Bioengineered Silkworm for Producing Cocoons with High Fibroin Content for Regenerated Fibroin Biomaterial-Based Applications
Abstract
1. Introduction
2. Results
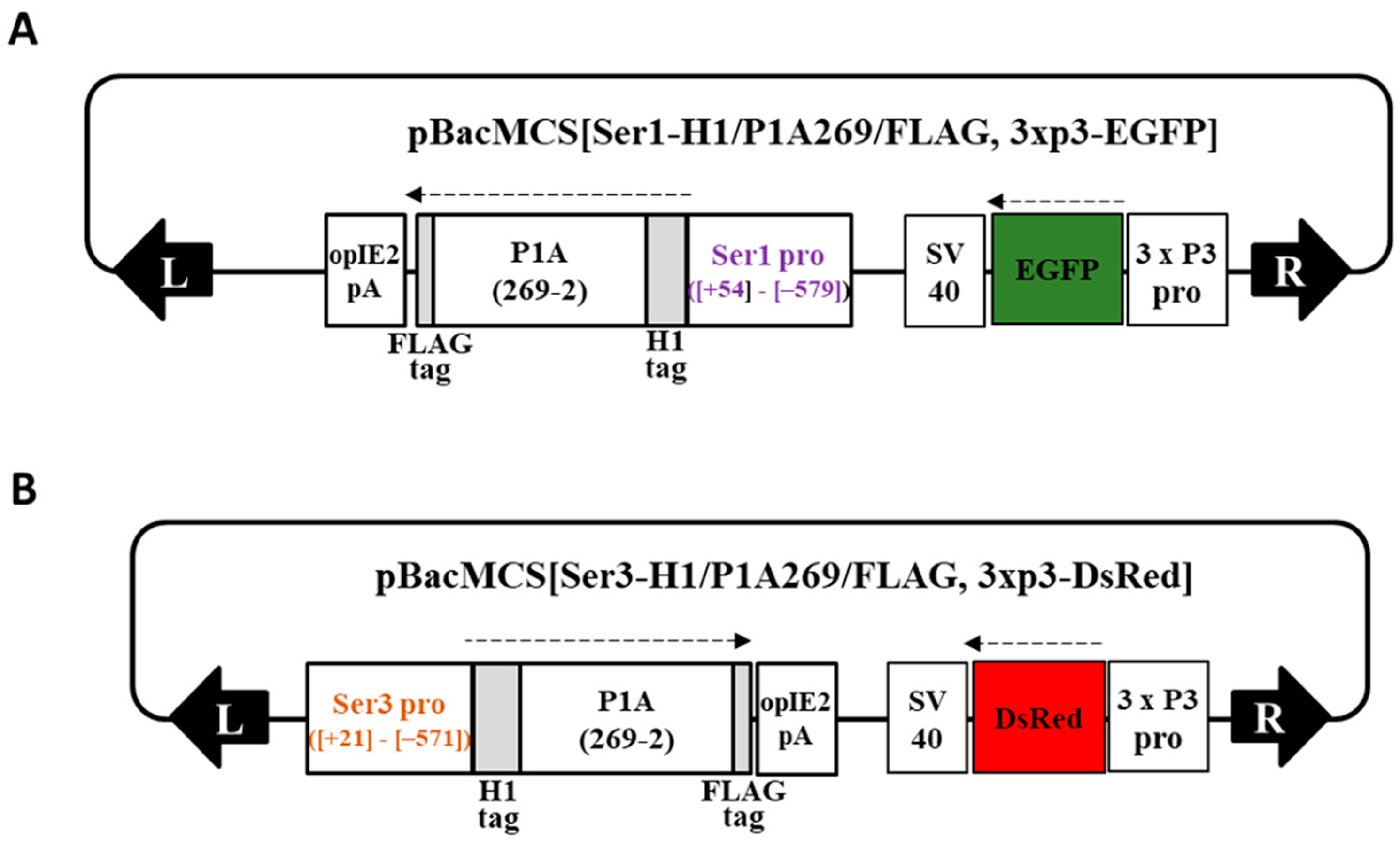
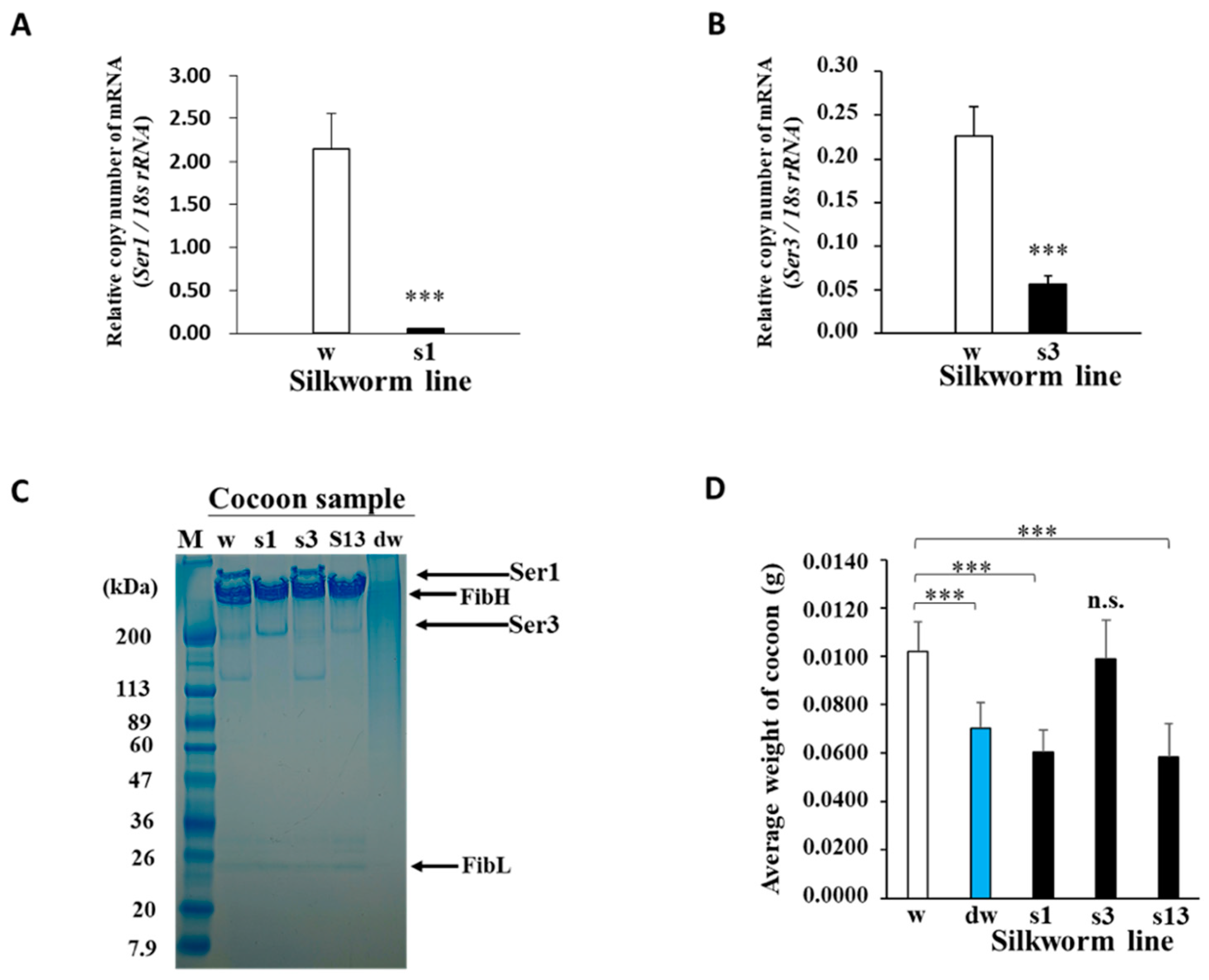
2.1. Generation of Bioengineered Silkworms Producing Cocoons with High Fibroin Content
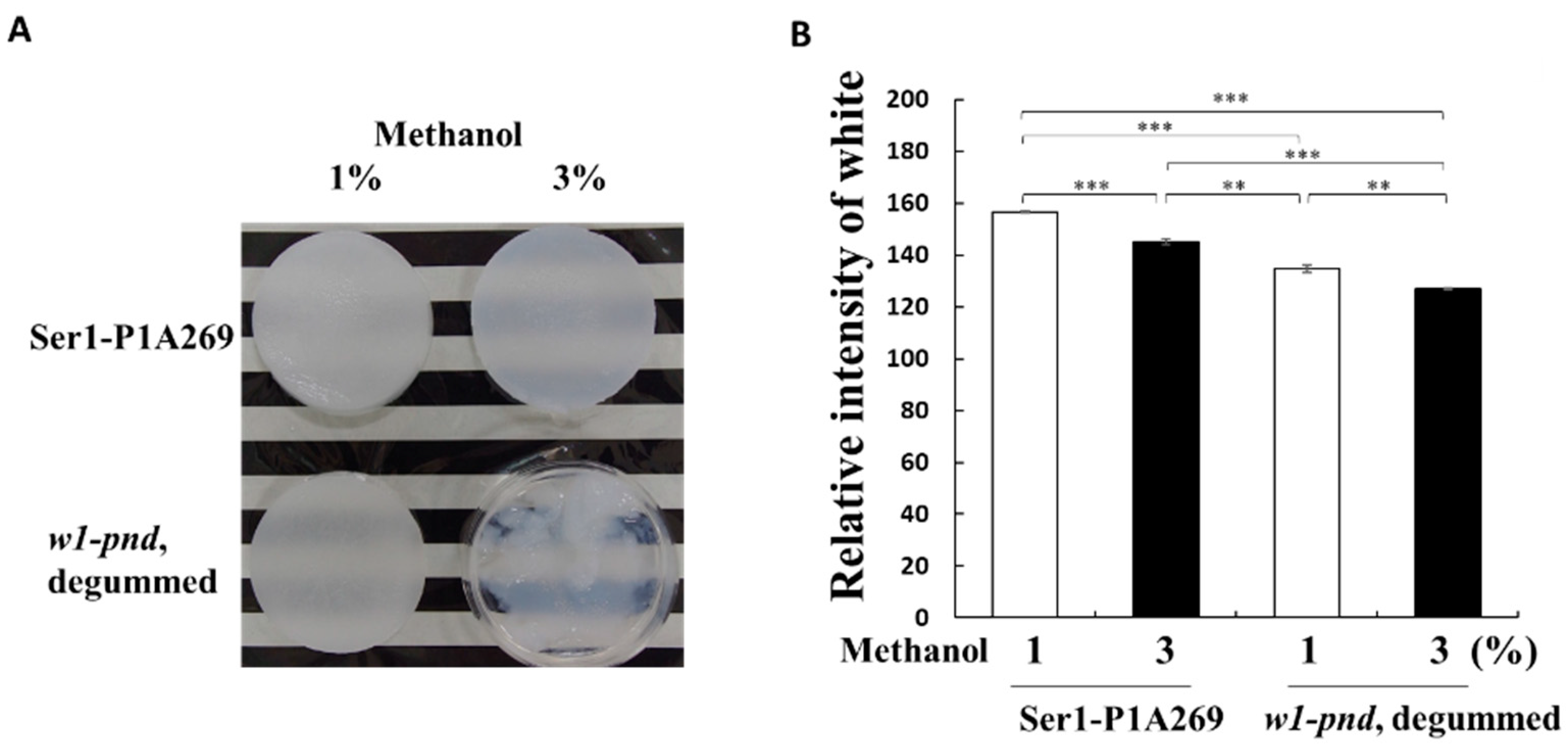
2.2. Recrystallization of Intact Fibroin from the Ser1-P1A269 Cocoon to Form a 3D Sponge
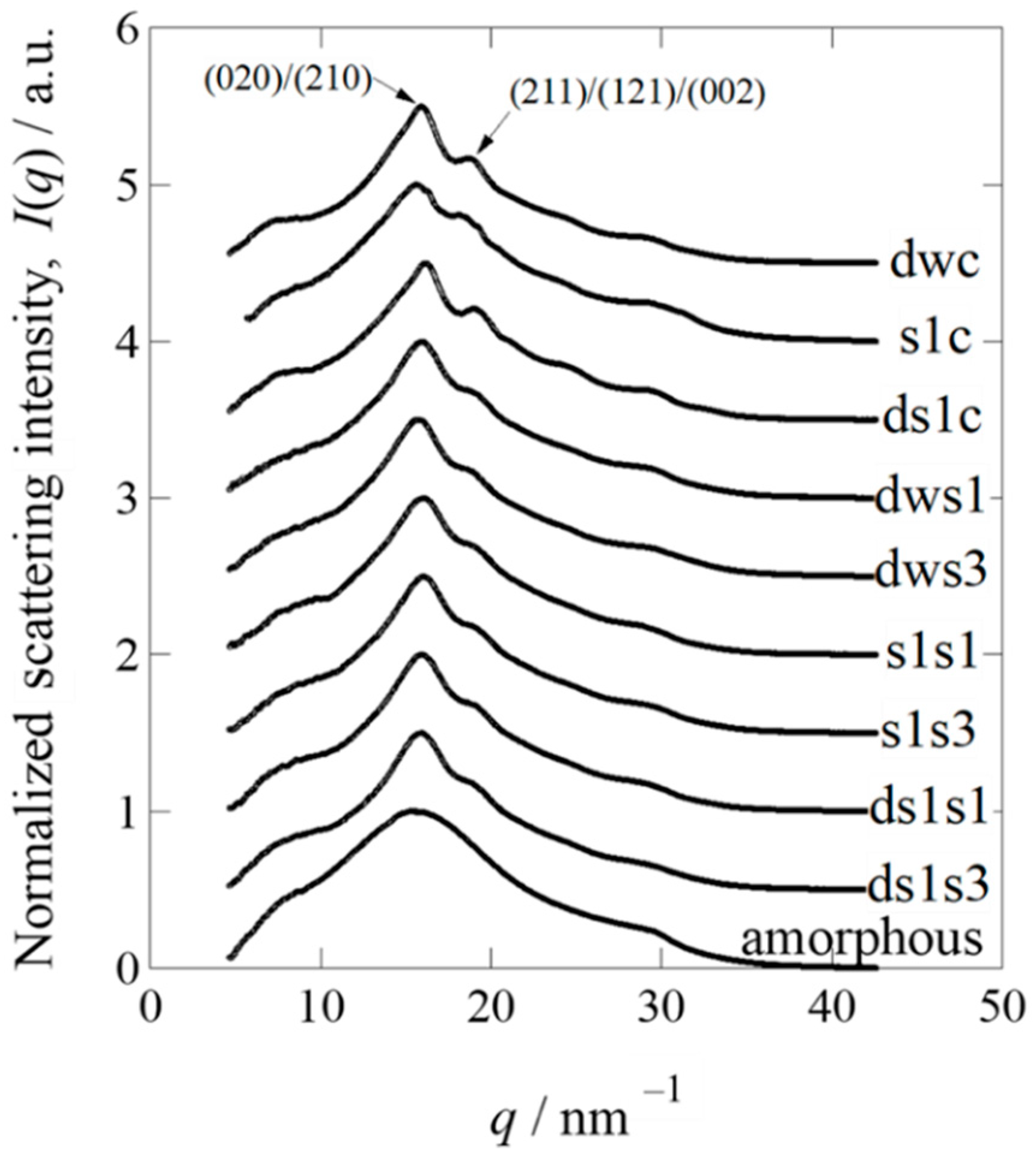
2.3. Wide-Angle X-ray Diffraction Analysis of Cocoons and Regenerated Fibroin Sponges
2.4. Proliferation and Differentiation of Chondrogenic Cells on the Fibroin Material
3. Discussion
4. Materials and Methods
4.1. Silkworm Strains and Cultured Cells
4.2. Generation of the Transgenic Silkworm Line Carrying MSG-Specific, Promoter-Driven P1A
4.3. SDS–PAGE Analysis of Proteins from Cocoons and Protein Band Analysis
4.4. Quantitative RT-PCR Analysis of MSG mRNA
4.5. Fibroin Sponge Preparation
4.6. Synchrotron WAXD Analysis
4.7. Cultivation of ATDC5 Cells on Fibroin Coats
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain and P25 with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef]
- Takasu, Y.; Yamada, H.; Tsubouchi, K. Isolation of three main sericin components from the cocoon of the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2002, 66, 2715–2718. [Google Scholar] [CrossRef]
- Teramoto, H.; Nakajima, K.; Takabayashi, C. Chemical modification of silk sericin in lithium chloride/dimethyl sulfoxide solvent with 4-cyanophenyl isocyanate. Biomacromolecules 2004, 5, 1392–1398. [Google Scholar] [CrossRef]
