Cannabinerol and NSC-34 Transcriptomic Analysis: Is the Dose Who Makes Neuronal Differentiation?
Abstract
1. Introduction
2. Results
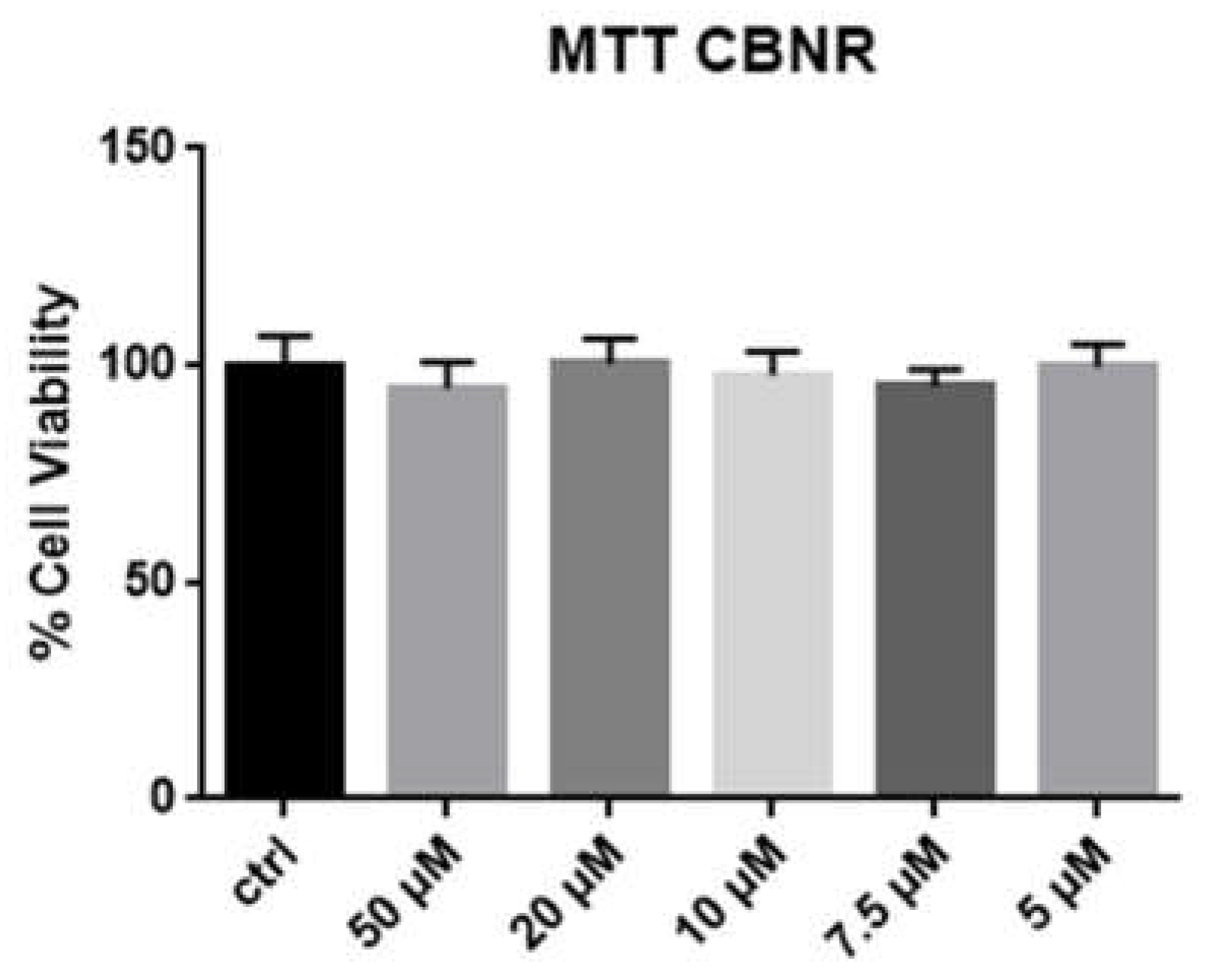
2.1. MTT Test
2.2. Transcriptomic Inspection
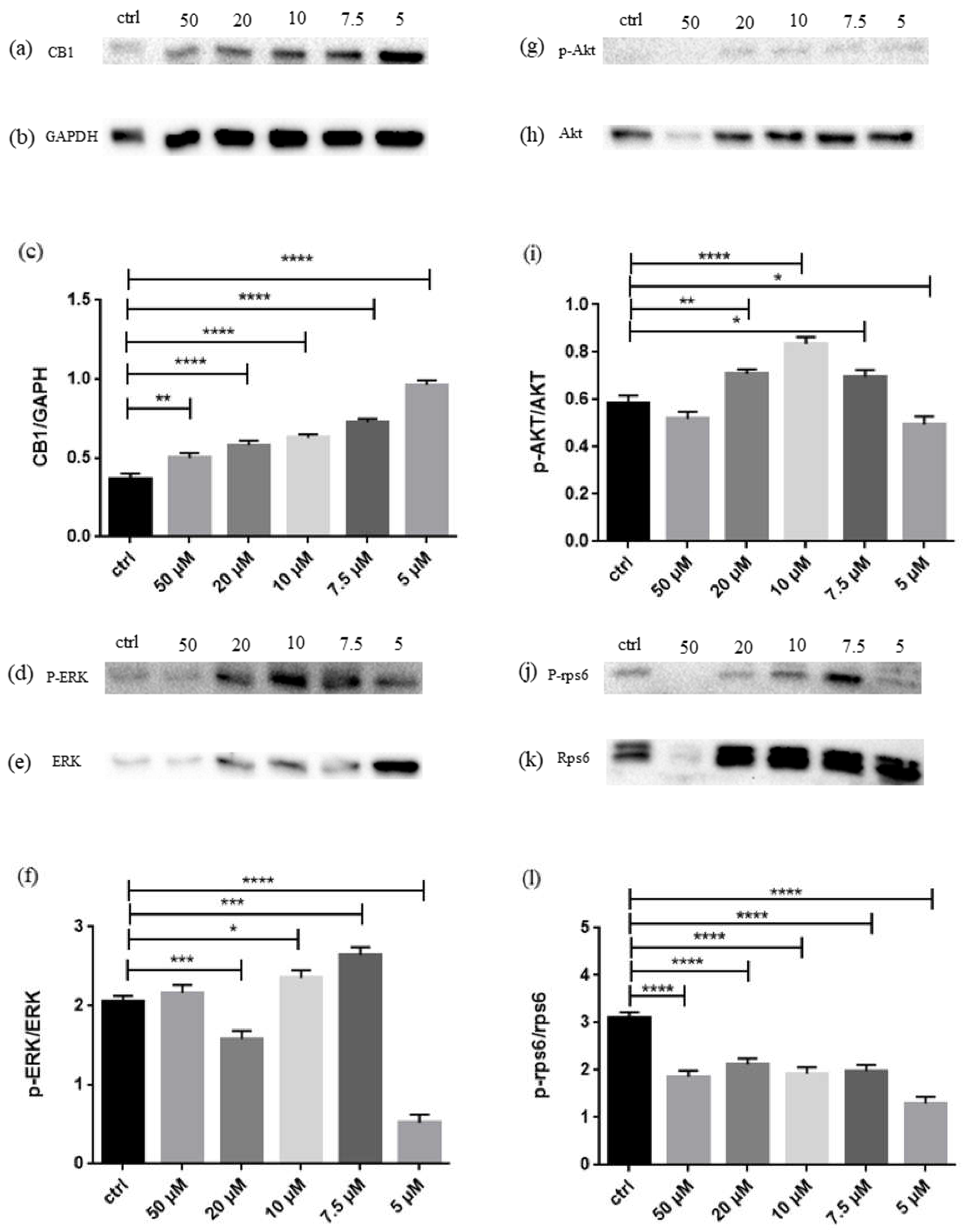
2.3. Western Blot
3. Discussion
4. Materials and Methods
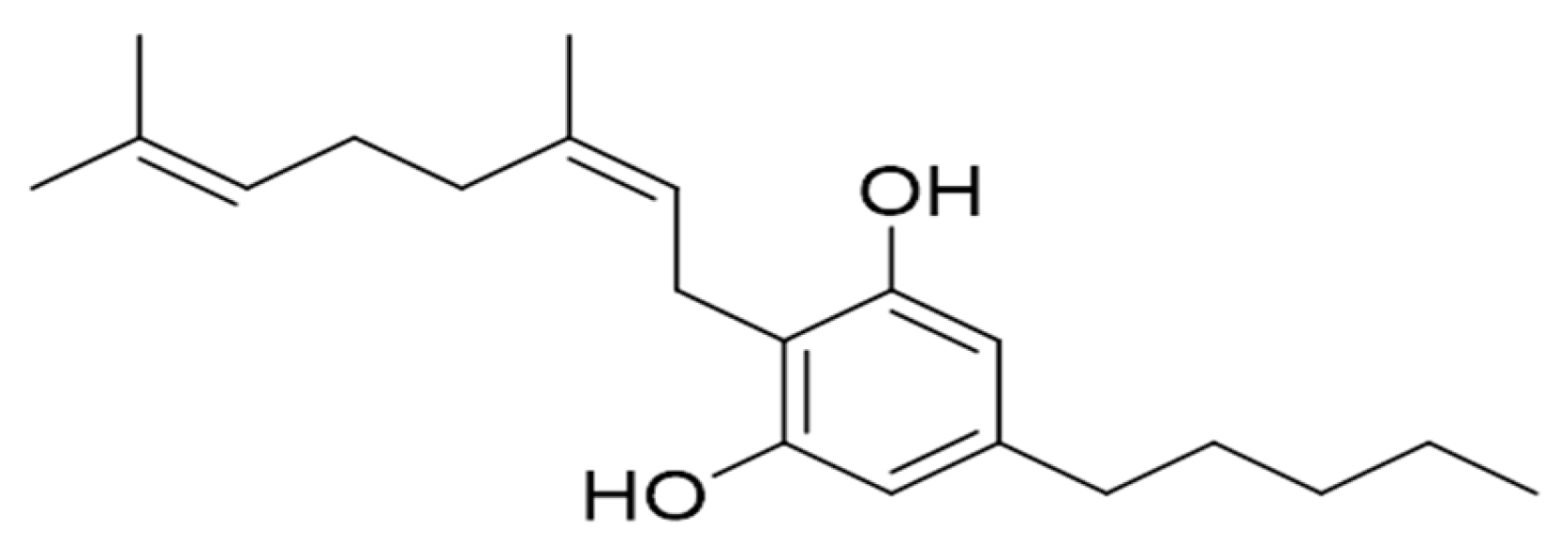
4.1. Obtaining CBNR from C. sativa
4.2. NSC-34 Colture and Treatment
4.3. MTT Test
4.4. Library Preparation and Bioinformatics Inspection
4.5. Western Blot Analyses
4.6. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Devane, W.A.; Dysarz, F.A., 3rd; Johnson, M.R.; Melvin, L.S.; Howlett, A.C. Determination and characterization of a cannabinoid receptor in rat brain. Mol. Pharmacol. 1988, 34, 605–613. [Google Scholar] [PubMed]
- Schilling, S.; Melzer, R.; McCabe, P.F. Cannabis sativa. Curr. Biol. 2020, 30, R8–R9. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, W.B. On the Preparations of the Indian Hemp, or Gunjah (Cannabis indica), Their Effects on the Animal System in Health, and Their Utility in the Treatment of Tetanus and Other Convulsive Diseases. Br. Foreign. Med. Rev. 1840, 10, 225–228. [Google Scholar]
- Rock, E.M.; Parker, L.A. Constituents of Cannabis Sativa. In Cannabinoids and Neuropsychiatric Disorders; Murillo-Rodriguez, E., Pandi-Perumal, S.R., Monti, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–13. [Google Scholar] [CrossRef]
- Pennypacker, S.D.; Romero-Sandoval, E.A. CBD and THC: Do They Complement Each Other Like Yin and Yang? Pharmacotherapy 2020, 40, 1152–1165. [Google Scholar] [CrossRef]
- Taura, F.; Morimoto, S.; Shoyama, Y. Cannabinerolic acid, a cannabinoid from Cannabis sativa. Phytochemistry 1995, 39, 457–458. [Google Scholar] [CrossRef]
- Radwan, M.M.; Wanas, A.S.; Chandra, S.; ElSohly, M.A. Natural Cannabinoids of Cannabis and Methods of Analysis. In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 161–182. [Google Scholar] [CrossRef]
