Synergistic Antibacterial Activity with Conventional Antibiotics and Mechanism of Action of Shikonin against Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Results
2.1. Synergistic Effects of SKN and Antibiotics against MRSA and Its Clinical Strain
2.2. Time Kill Assay
2.3. Effect of SKN on MRSA Cell Membrane Integrity and Permeability
2.4. Transmission Electron Microscopy (TEM)
2.5. The Anti-Biofilm Formation Activity of SKN
2.6. Effect of SKN on PBP2a Expression
2.7. Effect of SKN on Virulence Factor Expression
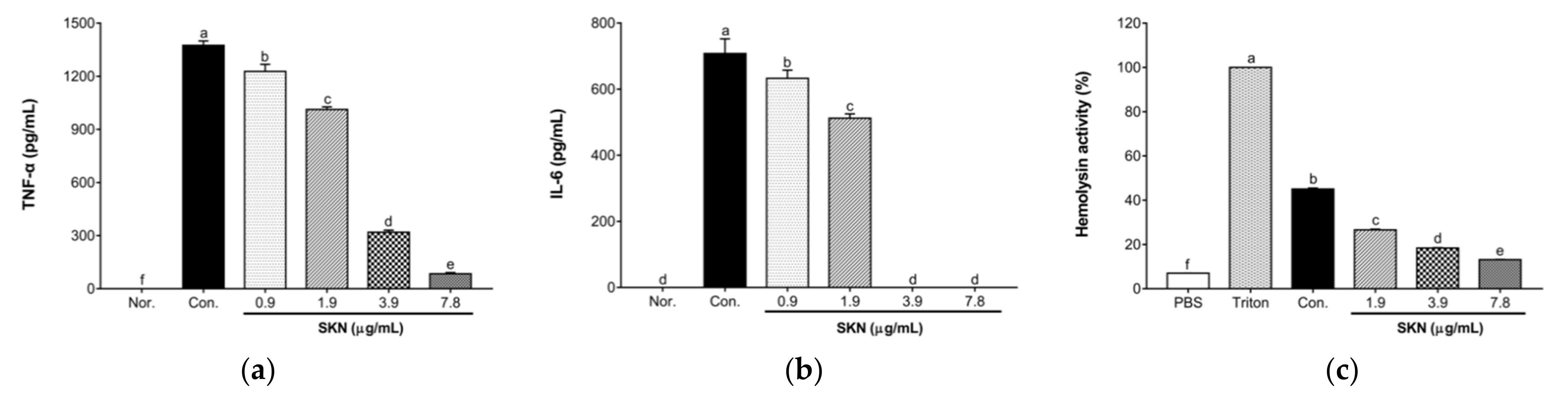
2.8. Effects of SKN in the Pro-Inflammatory Cytokine Release of 264.7 RAW Macrophages Stimulated by MRSA
2.9. Effect of SKN on the Hemolytic Activity of S. aureus Strains
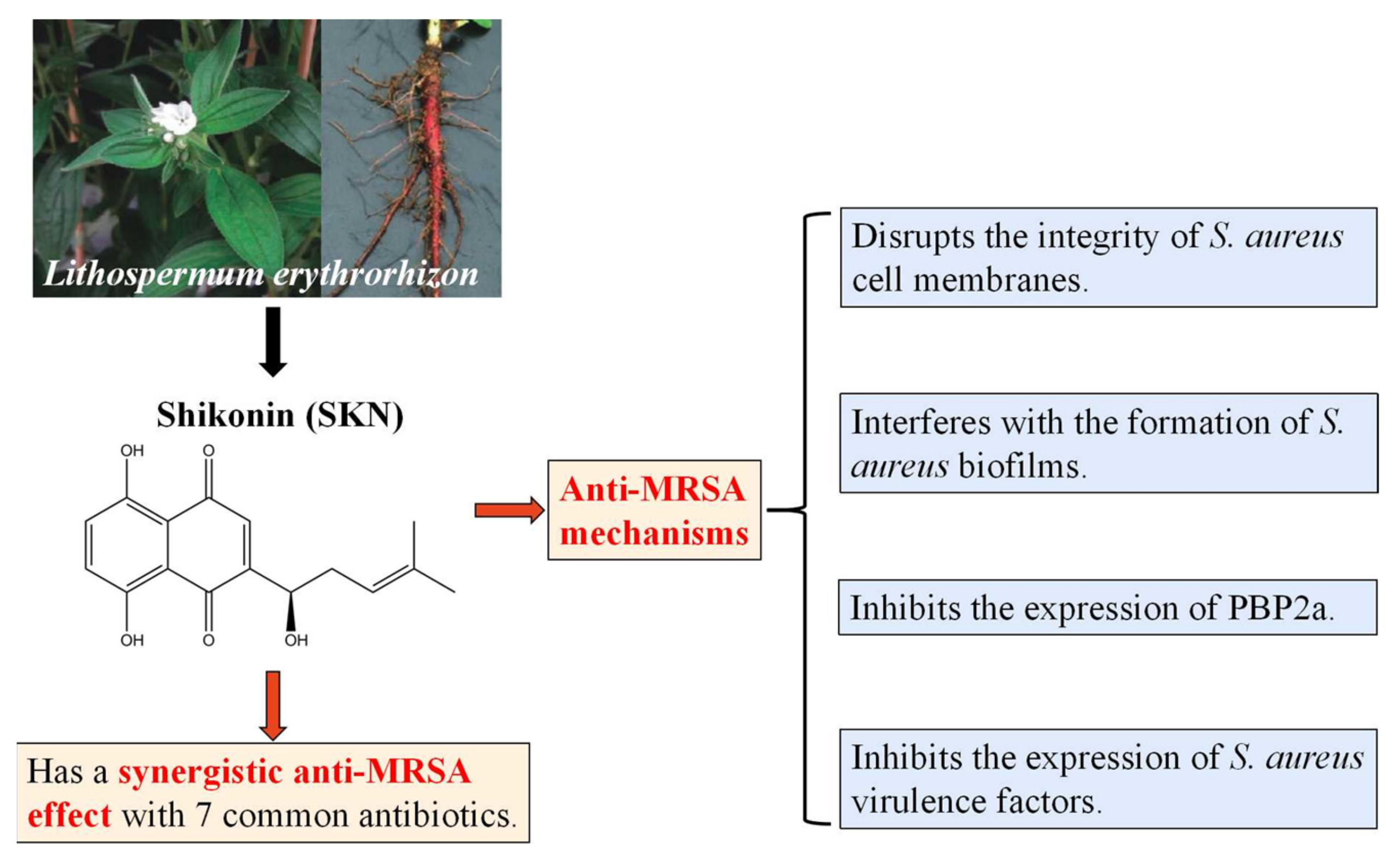
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Reagents
4.2. Checkerboard Dilution Assay
4.3. Time–Kill Assay
4.4. Nucleotide and Protein Leakage
4.5. Bacteriolysis Assay
4.6. Transmission Electron Microscopy (TEM)
4.7. Anti-Biofilm Formation Assay
4.8. qRT-PCR
4.9. Western Blot Assay
4.10. Enzyme-Linked Immunosorbent Assay (ELISA)
4.11. Hemolysis Assay
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Negrete-González, C.; Turrubiartes-Martínez, E.; Galicia-Cruz, O.G.; Noyola, D.E.; Martínez-Aguilar, G.; Pérez-González, L.F.; González-Amaro, R.; Niño-Moreno, P. High prevalence of t895 and t9364 spa types of methicillin-resistant Staphylococcus aureus in a tertiary-care hospital in Mexico: Different lineages of clonal complex 5. BMC Microbiol. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Kateete, D.P.; Bwanga, F.; Seni, J.; Mayanja, R.; Kigozi, E.; Mujuni, B.; Ashaba, F.K.; Baluku, H.; Najjuka, C.F.; Källander, K.; et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob. Resist. Infect. Control 2019, 8, 94. [Google Scholar] [CrossRef]
