Profile of TREM2-Derived circRNA and mRNA Variants in the Entorhinal Cortex of Alzheimer’s Disease Patients
Abstract
:1. Introduction
2. Results
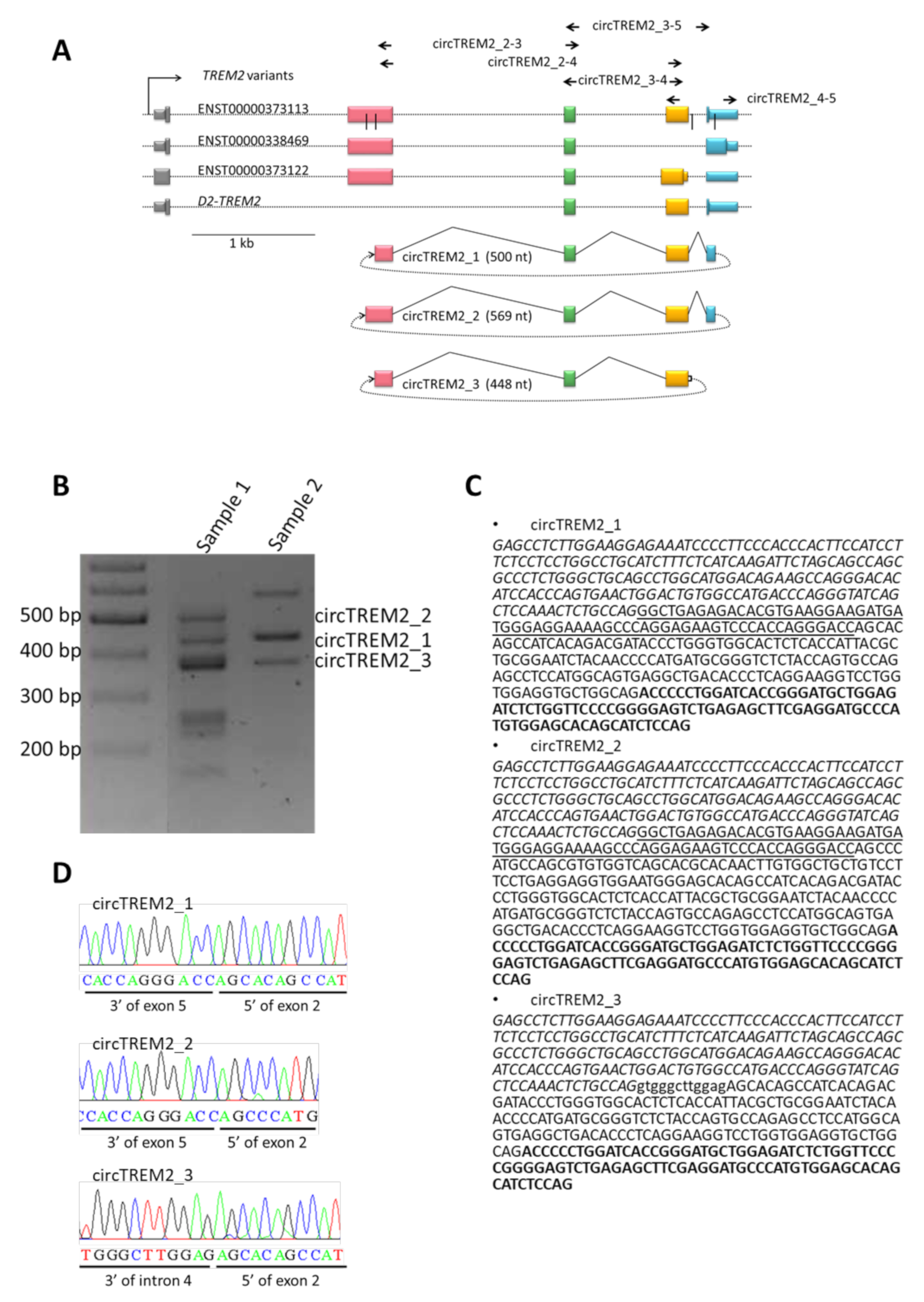
2.1. circTREM2 Identification in Entorhinal Human Cortex
2.2. circTREM2 in Human Microglia Cells
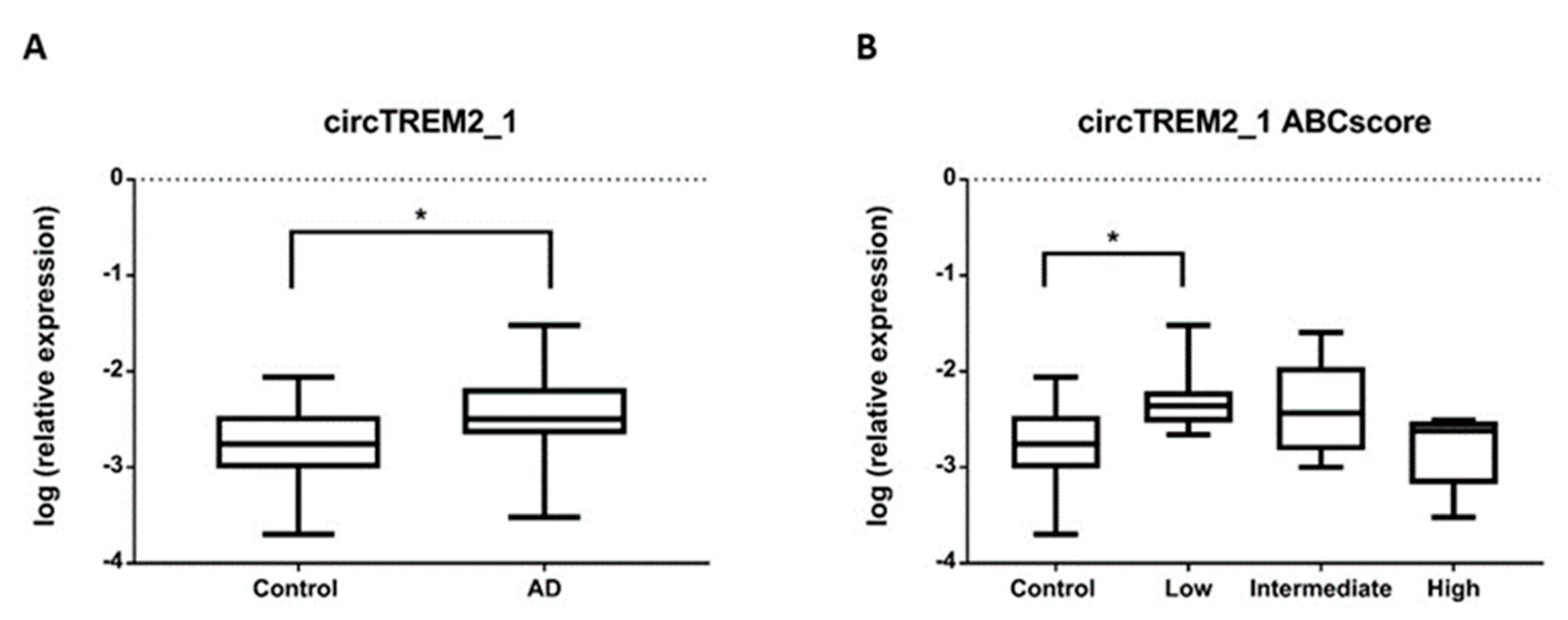
2.3. circTREM2 Differential Expression in AD Entorhinal Cortex
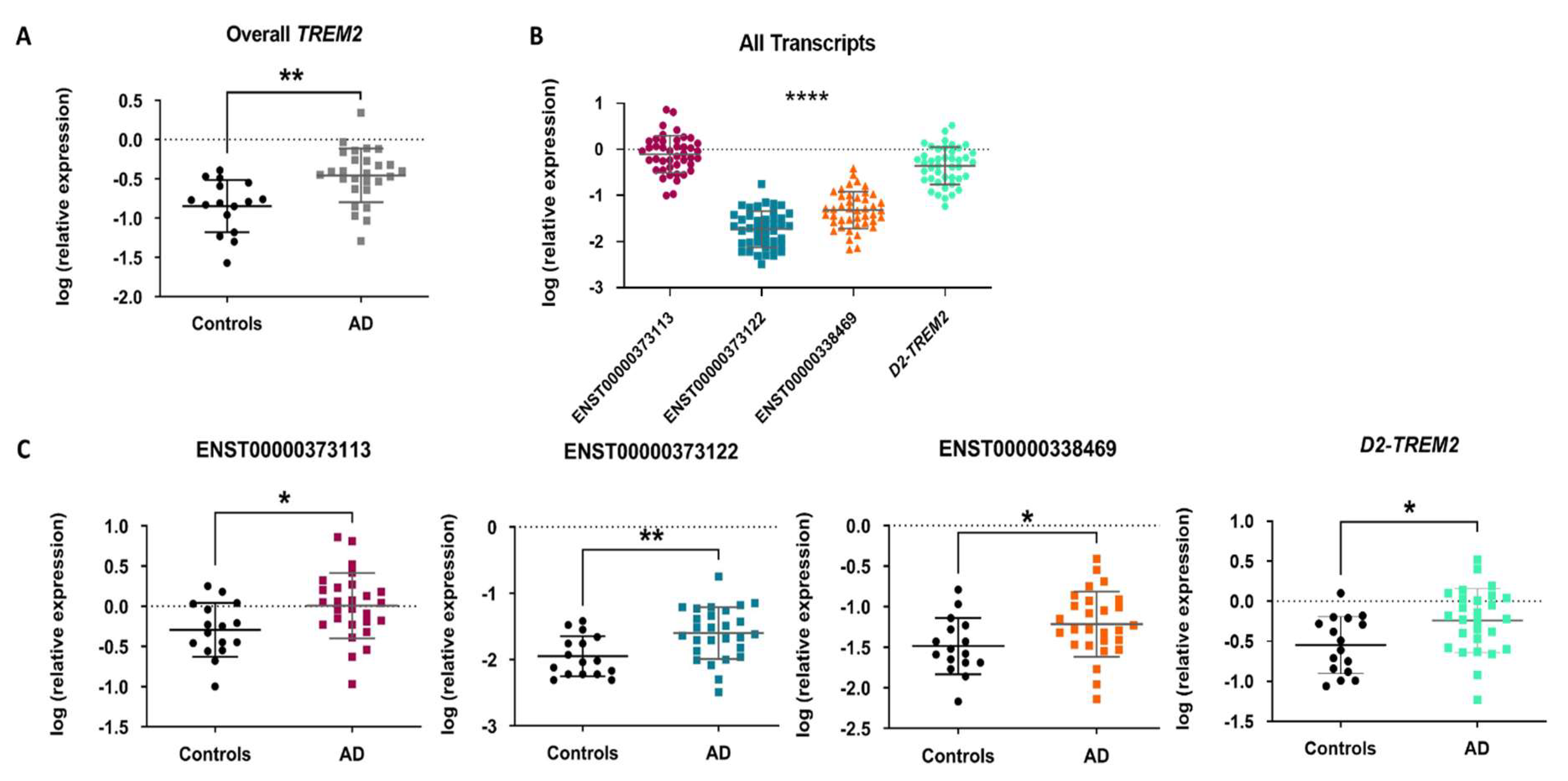
2.4. TREM2 mRNA Expression Profiling and Differential Expression in AD Entorhinal Cortex
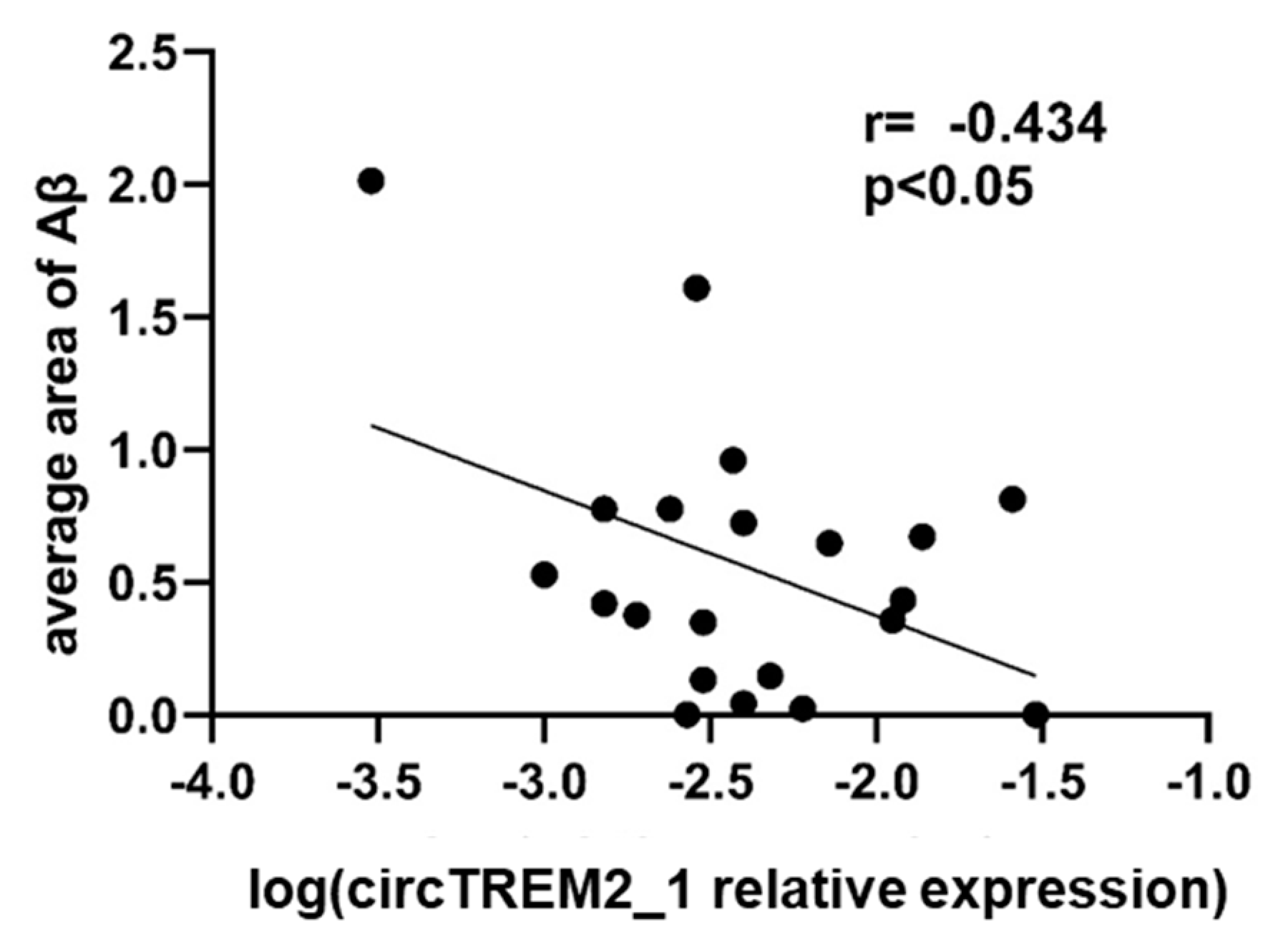
2.5. circTREM2, Overall TREM2, TREM2 Transcript Variants and Aβ Deposits
2.6. Prediction of miRNAs Binding Sites within circTREM2s
3. Discussion
4. Materials and Methods
4.1. Human Entorhinal Cortex Samples
4.2. Microglial Cell Culture
4.3. RNA Isolation and Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
4.4. Candidate Band Isolation and Sanger Sequencing
4.5. Real-Time Quantitative PCR (RT-qPCR) Assay
4.6. Quantitative Assessment of Aβ Deposits in Brain Tissues
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Vicens, Q.; Westhof, E. Biogenesis of Circular RNAs. Cell 2014, 159, 13–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. J. Genom. 2017, 2017, 6218353. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2014, 10, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [Green Version]
- Wilusz, J.E. Repetitive elements regulate circular RNA biogenesis. Mob. Genet. Elem. 2015, 5, 39–45. [Google Scholar] [CrossRef] [Green Version]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [Green Version]
- Hanan, M.; Soreq, H.; Kadener, S. circRNAs in the brain. RNA Biol. 2016, 14, 1028–1034. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Schuman, E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016, 39, 597–604. [Google Scholar] [CrossRef]
- Lipscombe, D.; Soto, E.J.L. Alternative splicing of neuronal genes: New mechanisms and new therapies. Curr. Opin. Neurobiol. 2019, 57, 26–31. [Google Scholar] [CrossRef]