- Elsohly, M.A.; Slade, D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Hanuš, L.O.; Levy, R.; De La Vega, D.; Katz, L.; Roman, M.; Tomíček, P. The main cannabinoids content in hashish samples seized in Israel and Czech Republic. Isr. J. Plant Sci. 2016, 63, 182–190. [Google Scholar] [CrossRef]
- Altman, J.; Das, G.D. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 1965, 124, 319–335. [Google Scholar] [CrossRef]
- Bond, A.M.; Ming, G.-L.; Song, H. Adult Mammalian Neural Stem Cells and Neurogenesis: Five Decades Later. Cell Stem Cell 2015, 17, 385–395. [Google Scholar] [CrossRef]
- Galve-Roperh, I.; Chiurchiù, V.; Díaz-Alonso, J.; Bari, M.; Guzmán, M.; Maccarrone, M. Cannabinoid receptor signaling in progenitor/stem cell proliferation and differentiation. Prog. Lipid Res. 2013, 52, 633–650. [Google Scholar] [CrossRef]
- Oddi, S.; Scipioni, L.; Maccarrone, M. Endocannabinoid system and adult neurogenesis: A focused review. Curr. Opin. Pharmacol. 2020, 50, 25–32. [Google Scholar] [CrossRef]
- Valeri, A.; Chiricosta, L.; Gugliandolo, A.; Pollastro, F.; Mazzon, E. Will Cannabigerol Trigger Neuroregeneration after a Spinal Cord Injury? An In Vitro Answer from NSC-34 Scratch-Injured Cells Transcriptome. Pharmaceuticals 2022, 15, 117. [Google Scholar] [CrossRef]
- O’Sullivan, S.E. An update on PPAR activation by cannabinoids. Br. J. Pharm. 2016, 173, 1899–1910. [Google Scholar] [CrossRef]
- Esposito, G.; Scuderi, C.; Valenza, M.; Togna, G.I.; Latina, V.; De Filippis, D.; Cipriano, M.; Carratù, M.R.; Iuvone, T.; Steardo, L. Cannabidiol reduces Aβ-induced neuroinflammation and promotes hippocampal neurogenesis through PPARγ involvement. PLoS ONE 2011, 6, e28668. [Google Scholar] [CrossRef]
- Compagnucci, C.; Di Siena, S.; Bustamante, M.B.; Di Giacomo, D.; Di Tommaso, M.; Maccarrone, M.; Grimaldi, P.; Sette, C. Type-1 (CB1) cannabinoid receptor promotes neuronal differentiation and maturation of neural stem cells. PLoS ONE 2013, 8, e54271. [Google Scholar] [CrossRef]