- Ike, B.; Ugwu, M.C.; Ikegbunam, M.N.; Nwobodo, D.; Ejikeugwu, C.; Gugu, T.; Esimone, C.O. Prevalence, Antibiogram and Molecular Characterization of Comunity-Acquired Methicillin-Resistant Staphylococcus aureus in AWKA, Anambra Nigeria. Open Microbiol. J. 2016, 10, 211–221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seni, J.; Bwanga, F.; Najjuka, C.F.; Makobore, P.; Okee, M.; Mshana, S.E.; Kidenya, B.R.; Joloba, M.L.; Kateete, D.P. Molecular Characterization of Staphylococcus aureus from Patients with Surgical Site Infections at Mulago Hospital in Kampala, Uganda. PLoS ONE 2013, 8, e66153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, X.; Zheng, Y.; Lv, Q.; Kong, D.; Ji, B.; Han, X.; Zhou, D.; Sun, Z.; Zhu, L.; Liu, P.; et al. Staphylococcus aureus N-terminus formylated δ-toxin tends to form amyloid fibrils, while the deformylated δ-toxin tends to form functional oligomer complexes. Virulence 2021, 12, 1418–1437. [Google Scholar] [CrossRef] [PubMed]
- Surewaard, B.G.; Nijland, R.; Spaan, A.N.; Kruijtzer, J.A.; de Haas, C.J.; van Strijp, J.A. Inactivation of Staphylococcal Phenol Soluble Modulins by Serum Lipoprotein Particles. PLoS Pathog. 2012, 8, e1002606. [Google Scholar] [CrossRef] [PubMed]
- Mairi, A.; Touati, A.; Lavigne, J.P. Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review. Toxins 2020, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Jafari Saraf, L.; Wilson, S.E. Telavancin, a new lipoglycopeptide antimicrobial, in complicated skin and soft tissue infections. Infect. Drug Resist. 2011, 4, 87–95. [Google Scholar]
- Song, Y.; Lunde, C.S.; Benton, B.M.; Wilkinson, B.J. Studies on the Mechanism of Telavancin Decreased Susceptibility in a Laboratory-Derived Mutant. Microb. Drug Resist. 2013, 19, 247–255. [Google Scholar] [CrossRef]
- Arjyal, C.; Kc, J.; Neupane, S. Prevalence of Methicillin-Resistant Staphylococcus aureus in Shrines. Int. J. Microbiol. 2020, 2020, 7981648. [Google Scholar] [CrossRef]