- Kludkiewicz, B.; Takasu, Y.; Fedric, R.; Tamura, T.; Sehnal, F.; Zurovec, M. Structure and expression of the silk adhesive protein Ser2 in Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 938–946. [Google Scholar] [CrossRef]
- Takasu, Y.; Hata, T.; Uchino, K.; Zhang, Q. Identification of Ser2 proteins as major sericin components in the non-cocoon silk of Bombyx mori. Insect Biochem. Mol. Biol. 2010, 40, 339–344. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, B. Biodegradation of silk biomaterials. Int. J. Mol. Sci. 2009, 31, 1514–1524. [Google Scholar] [CrossRef]
- Khademolqorani, S.; Tavanai, H.; Chronakis, I.S.; Boisen, A.; Ajallouejan, F. The determinant role of fabrication technique in final characteristics of scaffolds for tissue engineering applications: A focus on silk fibroin-based scaffolds. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 122, 111867. [Google Scholar] [CrossRef]
- Oral, C.B.; Yetiskin, B.; Okey, O. Stretchable silk fibroin hydrogels. Int. J. Biol. Macromol. 2020, 161, 1371–1380. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, T.; Jiang, C. Biomacromolecules as carriers in drug delivery and tissue engineering. Acta Pharm. Sinica B 2018, 8, 34–50. [Google Scholar] [CrossRef]
- Kotani, E.; Yamamoto, N.; Kobayashi, I.; Uchino, K.; Muto, S.; Ijiri, H.; Shimabukuro, J.; Tamura, T.; Sezutsu, H.; Mori, H. Cell proliferation by silk gut incorporating FGF-2 protein microcrystals. Sci. Rep. 2015, 5, 11051. [Google Scholar] [CrossRef]