- Aguareles, J.; Paraíso-Luna, J.; Palomares, B.; Bajo-Grañeras, R.; Navarrete, C.; Ruiz-Calvo, A.; García-Rincón, D.; García-Taboada, E.; Guzmán, M.; Muñoz, E.; et al. Oral administration of the cannabigerol derivative VCE-003.2 promotes subventricular zone neurogenesis and protects against mutant huntingtin-induced neurodegeneration. Transl. Neurodegener. 2019, 8, 9. [Google Scholar] [CrossRef]
- Shinjyo, N.; Di Marzo, V. The effect of cannabichromene on adult neural stem/progenitor cells. Neurochem. Int. 2013, 63, 432–437. [Google Scholar] [CrossRef]
- Cashman, N.R.; Durham, H.D.; Blusztajn, J.K.; Oda, K.; Tabira, T.; Shaw, I.T.; Dahrouge, S.; Antel, J.P. Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 1992, 194, 209–221. [Google Scholar] [CrossRef]
- Maier, O.; Böhm, J.; Dahm, M.; Brück, S.; Beyer, C.; Johann, S. Differentiated NSC-34 motoneuron-like cells as experimental model for cholinergic neurodegeneration. Neurochem. Int. 2013, 62, 1029–1038. [Google Scholar] [CrossRef]
- Dell’Anno, M.T.; Wang, X.; Onorati, M.; Li, M.; Talpo, F.; Sekine, Y.; Ma, S.; Liu, F.; Cafferty, W.B.J.; Sestan, N.; et al. Human neuroepithelial stem cell regional specificity enables spinal cord repair through a relay circuit. Nat. Commun. 2018, 9, 3419. [Google Scholar] [CrossRef]
- Kanakasabai, S.; Pestereva, E.; Chearwae, W.; Gupta, S.K.; Ansari, S.; Bright, J.J. PPARγ agonists promote oligodendrocyte differentiation of neural stem cells by modulating stemness and differentiation genes. PLoS ONE 2012, 7, e50500. [Google Scholar] [CrossRef]
- Zang, D.W.; Lopes, E.C.; Cheema, S.S. Loss of synaptophysin-positive boutons on lumbar motor neurons innervating the medial gastrocnemius muscle of the SOD1G93A G1H transgenic mouse model of ALS. J. Neurosci. Res. 2005, 79, 694–699. [Google Scholar] [CrossRef]
- Navarro, G.; Varani, K.; Lillo, A.; Vincenzi, F.; Rivas-Santisteban, R.; Raïch, I.; Reyes-Resina, I.; Ferreiro-Vera, C.; Borea, P.A.; Sánchez de Medina, V.; et al. Pharmacological data of cannabidiol- and cannabigerol-type phytocannabinoids acting on cannabinoid CB1, CB2 and CB1/CB2 heteromer receptors. Pharmacol. Res. 2020, 159, 104940. [Google Scholar] [CrossRef]
