- Mattis, S. Dementia Rating Scale: Professional Manual; Psychological Assessment Resources: Odessa, FL, USA, 1988. [Google Scholar]
- Morris, J.C. The clinical dementia rating (CDR): Current version and scoring rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef]
- World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders: Clinical Descriptions and Diagnostic Guidelines; World Health Organization: Geneva, Switzerland, 1992.
- Pijnenburg, Y.A.L. New diagnostic criteria for the behavioural variant of frontotemporal dementia. Eur. Neurol. Rev. 2011, 6, 234–237. [Google Scholar] [CrossRef]
- Peterson, R.S.; Touchon, J. Consensus in mild cognitive impairment. Research and practice in Alzheimers disease. EADS ADCS Jt. Meet 2005, 10, 24–32. [Google Scholar]
- Reisberg, B.; Ferris, S.H.; de Leon, M.J.; Crook, T. The Global Deterioration Scale for assessment of primary degenerative dementia. Am. J. Psychiatry 1982, 139, 1136–1139. [Google Scholar] [CrossRef]
- Percy, A.J.; Borchers, C.H. Detailed Method for Performing the ExSTA Approach in Quantitative Bottom-Up Plasma Proteomics. Methods Mol. Biol. 2021, 2228, 353–384. [Google Scholar] [CrossRef]
- Mohammed, Y.; Pan, J.; Zhang, S.; Han, J.; Borchers, C.H. ExSTA: External standard addition method for accurate high-throughput quantitation in targeted proteomics experiments. Proteom. Clin. Appl. 2018, 12, 1600180. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- MacLean, B.X.; Pratt, B.S.; Egertson, J.D.; MacCoss, M.J.; Smith, R.D.; Baker, E.S. Using skyline to analyze data-containing liquid chromatography, ion mobility spectrometry, and mass spectrometry dimensions. J. Am. Soc. Mass Spectrom. 2018, 29, 2182–2188. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 445, pp. 51–56. [Google Scholar]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Machine Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
| Control | MCI (nc/c) | AD (ps/s) | VD | FTD | |
|---|---|---|---|---|---|
| N | 39 | 32 (23/9) | 37 (13/24) | 6 | 11 |
| Age (years) | 67.6 ± 8.0 | 70.7 ± 7.4/76.1 ± 7.9 | 66.4 ± 4.6/78.4 ± 5.8 | 73.6 ± 7.8 | 64.4 ± 11.4 |
| Sex (%, F) | 69.2 | 70.6 (78.3/44.4) | 54.1 (57.1/52.2) | 66.7 | 63.6 |
| APOE (%, e4+) | 10.0 | 12.5/33.3 | 46.2/41.7 | 50.0 | 18.2 |
| e2/e3 | 17.5 | 12.5/11.1 | 0/8.3 | 16.7 | 45.5 |
| e3/e3 | 72.5 | 75/55.6 | 53.8/50.0 | 33.3 | 36.4 |
| e2/e4 | 2.5 | 0/0 | 0/4.2 | 0 | 0 |
| e3/e4 | 7.5 | 12.5/33.3 | 15.4/20.8 | 50.0 | 18.2 |
| e4/e4 | 0 | 0/0 | 30.8/16.7 | 0 | 0 |
| MMSE | 29.5 ± 0.7 | 28.7 ± 1.5/25.9 ± 4 | 14.7 ± 6.6/17.6 ± 4.8 | 22.0 ± 3.7 | 16.9 ± 8.6 |
| CDT | 9.9 ± 0.29 | 9.5 ± 1.1/8.6 ± 1.6 | 4.7 ± 2.6/5.5 ± 2.7 | 8.3 ± 1.7 | 5.7 ± 3.9 |