- Velasco, V.; Buyukcangaz, E.; Sherwood, J.S.; Stepan, R.M.; Koslofsky, R.J.; Logue, C.M. Characterization of Staphylococcus aureus from Humans and a Comparison with Isolates of Animal Origin, in North Dakota, United States. PLoS ONE 2015, 10, e0140497. [Google Scholar]
- Zhang, N.; Peng, F.; Wang, Y.; Yang, L.; Wu, F.; Wang, X.; Ye, C.; Han, B.; He, G. Shikonin induces colorectal carcinoma cells apoptosis and autophagy by targeting galectin-1/JNK signaling axis. Int. J. Biol. Sci. 2020, 16, 147–161. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, D.Y.; Kim, Y.B.; Lee, S.W.; Cha, S.W.; Park, H.W.; Kim, G.S.; Kwon, D.Y.; Lee, M.H.; Han, S.H. The Mechanism Underlying the Antibacterial Activity of Shikonin against Methicillin-Resistant Staphylococcus aureus. Evid. Based Complement. Alternat. Med. 2015, 2015, 520578. [Google Scholar] [CrossRef]
- Huang, C.; Hu, G. Shikonin suppresses proliferation and induces apoptosis in endometrioid endometrial cancer cells via modulating miR-106b/PTEN/AKT/mTOR signaling pathway. Biosci. Rep. 2018, 38, BSR20171546. [Google Scholar] [CrossRef]
- Zhang, F.Y.; Hu, Y.; Que, Z.Y.; Wang, P.; Liu, Y.H.; Wang, Z.H.; Xue, Y.X. Shikonin Inhibits the Migration and Invasion of Human Glioblastoma Cells by Targeting Phosphorylated β-Catenin and Phosphorylated PI3K/Akt: A Potential Mechanism for the Anti-Glioma Efficacy of a Traditional Chinese Herbal Medicine. Int. J. Mol. Sci. 2015, 16, 23823–23848. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, T.; Pan, Z.; Ge, X.; Sun, C.; Lu, C.; Chen, H.; Xiao, Z.; Zhang, B.; Dai, Y.; et al. Shikonin inhibits myeloid differentiation protein 2 to prevent LPS-induced acute lung injury. Br. J. Pharmacol. 2018, 175, 840–854. [Google Scholar] [CrossRef]