- Mehta, S.L.; Dempsey, R.J.; Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020, 186, 101746. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Muñoz-Culla, M.; Prada-Luengo, I.; Castillo-Triviño, T.; Olascoaga, J.; Otaegui, D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017, 26, 3564–3572. [Google Scholar] [CrossRef]
- Lee, E.-G.; Tulloch, J.; Chen, S.; Leong, L.; Saxton, A.D.; Kraemer, B.; Darvas, M.; Keene, C.D.; Shutes-David, A.; Todd, K.; et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer’s disease. PLoS ONE 2020, 15, e0227667. [Google Scholar] [CrossRef]
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019, 22, 1903–1912. [Google Scholar] [CrossRef]
- Mayeux, R.; Stern, Y. Epidemiology of Alzheimer disease. Cold Spring Harb Perspect. Med. 2012, 2, a006239. [Google Scholar] [CrossRef] [Green Version]
- Reitz, C.; Mayeux, R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 2014, 88, 640–651. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S. Molecular and Cellular Basis of Neurodegeneration in Alzheimer’s Disease. Mol. Cells 2017, 40, 613–620. [Google Scholar]
- Millan, M.J. The epigenetic dimension of Alzheimer’s disease: Causal, consequence, or curiosity? Dialogues Clin. Neurosci. 2014, 16, 373–393. [Google Scholar] [CrossRef]
- Millan, M.J. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: An integrative review. Prog. Neurobiol. 2017, 156, 1–68. [Google Scholar] [CrossRef]
- Qazi, T.J.; Quan, Z.; Mir, A.; Qing, H. Epigenetics in Alzheimer’s Disease: Perspective of DNA Methylation. Mol. Neurobiol. 2017, 55, 1026–1044. [Google Scholar] [CrossRef]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 2018, 9, wrna.1463. [Google Scholar] [CrossRef]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.-Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, aam8526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes 2016, 7, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welden, J.R.; van Doorn, J.; Nelson, P.T.; Stamm, S. The human MAPT locus generates circular RNAs. Biochim. Biophys. Acta 2018, 1864, 2753–2760. [Google Scholar] [CrossRef]
