Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina
Abstract
1. Introduction
2. Results
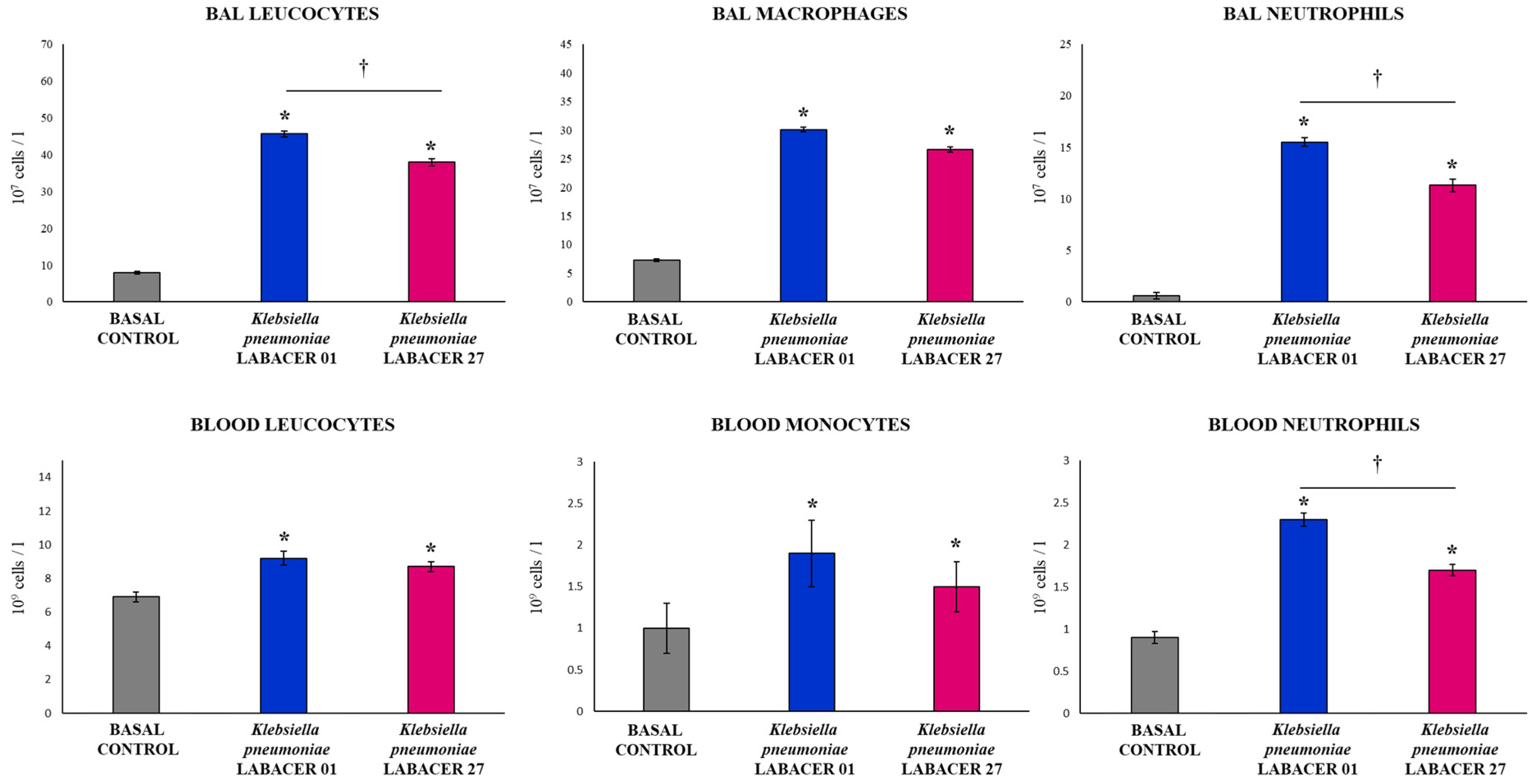