- Gara, R.K.; Srivastava, V.K.; Duggal, S.; Bagga, J.K.; Bhatt, M.; Sanyal, S.; Mishra, D.P. Shikonin selectively induces apoptosis in human prostate cancer cells through the endoplasmic reticulum stress and mitochondrial apoptotic pathway. J. Biomed. Sci. 2015, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ni, J.; Gao, Y.; Zhang, J.; Liu, X.; Chen, Y.; Chen, Z.; Wu, Y. Integrated proteomics and metabolomics reveals the comprehensive characterization of antitumor mechanism underlying Shikonin on colon cancer patient-derived xenograft model. Sci. Rep. 2020, 10, 14092. [Google Scholar] [CrossRef]
- Lu, B.; Gong, X.; Wang, Z.Q.; Ding, Y.; Wang, C.; Luo, T.F.; Piao, M.H.; Meng, F.K.; Chi, G.F.; Luo, Y.N.; et al. Shikonin induces glioma cell necroptosis in vitro by ROS overproduction and promoting RIP1/RIP3 necrosome formation. Acta Pharmacol. Sin. 2017, 38, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Liu, B.; Shao, Y.; Hua, L.; Fu, R.; Wang, H.; Wang, T.; Qi, W.; Shao, Z.; Liu, C. Effects of Shikonin on the Functions of Myeloid Dendritic Cells in a Mouse Model of Severe Aplastic Anemia. Mediators Inflamm. 2020, 2020, 9025705. [Google Scholar] [CrossRef] [PubMed]
- Otero, L.H.; Rojas-Altuve, A.; Llarrull, L.I.; Carrasco-López, C.; Kumarasiri, M.; Lastochkin, E.; Fishovitz, J.; Dawley, M.; Hesek, D.; Lee, M.; et al. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl. Acad. Sci. USA 2013, 110, 16808–16813. [Google Scholar] [CrossRef]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational dynamics in penicillin-binding Protein 2a of methicillin-resistant Staphylococcus aureus, allosteric communication network and enablement of catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef]
- Janardhanan, J.; Bouley, R.; Martínez-Caballero, S.; Peng, Z.; Batuecas-Mordillo, M.; Meisel, J.E.; Ding, D.; Schroeder, V.A.; Wolter, W.R.; Mahasenan, K.V.; et al. The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e02637-18. [Google Scholar] [CrossRef]