- Mo, D.; Li, X.; Raabe, C.; Rozhdestvensky, T.; Skryabin, B.; Brosius, J. Circular RNA Encoded Amyloid Beta Peptides—A Novel Putative Player in Alzheimer’s Disease. Cells 2020, 9, 2196. [Google Scholar] [CrossRef]
- Mo, D.; Li, X.; Raabe, C.A.; Cui, D.; Vollmar, J.-F.; Rozhdestvensky, T.S.; Skryabin, B.V.; Brosius, J. A universal approach to investigate circRNA protein coding function. Sci. Rep. 2019, 9, 11684. [Google Scholar] [CrossRef] [Green Version]
- Cervera-Carles, L.; Dols-Icardo, O.; Molina-Porcel, L.; Alcolea, D.; Cervantes-Gonzalez, A.; Muñoz-Llahuna, L.; Clarimon, J. Assessing circular RNAs in Alzheimer’s disease and frontotemporal lobar degeneration. Neurobiol. Aging 2020, 92, 7–11. [Google Scholar] [CrossRef]
- Lo, I.; Hill, J.; Vilhjálmsson, B.J.; Kjems, J. Linking the association between circRNAs and Alzheimer’s disease progression by multi-tissue circular RNA characterization. RNA Biol. 2020, 17, 1789–1797. [Google Scholar] [CrossRef]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Efthymiou, A.G.; Goate, A.M. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 2017, 12, 43. [Google Scholar] [CrossRef]
- Jin, S.C.; Benitez, B.A.; Karch, C.M.; Cooper, B.; Skorupa, T.; Carrell, D.; Norton, J.B.; Hsu, S.; Harari, O.; Cai, Y.; et al. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum. Mol. Genet. 2014, 23, 5838–5846. [Google Scholar] [CrossRef] [Green Version]