- Maruta, R.; Takaki, K.; Yamaji, Y.; Sezutsu, H.; Mori, H.; Kotani, E. Effects of transgenic silk materials that incorporate FGF-7 protein microcrystals on the proliferation and differentiation of human keratinocytes. FASEB Bioadv. 2020, 2, 734–744. [Google Scholar] [CrossRef]
- Tamada, Y. New process to form a silk fibroin porous 3-D structure. Biomacromolecules 2005, 6, 3100–3106. [Google Scholar] [CrossRef]
- Hashimoto, T.; Kojima, K.; Otaka, A.; Takeda, Y.S.; Tomita, N.; Tamada, Y. Quantification evaluation of fibroblast migration on a silk fibroin surface and TGF β1gene expression. J. Biomater. Sci. Polym. Ed. 2013, 24, 158–169. [Google Scholar] [CrossRef]
- Hirakata, E.; Tomita, N.; Tamada, Y.; Suguro, T.; Nakajima, M.; Kambe, Y.; Yamada, K.; Yamamoto, K.; Kawakami, M.; Otaka, A.; et al. Early tissue formation on whole-area osterchondral defect of rabbit patella by covering with fibroin sponge. J. Biomed. Mater. Res. B Appl. Biomater. 2016, 104, 1474–1482. [Google Scholar] [CrossRef]
- Yagi, T.; Sato, M.; Nakazawa, Y.; Tanaka, K.; Sata, M.; Itoh, K.; Takagi, Y.; Asakura, T. Preparation of double-raschel knitted silk vascular grafts and evaluation of short-term function in a rat abdominal aorta. J. Artif. Organs 2011, 14, 89–99. [Google Scholar] [CrossRef]
- Kuboyama, N.; Kiba, H.; Arai, K.; Uchida, R.; Tanimoto, Y.; Bhawal, U.K.; Abiko, Y.; Miyamoto, S.; Knight, D.; Asakura, T.; et al. Silk fibroin-based scaffolds for bone regeneration. J. Biomed. Mater. Res. B Appl. Biomater. 2013, 101, 295–302. [Google Scholar] [CrossRef]