- Fuda, C.; Hesek, D.; Lee, M.; Morio, K.; Nowak, T.; Mobashery, S. Activation for Catalysis of Penicillin-Binding Protein 2a from Methicillin-Resistant Staphylococcus aureus by Bacterial Cell Wall. J. Am. Chem. Soc. 2005, 127, 2056–2057. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Baek, K.H. Antibacterial mechanism of the action of Enteromorpha linza L. essential oil against Escherichia coli and Salmonella Typhimurium. Bot. Stud. 2015, 56, 13. [Google Scholar] [CrossRef]
- Ahmad, S.J.; Lian, H.H.; Basri, D.F.; Zin, N.M. Mode of action of Endophytic Streptomyces sp., SUK 25 extracts Against MRSA.; Microscopic, Biochemical and Time-Kill Analysis. Int. J. Pharm. Sci. Rev. Res. 2015, 30, 11–17. [Google Scholar]
- Abbas, H.A.; Atallah, H.; El-Sayed, M.A.; El-Ganiny, A.M. Diclofenac mitigates virulence of multidrug-resistant Staphylococcus aureus. Arch. Microbiol. 2020, 202, 2751–2760. [Google Scholar] [CrossRef]
- Jin, Y.; Li, M.; Shang, Y.; Liu, L.; Shen, X.; Lv, Z.; Hao, Z.; Duan, J.; Wu, Y.; Chen, C.; et al. Sub-Inhibitory Concentrations of Mupirocin Strongly Inhibit Alpha-Toxin Production in High-Level Mupirocin-Resistant MRSA by Down-Regulating agr, saeRS, and sarA. Front. Microbiol. 2018, 9, 993. [Google Scholar] [CrossRef]
- Li, Q.Q.; Luo, J.; Liu, X.Q.; Kang, O.H.; Kwon, D.Y. Eleutheroside K Isolated from Acanthopanax henryi (Oliv.) Harms Inhibits the Expression of Virulence-Related Exoproteins in Methicillin-Resistant Staphylococcus aureus. Curr. Microbiol. 2021, 78, 3980–3988. [Google Scholar] [CrossRef]
- Liu, C.; Huang, H.; Zhou, Q.; Liu, B.; Wang, Y.; Li, P.; Liao, K.; Su, W. Pithecellobium clypearia extract enriched in gallic acid and luteolin has antibacterial activity against MRSA and reduces resistance to erythromycin, ceftriaxone sodium and levofloxacin. J. Appl. Microbiol. 2020, 129, 848–859. [Google Scholar] [CrossRef]