- Del-Aguila, J.L.; Benitez, B.A.; Li, Z.; Dube, U.; Mihindukulasuriya, K.A.; Budde, J.P.; Farias, F.H.G.; Fernández, M.V.; Ibanez, L.; Jiang, S.; et al. TREM2 brain transcript-specific studies in AD and TREM2 mutation carriers. Mol. Neurodegener. 2019, 14, 18. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, B.; Li, Y.; Li, X.; Chen, X.; Zhang, Y.-W. TREM2 in Alzheimer’s Disease: Microglial Survival and Energy Metabolism. Front. Aging Neurosci. 2018, 10, 395. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Allen, M.; Sakae, N.; Ertekin-Taner, N.; Graff-Radford, N.R.; Dickson, D.W.; Younkin, S.G.; Sevlever, D. Expression and processing analyses of wild type and p.R47H TREM2 variant in Alzheimer’s disease brains. Mol. Neurodegener. 2016, 11, 72. [Google Scholar] [CrossRef] [Green Version]
- Casper, J.; Zweig, A.S.; Villarreal, C.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Karolchik, D.; et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2017, 46, D762–D769. [Google Scholar] [CrossRef] [Green Version]
- Yanaizu, M.; Sakai, K.; Tosaki, Y.; Kino, Y.; Satoh, J.-I. Small nuclear RNA-mediated modulation of splicing reveals a therapeutic strategy for a TREM2 mutation and its post-transcriptional regulation. Sci. Rep. 2018, 8, 6937. [Google Scholar] [CrossRef] [Green Version]
- Martiskainen, H.; Viswanathan, J.; Nykanen, N.P.; Kurki, M.; Helisalmi, S.; Natunen, T.; Sarajärvi, T.; Kurkinen, K.M.A.; Pursiheimo, J.; Rauramaa, T.; et al. Transcriptomics and mechanistic elucidation of Alzheimer’s disease risk genes in the brain and in vitro models. Neurobiol. Aging 2015, 36, 1221.e15–1221.e28. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Na, Y.; Koh, I.; Nho, K.; Lee, Y. Alternative Splicing Regulation of Low-Frequency Genetic Variants in Exon 2 of TREM2 in Alzheimer’s Disease by Splicing-Based Aggregation. Int. J. Mol. Sci. 2021, 22, 9865. [Google Scholar] [CrossRef] [PubMed]
- Kiianitsa, K.; Kurtz, I.; Beeman, N.; Matsushita, M.; Chien, W.; Raskind, W.H.; Korvatska, O. Novel TREM2 splicing isoform that lacks the V-set immunoglobulin domain is abundant in the human brain. J. Leukoc. Biol. 2021, 110, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Shaw, B.C.; Snider, H.C.; Turner, A.K.; Zajac, D.J.; Simpson, J.F.; Estus, S. An Alternatively Spliced TREM2 Isoform Lacking the Ligand Binding Domain is Expressed in Human Brain. J. Alzheimer’s Dis. 2022, 87, 1647–1657. [Google Scholar] [CrossRef]