- Moreno-Martet, M.; Mestre, L.; Loría, F.; Guaza, C.; Fernández-Ruiz, J.; de Lago, E. Identification of receptors and enzymes for endocannabinoids in NSC-34 cells: Relevance for in vitro studies with cannabinoids in motor neuron diseases. Neurosci. Lett. 2012, 508, 67–72. [Google Scholar] [CrossRef][Green Version]
- Vadodaria, K.C.; Brakebusch, C.; Suter, U.; Jessberger, S. Stage-specific functions of the small Rho GTPases Cdc42 and Rac1 for adult hippocampal neurogenesis. J. Neurosci. 2013, 33, 1179–1189. [Google Scholar] [CrossRef]
- Gehman, L.T.; Meera, P.; Stoilov, P.; Shiue, L.; O’Brien, J.E.; Meisler, M.H.; Ares, M.; Otis, T.S.; Black, D.L. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012, 26, 445–460. [Google Scholar] [CrossRef]
- Cho, A.; Tang, Y.; Davila, J.; Deng, S.; Chen, L.; Miller, E.; Wernig, M.; Graef, I.A. Calcineurin signaling regulates neural induction through antagonizing the BMP pathway. Neuron 2014, 82, 109–124. [Google Scholar] [CrossRef]
- Nguyen, M.D.; Mushynski, W.E.; Julien, J.P. Cycling at the interface between neurodevelopment and neurodegeneration. Cell Death Differ. 2002, 9, 1294–1306. [Google Scholar] [CrossRef]
- Allnutt, A.B.; Waters, A.K.; Kesari, S.; Yenugonda, V.M. Physiological and Pathological Roles of Cdk5: Potential Directions for Therapeutic Targeting in Neurodegenerative Disease. ACS Chem. Neurosci. 2020, 11, 1218–1230. [Google Scholar] [CrossRef]
- Li, W.; Allen, M.E.; Rui, Y.; Ku, L.; Liu, G.; Bankston, A.N.; Zheng, J.Q.; Feng, Y. p39 Is Responsible for Increasing Cdk5 Activity during Postnatal Neuron Differentiation and Governs Neuronal Network Formation and Epileptic Responses. J. Neurosci. 2016, 36, 11283–11294. [Google Scholar] [CrossRef]