- Wu, S.C.; Han, F.; Song, M.R.; Chen, S.; Li, Q.; Zhang, Q.; Zhu, K.; Shen, J.Z. Natural Flavones from morus alba against Methicillin-Resistant Staphylococcus aureus via Targeting the Proton Motive Force and Membrane Permeability. J. Agric. Food Chem. 2019, 67, 10222–10234. [Google Scholar] [CrossRef] [PubMed]
- Meah, M.S.; Lertcanawanichakul, M.; Pedpradab, P.; Lin, W.; Zhu, K.; Li, G.; Panichayupakaranant, P. Synergistic effect on anti-methicillin-resistant Staphylococcus aureus among combinations of α-mangostin-rich extract, lawsone methyl ether and ampicillin. Lett. Appl. Microbiol. 2020, 71, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Bahuguna, A.; Kumar, P.; Bajpai, V.K.; Kang, S.C. Antimicrobial Potential of Carvacrol against Uropathogenic Escherichia coli via Membrane Disruption, Depolarization, and Reactive Oxygen Species Generation. Front. Microbiol. 2017, 8, 2421. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.K.; Yusoff, K.; Ajat, M.; Wee, C.Y.; Yap, P.S.; Lim, S.H.; Lai, K.S. Combinatorial Antimicrobial Efficacy and Mechanism of Linalool Against Clinically Relevant Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 635016. [Google Scholar] [CrossRef] [PubMed]
- Lemos, A.S.O.; Campos, L.M.; Melo, L.; Guedes, M.; Oliveira, L.G.; Silva, T.P.; Melo, R.; Rocha, V.N.; Aguiar, J.; Apolônio, A.; et al. Antibacterial and Antibiofilm Activities of Psychorubrin, a Pyranonaphthoquinone Isolated from Mitracarpus frigidus (Rubiaceae). Front. Microbiol. 2018, 9, 724. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Kang, O.H.; Kwon, D.Y. Study on demethoxycurcumin as a promising approach to reverse methicillin-resistance of Staphylococcus aureus. Int. J. Mol. Sci. 2021, 22, 3778. [Google Scholar] [CrossRef]