- Celarain, N.; De Gordoa, J.S.-R.; Zelaya, M.V.; Roldán, M.; Larumbe, R.; Pulido, L.; Echavarri, C.; Mendioroz, M. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer’s disease hippocampus. Clin. Epigenetics 2016, 8, 37. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.R.; Smith, R.G.; Condliffe, D.; Hannon, E.; Schalkwyk, L.; Mill, J.; Lunnon, K. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol. Aging 2016, 47, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, Y.; Yoshino, Y.; Yamazaki, K.; Sao, T.; Mori, Y.; Ochi, S.; Yoshida, T.; Mori, T.; Iga, J.; Ueno, S. DNA methylation changes at TREM2 intron 1 and TREM2 mRNA expression in patients with Alzheimer’s disease. J. Psychiatr. Res. 2017, 92, 74–80. [Google Scholar] [CrossRef]
- Yoshino, Y.; Ozaki, Y.; Yamazaki, K.; Sao, T.; Mori, Y.; Ochi, S.; Iga, J.-I.; Ueno, S.-I. DNA Methylation Changes in Intron 1 of Triggering Receptor Expressed on Myeloid Cell 2 in Japanese Schizophrenia Subjects. Front. Neurosci. 2017, 11, 275. [Google Scholar] [CrossRef] [Green Version]
- Hashemiaghdam, A.; Mroczek, M. Microglia heterogeneity and neurodegeneration: The emerging paradigm of the role of immunity in Alzheimer’s disease. J. Neuroimmunol. 2020, 34, 577185. [Google Scholar] [CrossRef]
- Lue, L.F.; Schmitz, C.T.; Serrano, G.; Sue, L.I.; Beach, T.G.; Walker, D.G. TREM2 Protein Expression Changes Correlate with Alzheimer’s Disease Neurodegenerative Pathologies in Post-Mortem Temporal Cortices. Brain Pathol. 2015, 25, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Lessard, C.B.; Malnik, S.L.; Zhou, Y.; Ladd, T.B.; E Cruz, P.; Ran, Y.; E Mahan, T.; Chakrabaty, P.; Holtzman, D.M.; Ulrich, J.D.; et al. High-affinity interactions and signal transduction between Aβ oligomers and TREM 2. EMBO Mol. Med. 2018, 10, e201809027. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. Deficits in the miRNA-34a-regulated endogenous TREM2 phagocytosis sensor-receptor in Alzheimer’s disease (AD)—An update. Front. Aging Neurosci. 2014, 6, 116. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Wang, Y. TREM2 variants: New keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 2016, 17, 201–207. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, C.; Wu, C.; Liu, J.; Yu, H.; Zhou, X.; Zhang, J.; Wang, X.; He, S.; Xu, X.; et al. miR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling pathway. Stem Cell Res. Ther. 2020, 11, 62. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, C.; Chen, K.; Wang, T.; Xing, J.; Zhang, X.; Wang, X. MiR-765 functions as a tumour suppressor and eliminates lipids in clear cell renal cell carcinoma by downregulating PLP2. eBioMedicine 2020, 51, 102622. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liao, Y.; Gao, L.; Zhuang, T.; Zhu, H.; Ge, J. Coronary Serum Exosomes Derived from Patients with Myocardial Ischemia Regulate Angiogenesis through the miR-939-mediated Nitric Oxide Signaling Pathway. Theranostics 2018, 8, 2079–2093. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.L.; Koike, M.; Spusta, S.C.; Niemi, E.C.; Yenari, M.; Nakamura, M.C.; Seaman, W.E. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 2009, 109, 1144–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefano, L.; Racchetti, G.; Bianco, F.; Passini, N.; Gupta, R.S.; Bordignon, P.P.; Meldolesi, J. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J. Neurochem. 2009, 110, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Rochford, C.D.; Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005, 201, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Yeh, F.L.; Hansen, D.V.; Sheng, M. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med. 2017, 23, 512–533. [Google Scholar] [CrossRef]