| BNT | 53.3 ± 1.64 | 51 ± 3.4/45.7 ± 5.7 | 23.6 ± 16/26.5 ± 17.2 | 38.5 ± 7.2 | 18.7 ± 19.7 |
| LMWT | |||||
| NM | 7.98 ± 1.14 | 7.7 ± 1.2/5.7 ± 1.9 | 2.3 ± 1.9/3.4 ± 2.2 | 5.3 ± 1.7 | 3.1 ± 2.8 |
| DM | 6.81 ± 1.71 | 6.4 ± 1.9/4.5 ± 2.7 | 0.26 ± 0.6/1.6 ± 2 | 3.1 ± 1.6 | 1.9 ± 3.0 |
| MDRS | |||||
| Sound associations | 17.9 ± 3.4 | 15.4 ± 4/12.5 ± 5.6 | 6.1 ± 4.7/6.5 ± 4.6 | 12.3 ± 3.0 | 3.3 ± 4.1 |
| Categorial associations | 19.8 ± 1.7 | 17.1 ± 4.4/12.3 ± 5.3 | 7.6 ± 5.4/7.0 ± 5.2 | 12.8 ± 2.9 | 3.6 ± 4.1 |
| Cardiovascular diseases (%) | 65.8 | 70.8/55.6 | 71.4/87.0 | 100.0 | 63.6 |
| Diabetes mellitus (%) | 0.0 | 12.5/11.1 | 7.1/13.0 | 33.3 | 9.1 |
| Gastrointestinal pathologies (%) | 21.1 | 16.7/44.4 | 35.7/26.1 | 66.7 | 36.4 |
| Genitourinary pathologies (%) | 15.8 | 25.0/55.5 | 13.3/39.1 | 50.0 | 9.1 |
| Nicergoline usage (%) | 0 | 13.8/18.2 | 10.5/7.1 | 0 | 26.7 |
| Choline alfoscerate usage (%) | 0 | 65.5/63.6 | 15.8/28.6 | 0 | 0 |
| Donepezil usage (%) | 0 | 0/9.1 | 26.3/28.6 | 25 | 26.7 |
| Memantine usage (%) | 0 | 3.4/0 | 73.7/67.9 | 50 | 73.3 |
| Rivastigmine usage (%) | 0 | 0/0 | 26.3/21.4 | 0 | 0 |
| Quetiapine usage (%) | 0 | 0/0 | 10.5/17.9 | 0 | 33.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kononikhin, A.S.; Zakharova, N.V.; Semenov, S.D.; Bugrova, A.E.; Brzhozovskiy, A.G.; Indeykina, M.I.; Fedorova, Y.B.; Kolykhalov, I.V.; Strelnikova, P.A.; Ikonnikova, A.Y.; et al. Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning. Int. J. Mol. Sci. 2022, 23, 7907. https://doi.org/10.3390/ijms23147907
Kononikhin AS, Zakharova NV, Semenov SD, Bugrova AE, Brzhozovskiy AG, Indeykina MI, Fedorova YB, Kolykhalov IV, Strelnikova PA, Ikonnikova AY, et al. Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning. International Journal of Molecular Sciences. 2022; 23(14):7907. https://doi.org/10.3390/ijms23147907
Chicago/Turabian StyleKononikhin, Alexey S., Natalia V. Zakharova, Savva D. Semenov, Anna E. Bugrova, Alexander G. Brzhozovskiy, Maria I. Indeykina, Yana B. Fedorova, Igor V. Kolykhalov, Polina A. Strelnikova, Anna Yu. Ikonnikova, and et al. 2022. "Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning" International Journal of Molecular Sciences 23, no. 14: 7907. https://doi.org/10.3390/ijms23147907
APA StyleKononikhin, A. S., Zakharova, N. V., Semenov, S. D., Bugrova, A. E., Brzhozovskiy, A. G., Indeykina, M. I., Fedorova, Y. B., Kolykhalov, I. V., Strelnikova, P. A., Ikonnikova, A. Y., Gryadunov, D. A., Gavrilova, S. I., & Nikolaev, E. N. (2022). Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning. International Journal of Molecular Sciences, 23(14), 7907. https://doi.org/10.3390/ijms23147907