- Li, Q.Q.; Luo, J.; Liu, X.Q.; Kwon, D.Y.; Kang, O.H. Eleutheroside K isolated from Acanthopanax henryi (Oliv.) Harms suppresses methicillin resistance of Staphylococcus aureus. Lett. Appl. Microbiol. 2020, 72, 669–676. [Google Scholar] [CrossRef]
- Duan, J.; Li, M.; Hao, Z.; Shen, X.; Liu, L.; Jin, Y.; Wang, S.; Guo, Y.; Yang, L.; Wang, L.; et al. Subinhibitory concentrations of resveratrol reduce alpha-hemolysin production in Staphylococcus aureus isolates by downregulating saeRS. Emerg. Microbes Infect. 2018, 7, 136. [Google Scholar] [CrossRef]
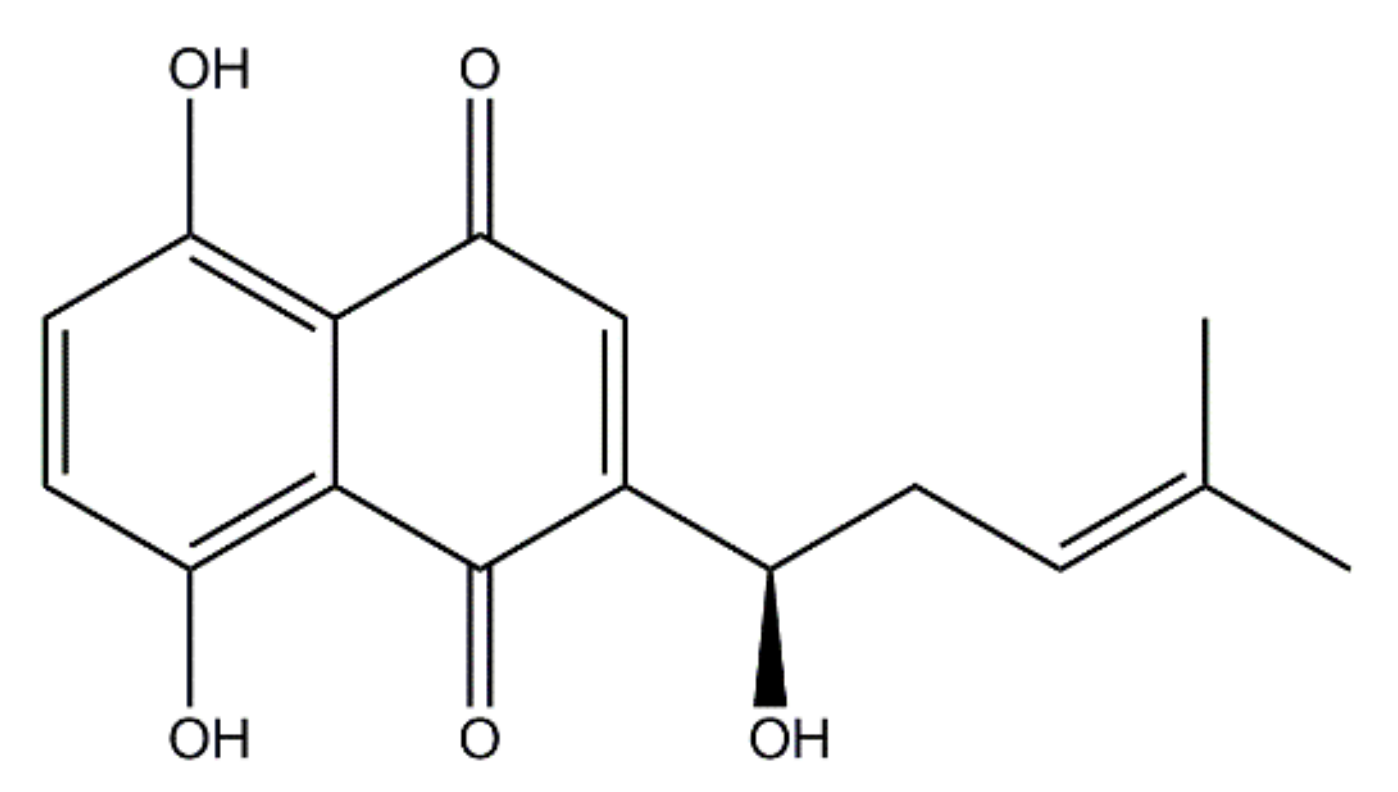
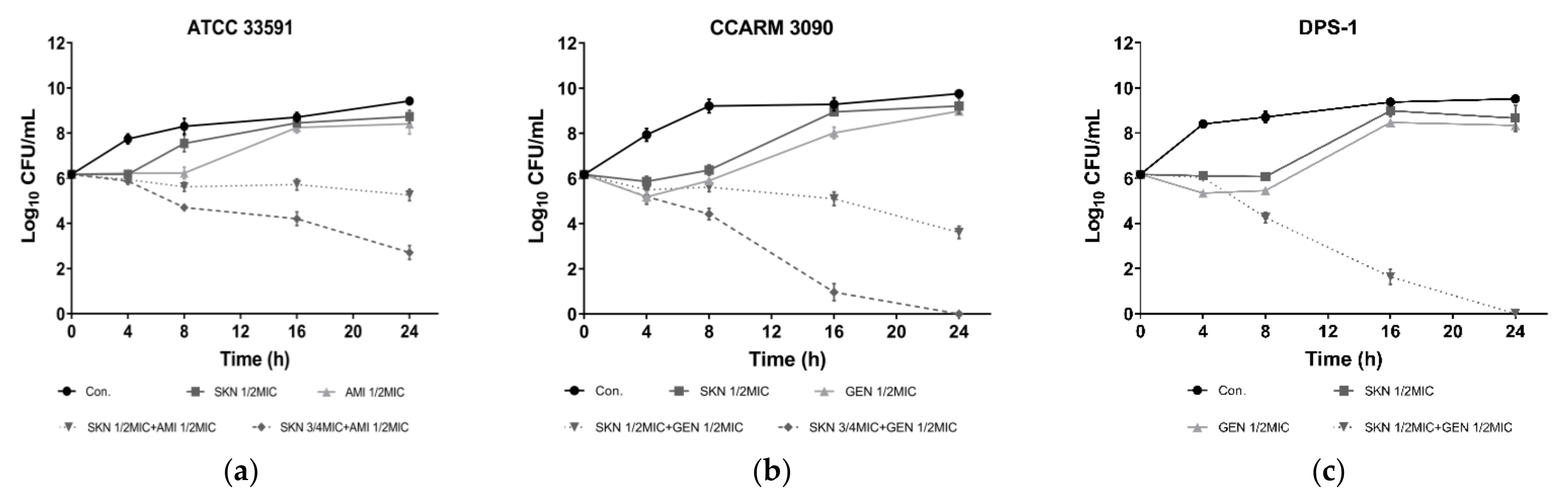
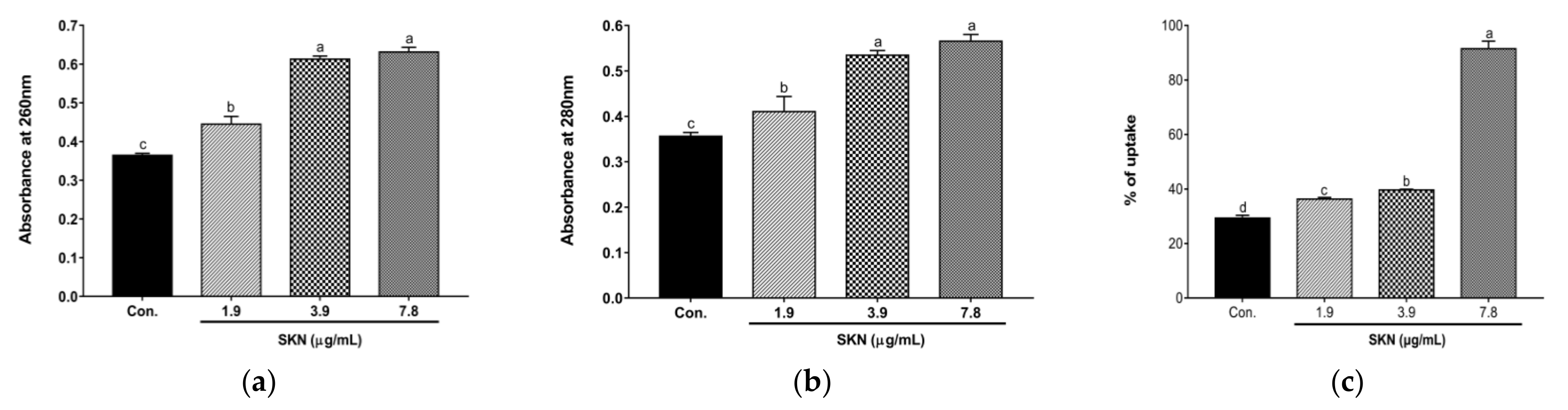
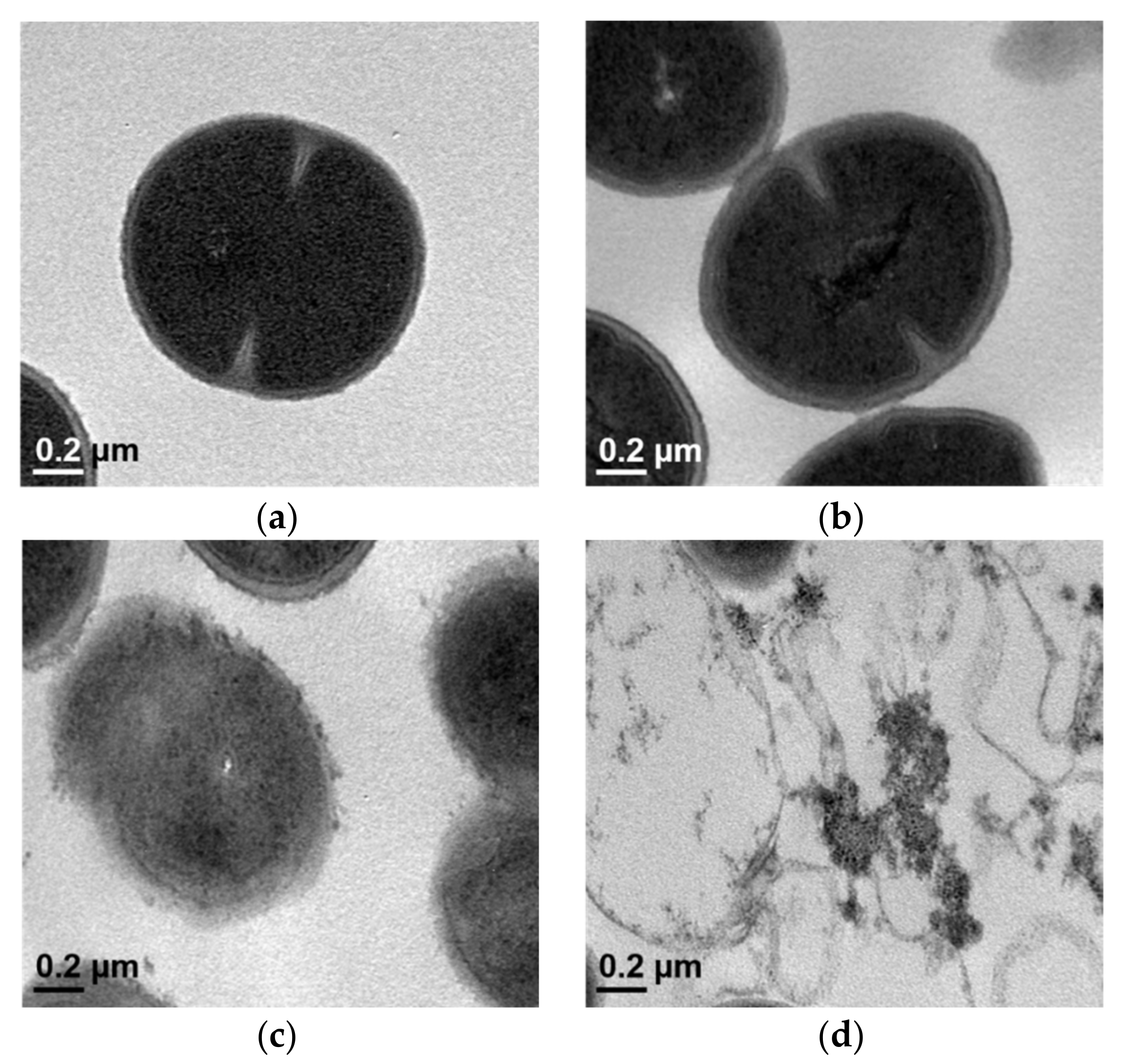



| Agents | ATCC 33591 | CCARM 3090 | DPS-1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | Fold | FICI | MIC (µg/mL) | Fold | FICI | MIC (µg/mL) | Fold | FICI | ||||
| Alone | Comb. | Alone | Comb. | Alone | Comb. | |||||||
| SKN | 15.6 | 7.8 | 2 | 1 | 125 | 7.8 | 16 | 0.56 | 15.6 | 3.9 | 4 | 0.5 |
| OXA | 31.25 | 15.6 | 2 | 250 | 125 | 2 | 31.25 | 7.8 | 4 | |||
| SKN | 15.6 | 3.9 | 4 | 0.5 | 125 | 7.8 | 16 | 0.19 | 15.6 | 1.9 | 8 | 0.37 |
| GEN | 7.8 | 1.9 | 4 | 250 | 31.25 | 8 | 125 | 31.25 | 4 | |||
| SKN | 15.6 | 1.9 | 8 | 0.37 | 125 | 7.8 | 16 | 0.31 | 15.6 | 1.9 | 8 | 0.37 |
| AMI | 62.5 | 15.6 | 4 | 7.8 | 1.9 | 4 | 62.5 | 15.6 | 4 | |||
| SKN | 15.6 | 0.9 | 16 | 0.31 | 125 | 31.25 | 4 | 0.5 | 15.6 | 0.9 | 16 | 0.56 |
| AMO | 250 | 62.5 | 4 | 62.5 | 15.6 | 4 | 62.5 | 31.25 | 2 | |||
| SKN | 15.6 | 0.9 | 16 | 0.56 | 125 | 7.8 | 16 | 0.31 | 15.6 | 1.9 | 8 | 0.37 |
| CEFO | 31.25 | 15.6 | 2 | 62.5 | 15.6 | 4 | 31.25 | 7.8 | 4 | |||
| SKN | 15.6 | 3.9 | 4 | 0.75 | 125 | 7.8 | 16 | 0.31 | 15.6 | 0.9 | 16 | 0.31 |
| CEFT | 125 | 62.5 | 2 | 1,000 | 250 | 4 | 250 | 62.5 | 4 | |||
| SKN | 15.6 | 3.9 | 4 | 0.75 | 125 | 7.8 | 16 | 0.56 | 15.6 | 0.9 | 16 | 0.31 |
| CHL | 125 | 62.5 | 2 | 7.8 | 3.9 | 2 | 250 | 62.5 | 4 | |||