- Jaber, V.; Zhao, Y.; Lukiw, W.J. Alterations in micro RNA-messenger RNA (miRNA-mRNA) Coupled Signaling Networks in Sporadic Alzheimer’s Disease (AD) Hippocampal CA1. J. Alzheimers Dis. Parkinsonism 2017, 7, 312. [Google Scholar]
- Zhao, Y.; Lukiw, W.J. TREM2 signaling, miRNA-34a and the extinction of phagocytosis. Front. Cell. Neurosci. 2013, 7, 131. [Google Scholar] [CrossRef] [Green Version]
- Gruner, H.; Cortés-López, M.; Cooper, D.A.; Bauer, M.; Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016, 6, 38907. [Google Scholar] [CrossRef]
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2016, 54, 5156–5165. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Constantin, L. Circular RNAs and Neuronal Development. Circ. RNAs 2018, 1087, 205–213. [Google Scholar] [CrossRef]
- Van Rossum, D.; Verheijen, B.M.; Pasterkamp, R.J. Circular RNAs: Novel Regulators of Neuronal Development. Front. Mol. Neurosci. 2016, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Bell, J.E.; Alafuzoff, I.; Al-Sarraj, S.; Arzberger, T.; Bogdanovic, N.; Budka, H.; Dexter, D.T.; Falkai, P.; Ferrer, I.; Gelpi, E.; et al. Management of a twenty-first century brain bank: Experience in the BrainNet Europe consortium. Acta Neuropathol. 2008, 115, 497–507. [Google Scholar] [CrossRef]
- Montine, T.J.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; Mirra, S.S.; et al. National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 2011, 123, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Mirra, S.S.; Heyman, A.; McKeel, D.; Sumi, S.M.; Crain, B.J.; Brownlee, L.M.; Vogel, F.S.; Hughes, J.P.; van Belle, G.; Berg, L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 1991, 41, 479–486. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
| miRNAs Predicted to Join | circTREM2_1 | circTREM2_2 | circTREM2_3 |
|---|---|---|---|