- Sugihara, A.; Sugiura, K.; Morita, H.; Ninagawa, T.; Tubouchi, K.; Tobe, R.; Izumiya, M.; Horio, T.; Abraham, N.G.; Ikehara, S. Promotive effects of a silk film on epidermal recovery from full-thickness skin wounds. Proc. Soc. Exp. Biol. Med. 2000, 225, 58–64. [Google Scholar] [CrossRef]
- Kundu, B.; Rajkhowa, R.; Kundu, C.; Wang, X. Silk fibroin biomaterials for tissue regenerations. Adv. Drug Deliv. Rev. 2013, 65, 457–470. [Google Scholar] [CrossRef]
- Yamamoto, M.; Takahashi-Nakaguchi, A.; Matsushima-Hibiya, Y.; Nakano, T.; Totsuka, Y.; Imanishi, S.; Mitsuhashi, J.; Watanabe, M.; Nakagama, H.; Sugimura, T.; et al. Nucleotide sequence and chromosomal localization of the gene for pierisin-1, a DNA-ribosylating protein, in the cabbage butterfly Pieris rapae. Genetica 2011, 139, 1251–1258. [Google Scholar] [CrossRef]
- Otsuki, R.; Yamamoto, M.; Matsumoto, E.; Iwamoto, S.; Sezutsu, H.; Suzui, M.; Takaki, K.; Wakabayashi, K.; Mori, H.; Kotani, E. Bioengineered silkworms with butterfly cytotoxin-modified silk glands produce sericin cocoons with a utility for a new biomaterial. Proc. Natl. Acad. Sci. USA 2017, 114, 6740–6745. [Google Scholar] [CrossRef]
- Koyama, K.; Wakabayashi, K.; Masutani, M.; Koiwai, K.; Watanabe, M.; Yamazaki, S.; Kono, T.; Miki, K.; Sugiura, T. Presence in Pieris rapae of cytotoxic activity against human carcinoma cells. Jpn. J. Cancer Res. 1996, 87, 1259–1262. [Google Scholar] [CrossRef]
- Watanabe, M.; Kono, T.; Matsushima-Hibiya, Y.; Kanazawa, T.; Nishisaka, N.; Kishimoto, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: Possible involvement of ADP-ribosylation in its activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10608–10613. [Google Scholar] [CrossRef]