- Peng, Z.; Li, X.; Fu, M.; Zhu, K.; Long, L.; Zhao, X.; Chen, Q.; Deng, D.Y.B.; Wan, Y. Inhibition of Notch1 signaling promotes neuronal differentiation and improves functional recovery in spinal cord injury through suppressing the activation of Ras homolog family member A. J. Neurochem. 2019, 150, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.C.; Tönges, L.; Barski, E.; Michel, U.; Bähr, M.; Lingor, P. ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS. Cell Death Dis. 2014, 5, e1225. [Google Scholar] [CrossRef] [PubMed]
- Nicole, O.; Pacary, E. CaMKIIβ in Neuronal Development and Plasticity: An Emerging Candidate in Brain Diseases. Int. J. Mol. Sci. 2020, 21, 7272. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, C.; Chiarlone, A.; Bellocchio, L.; Resel, E.; Pruunsild, P.; García-Rincón, D.; Sendtner, M.; Timmusk, T.; Lutz, B.; Galve-Roperh, I.; et al. The CB1 cannabinoid receptor signals striatal neuroprotection via a PI3K/Akt/mTORC1/BDNF pathway. Cell Death Differ. 2015, 22, 1618–1629. [Google Scholar] [CrossRef]
- Bathina, S.; Das, U.N. Brain-derived neurotrophic factor and its clinical implications. Arch. Med. Sci. 2015, 11, 1164–1178. [Google Scholar] [CrossRef]
- Ferreira, F.F.; Ribeiro, F.F.; Rodrigues, R.S.; Sebastião, A.M.; Xapelli, S. Brain-Derived Neurotrophic Factor (BDNF) Role in Cannabinoid-Mediated Neurogenesis. Front. Cell. Neurosci. 2018, 12, 441. [Google Scholar] [CrossRef]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Pizzicannella, J.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Extracellular Vesicles Derived from Human Gingival Mesenchymal Stem Cells: A Transcriptomic Analysis. Genes 2020, 11, 118. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
| Gene | Ctrl vs. CBNR50 | Ctrl vs. CBNR20 | Ctrl vs. CBNR10 | Ctrl vs. CBNR7.5 | Ctrl vs. CBNR5 |
|---|---|---|---|---|---|
| Akt1 | 0 | 0.14 | 0.23 | 0 | 0.09 |
| Bdnf | 0.91 | 1.16 | 0 | 0 | 0 |
| Capn1 | 0.63 | 0.45 | 0 | 1.01 | 0.33 |
| Camk2b | −1.18 | −0.64 | −1.06 | −0.68 | −0.89 |
| Cdc42 | −0.20 | −0.25 | 0 | 0.30 | 0.10 |
| Cdk5r1 | 0.36 | 0 | 0 | 0.63 | −0.47 |
| Cdk5r2 | 0.40 | 0.31 | 0.52 | 0.53 | 0 |
| Creb1 | 0.18 | 0 | 0 | 0.42 | 0.19 |
| Limk1 | 0.23 | 0 | −0.28 | −0.61 | −0.20 |
| Map2 | 0 | 0 | 0 | 0.91 | 0 |