| hsa-miR-765 | Yes | Yes | No |
| hsa-miR-11181-3p | Yes | Yes | No |
| hsa-miR-6890-3p | Yes | Yes | Yes |
| hsa-miR-6766-5p | Yes | Yes | No |
| hsa-miR-6756-5p | Yes | Yes | No |
| hsa-miR-653-3p | Yes | Yes | Yes |
| hsa-miR-6770-5p | Yes | Yes | No |
| hsa-miR-6131 | Yes | No | No |
| hsa-miR-6762-3p | Yes | Yes | No |
| hsa-miR-4783-3p | Yes | Yes | Yes |
| hsa-miR-2392 | Yes | Yes | No |
| hsa-miR-4483 | Yes | Yes | No |
| hsa-miR-6745 | Yes | Yes | No |
| hsa-miR-6749-3p | No | No | Yes |
| hsa-miR-939-3p | No | No | Yes |
| hsa-miR-3657 | No | No | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urdánoz-Casado, A.; de Gordoa, J.S.-R.; Robles, M.; Roldan, M.; Zelaya, M.V.; Blanco-Luquin, I.; Mendioroz, M. Profile of TREM2-Derived circRNA and mRNA Variants in the Entorhinal Cortex of Alzheimer’s Disease Patients. Int. J. Mol. Sci. 2022, 23, 7682. https://doi.org/10.3390/ijms23147682
Urdánoz-Casado A, de Gordoa JS-R, Robles M, Roldan M, Zelaya MV, Blanco-Luquin I, Mendioroz M. Profile of TREM2-Derived circRNA and mRNA Variants in the Entorhinal Cortex of Alzheimer’s Disease Patients. International Journal of Molecular Sciences. 2022; 23(14):7682. https://doi.org/10.3390/ijms23147682
Chicago/Turabian StyleUrdánoz-Casado, Amaya, Javier Sánchez-Ruiz de Gordoa, Maitane Robles, Miren Roldan, María Victoria Zelaya, Idoia Blanco-Luquin, and Maite Mendioroz. 2022. "Profile of TREM2-Derived circRNA and mRNA Variants in the Entorhinal Cortex of Alzheimer’s Disease Patients" International Journal of Molecular Sciences 23, no. 14: 7682. https://doi.org/10.3390/ijms23147682
APA StyleUrdánoz-Casado, A., de Gordoa, J. S.-R., Robles, M., Roldan, M., Zelaya, M. V., Blanco-Luquin, I., & Mendioroz, M. (2022). Profile of TREM2-Derived circRNA and mRNA Variants in the Entorhinal Cortex of Alzheimer’s Disease Patients. International Journal of Molecular Sciences, 23(14), 7682. https://doi.org/10.3390/ijms23147682