- Takamura-Enya, T.; Watanabe, M.; Totsuka, Y.; Kanazawa, T.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc. Natl. Acad. Sci. USA 2001, 98, 12414–12419. [Google Scholar] [CrossRef]
- Kanazawa, K.; Watanabe, M.; Matsushima-Hibiya, Y.; Kono, T.; Tanaka, N.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Distinct roles for the N- and C-terminal regions in the cytotoxicity of pierisin-1, a putative ADP-ribosylating toxin from cabbage butterfly, against mammalian cells. Proc. Natl. Acad. Sci. USA 2001, 98, 2226–2231. [Google Scholar] [CrossRef]
- Campbell, A.E.; Bennett, D. Targeting protein function: The expanding toolkit for conditional disruption. Biochem. J. 2016, 473, 2573–2589. [Google Scholar] [CrossRef][Green Version]
- Lander, E.S. The heroes of CRISPR. Cell 2016, 164, 18–28. [Google Scholar] [CrossRef]
- Jiao, Z.; Song, Y.; Jin, Y.; Zhang, C.; Peng, D.; Chen, Z.; Chang, P.; Kundu, S.C.; Wang, G.; Wang, Z.; et al. In vivo characterizations of the immune properties of sericin: An ancient material with emerging value in biomedical applications. Macromol. Biosci. 2017, 17, 1700229. [Google Scholar] [CrossRef]
- Shukunami, C.; Shigeno, C.; Atsumi, T.; Ishizeki, K.; Suzuki, F.; Hiraki, Y. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: Differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-dependent peptide receptor. J. Cell Biol. 1996, 133, 457–468. [Google Scholar] [CrossRef]