| Mapk8 | 0 | 0 | 0 | 0.35 | 0.28 |
| Nes | −0.79 | 0.66 | 0 | 0 | 0 |
| Neurod1 | 0 | 0 | 0 | 0.40 | 0 |
| Ppp3cb | −0.15 | 0 | 0.16 | 0.25 | 0.14 |
| Ppp3cc | 0 | 0 | 0 | 0.40 | 0 |
| Rbfox2 | 0 | 0 | 0.19 | 0.20 | 0.16 |
| Rock1 | −0.28 | −0.22 | −0.25 | 0 | 0 |
| Rock2 | −0.16 | 0 | 0 | 0.33 | 0.11 |
| Rps6 | −0.16 | 0 | −1.27 | −1.68 | −1.44 |
| Syp | 0.52 | 0.66 | 0.46 | −1.04 | 0.57 |
| Vim | −0.39 | −0.09 | 0 | −0.11 | 0 |
| Gene | Ctrl vs. CBNR50 | Ctrl vs. CBNR20 | Ctrl vs. CBNR10 | Ctrl vs. CBNR7.5 | Ctrl vs. CBNR5 |
|---|---|---|---|---|---|
| Cnr1 | 0.25 | 0.38 | 0.44 | 0.26 | 0.33 |
| Pparg | 0 | −0.73 | 0 | 0.69 | 0 |
| Trpv2 | 0 | 0 | −1.14 | 0 | −1.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valeri, A.; Chiricosta, L.; Gugliandolo, A.; Pollastro, F.; Salamone, S.; Zingale, V.D.; Silvestro, S.; Mazzon, E. Cannabinerol and NSC-34 Transcriptomic Analysis: Is the Dose Who Makes Neuronal Differentiation? Int. J. Mol. Sci. 2022, 23, 7541. https://doi.org/10.3390/ijms23147541
Valeri A, Chiricosta L, Gugliandolo A, Pollastro F, Salamone S, Zingale VD, Silvestro S, Mazzon E. Cannabinerol and NSC-34 Transcriptomic Analysis: Is the Dose Who Makes Neuronal Differentiation? International Journal of Molecular Sciences. 2022; 23(14):7541. https://doi.org/10.3390/ijms23147541
Chicago/Turabian StyleValeri, Andrea, Luigi Chiricosta, Agnese Gugliandolo, Federica Pollastro, Stefano Salamone, Valeria Domenica Zingale, Serena Silvestro, and Emanuela Mazzon. 2022. "Cannabinerol and NSC-34 Transcriptomic Analysis: Is the Dose Who Makes Neuronal Differentiation?" International Journal of Molecular Sciences 23, no. 14: 7541. https://doi.org/10.3390/ijms23147541
APA StyleValeri, A., Chiricosta, L., Gugliandolo, A., Pollastro, F., Salamone, S., Zingale, V. D., Silvestro, S., & Mazzon, E. (2022). Cannabinerol and NSC-34 Transcriptomic Analysis: Is the Dose Who Makes Neuronal Differentiation? International Journal of Molecular Sciences, 23(14), 7541. https://doi.org/10.3390/ijms23147541