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| icaA | CAATACTATTTCGGGTGTCTTCACTCT | CAAGAAACTGCAATATCTTCGGTAATCAT |
| fnbA | AACTGCACAACCAGCAAATG | TTGAGGTTGTGTCGTTTCCTT |
| mecA | GCAATCGCTAAAGAACTAAG | AATGGGACCAACATAACCTA |
| mecR1 | ACACGACTTCTTCGGTTAG | GTACAATTTGGGATTTCACT |
| hla | TTGGTGCAAATGTTTC | TCACTTTCCAGCCTACT |
| sea | ATGGTGCTTATTATGGTTATC | CGTTTCCAAAGGTACTGTATT |
| agrA | TGATAATCCTTATGAGGTGCTT | CACTGTGACTCGTAACGAAAA |
| RNAIII | GCACTGAGTCCAAGGAAACTAAC | AAGCCATCCCAACTTAATAACC |
| saeS | TGTATTTAAAGTGATAATATGAGTC | CTTAGCCCATGATTTAAAAACACC |
| sigB | AAGTGATTCGTAAGGACGTCT | TCGATAACTATAACCAAAGCCT |
| 16S | ACTCCTACGGGAGGCAGCAG | ATTACCGCGGCTGCTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.-Q.; Chae, H.-S.; Kang, O.-H.; Kwon, D.-Y. Synergistic Antibacterial Activity with Conventional Antibiotics and Mechanism of Action of Shikonin against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 7551. https://doi.org/10.3390/ijms23147551
Li Q-Q, Chae H-S, Kang O-H, Kwon D-Y. Synergistic Antibacterial Activity with Conventional Antibiotics and Mechanism of Action of Shikonin against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences. 2022; 23(14):7551. https://doi.org/10.3390/ijms23147551
Chicago/Turabian StyleLi, Qian-Qian, Hee-Sung Chae, Ok-Hwa Kang, and Dong-Yeul Kwon. 2022. "Synergistic Antibacterial Activity with Conventional Antibiotics and Mechanism of Action of Shikonin against Methicillin-Resistant Staphylococcus aureus" International Journal of Molecular Sciences 23, no. 14: 7551. https://doi.org/10.3390/ijms23147551
APA StyleLi, Q.-Q., Chae, H.-S., Kang, O.-H., & Kwon, D.-Y. (2022). Synergistic Antibacterial Activity with Conventional Antibiotics and Mechanism of Action of Shikonin against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences, 23(14), 7551. https://doi.org/10.3390/ijms23147551