- Wang, F.; Xu, H.; Yuan, L.; Ma, S.; Wang, Y.; Duan, X.; Duan, J.; Xiang, Z.; Xia, Q. An optimized sericin-1 expression system for mass-producing recombinant proteins in the middle silk glands of transgenic silkworms. Transgenic Res. 2013, 22, 925–938. [Google Scholar] [CrossRef]
- Tatematsu, K.; Kobayashi, I.; Uchino, K.; Sezutsu, H.; Iizuka, T.; Yoneyama, N.; Tamura, T. Construction of a binary transgenic gene expression system for recombinant protein production in the middle silk gland of the silkworm Bombyx mori. Transgenic Res. 2010, 19, 473–487. [Google Scholar] [CrossRef]
- Takagi, H.; Igarashi, N.; Mori, T.; Saijo, S.; Ohta, H.; Nagatani, Y.; Kosuge, T.; Shimizu, N. Upgrade of small angle x-ray scattering beamline BL-6A at the photon factory. AIP Conf. Proc. 2016, 1741, 030018. [Google Scholar]
- Shimizu, N.; Mori, T.; Igarashi, N.; Ohta, H.; Nagatani, Y.; Kosuge, T.; Ito, K. Refurbishing of Small-Angle X-ray Scattering Beamline, BL-6A at the Photon Factory. J. Phys. Conf. Ser. 2013, 425, 202008. [Google Scholar] [CrossRef]
- Matsumoto, G.; Ueda, T.; Shimoyama, J.; Ijiri, H.; Omi, Y.; Yube, H.; Sugita, Y.; Kubo, K.; Maeda, H.; Kinoshita, Y.; et al. Bone regeneration by polyhedral microcrystals from silkworm virus. Sci. Rep. 2012, 2, 935. [Google Scholar] [CrossRef]
- Groslambert, J.; Prokhorova, E.; Ahel, I. ADP-ribosylation of DNA and RNA. DNA Repair 2021, 105, 103144. [Google Scholar] [CrossRef]
- Ling, S.; Qin, Z.; Li, C.; Huang, W.; Kaplan, D.L.; Buehler, M.J. Polymorphic regenerated silk fibers assembled through bioinspired spinning. Nat. Commun. 2017, 8, 1387. [Google Scholar] [CrossRef]
- Kataoka, K.; Uematsu, I. On the viscosity of liquid silk in the silk gland. Kobunshi Ronbunshu 1977, 34, 7–13. [Google Scholar] [CrossRef]
- Jin, H.-J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef]
- Umehara, C.; Lian, A.A.; Funahashi, Y.; Takaki, K.; Maruta, R.; Ohmaru, Y.; Okahisa, Y.; Aoki, T.; Mori, H.; Kotani, E. Proliferation of mouse embryonic stem cells on substrate coated with intact silkworm sericin. J. Tex. Inst. 2021, 113. ahead of print. [Google Scholar] [CrossRef]
- Sakudoh, T.; Sezutsu, H.; Nakashima, T.; Kobayashi, I.; Fujimoto, H.; Uchino, K.; Banno, Y.; Iwano, H.; Maekawa, H.; Tamura, T.; et al. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc. Natl. Acad. Sci. USA 2007, 104, 8941–8946. [Google Scholar] [CrossRef]
- Tamura, T.; Thibert, C.; Royer, C.; Kanda, T.; Abraham, E.; Kamba, M.; Komoto, N.; Thomas, J.L.; Mauchamp, B.; Chavancy, G.; et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 2000, 18, 81–84. [Google Scholar] [CrossRef]
- Kobayashi, I.; Uchino, K.; Sezutsu, H.; Iizuka, T.; Tamura, T. Development of a new piggyBac vector for generating transgenic silkworms using the kynurenine 3-mono oxygenase gene. J. Insect Biotechnol. Sericol. 2007, 76, 145–148. [Google Scholar]

| (020)/(210) ** | (211)/(121)/(002) | ||||
|---|---|---|---|---|---|
| Samples | Crystallinity | Peak Position/nm−1 | HWHM/nm−1 | Peak Position/nm−1 | HWHM/nm−1 |
| dwc | 0.32 * | 15.67 | 1.54 | 18.71 | 0.81 |
| s1c | 0.26 | 15.45 | 1.83 | 19.15 | 0.59 |
| ds1c | 0.26 | 15.94 | 1.39 | 19.00 | 0.50 |
| dws1 | 0.28 | 15.82 | 1.59 | 19.33 | 1.18 |
| dws3 | 0.24 | 15.60 | 1.53 | 19.13 | 1.03 |
| s1s1 | 0.27 | 15.87 | 1.71 | 19.45 | 1.45 |
| s1s3 | 0.24 | 15.84 | 1.51 | 19.36 | 1.40 |
| ds1s1 | 0.28 | 15.94 | 1.61 | 19.45 | 1.33 |
| ds1s1 | 0.25 | 15.79 | 1.56 | 19.31 | 1.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamano, M.; Hirose, R.; Lye, P.Y.; Takaki, K.; Maruta, R.; On Liew, M.W.; Sakurai, S.; Mori, H.; Kotani, E. Bioengineered Silkworm for Producing Cocoons with High Fibroin Content for Regenerated Fibroin Biomaterial-Based Applications. Int. J. Mol. Sci. 2022, 23, 7433. https://doi.org/10.3390/ijms23137433
Yamano M, Hirose R, Lye PY, Takaki K, Maruta R, On Liew MW, Sakurai S, Mori H, Kotani E. Bioengineered Silkworm for Producing Cocoons with High Fibroin Content for Regenerated Fibroin Biomaterial-Based Applications. International Journal of Molecular Sciences. 2022; 23(13):7433. https://doi.org/10.3390/ijms23137433
Chicago/Turabian StyleYamano, Mana, Ryoko Hirose, Ping Ying Lye, Keiko Takaki, Rina Maruta, Mervyn Wing On Liew, Shinichi Sakurai, Hajime Mori, and Eiji Kotani. 2022. "Bioengineered Silkworm for Producing Cocoons with High Fibroin Content for Regenerated Fibroin Biomaterial-Based Applications" International Journal of Molecular Sciences 23, no. 13: 7433. https://doi.org/10.3390/ijms23137433
APA StyleYamano, M., Hirose, R., Lye, P. Y., Takaki, K., Maruta, R., On Liew, M. W., Sakurai, S., Mori, H., & Kotani, E. (2022). Bioengineered Silkworm for Producing Cocoons with High Fibroin Content for Regenerated Fibroin Biomaterial-Based Applications. International Journal of Molecular Sciences, 23(13), 7433. https://doi.org/10.3390/ijms23137433