2.1. K. pneumoniae LABACER 01 and LABACER 27 Infect the Respiratory Tract of Immunocompetent Adult Mice
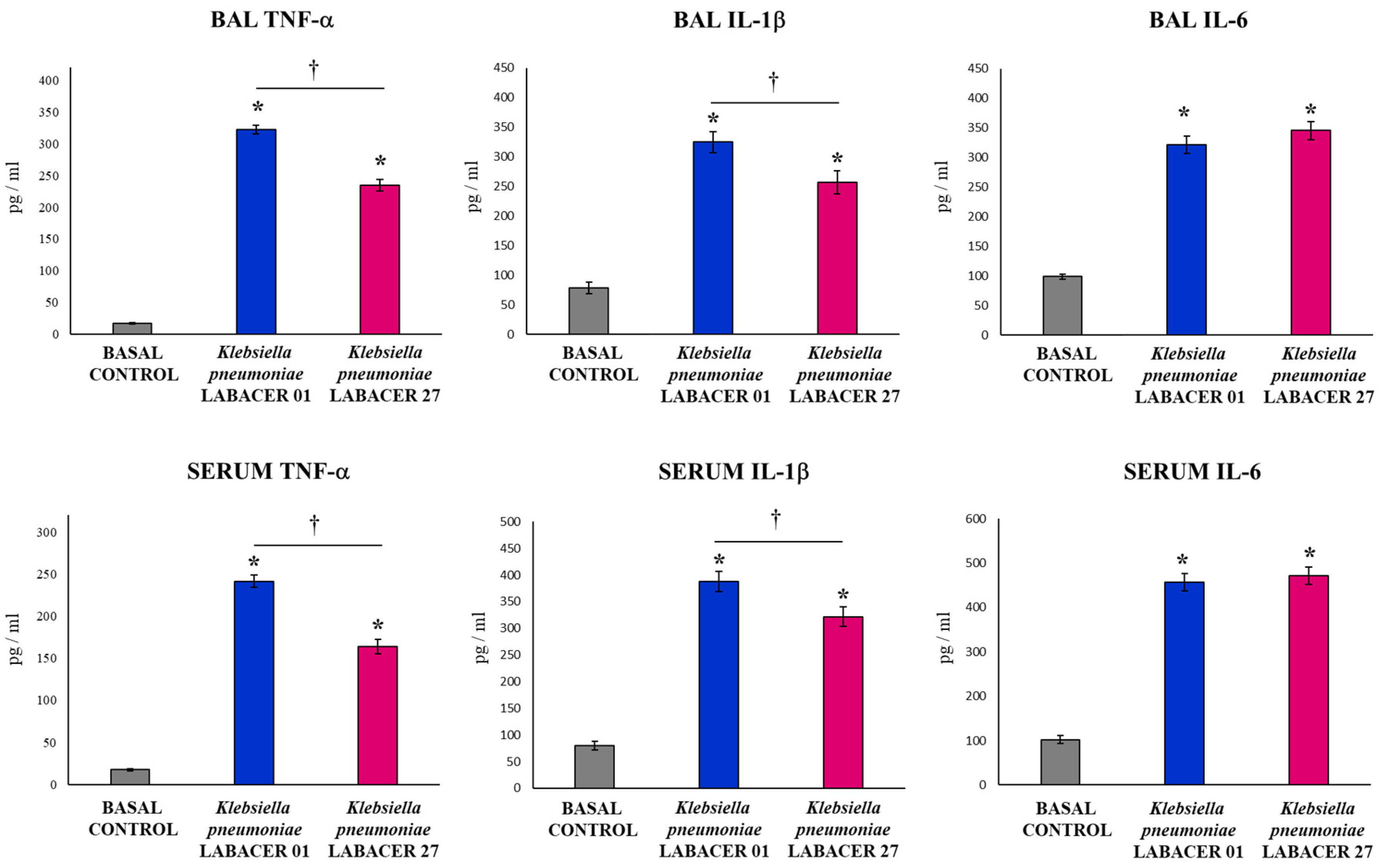
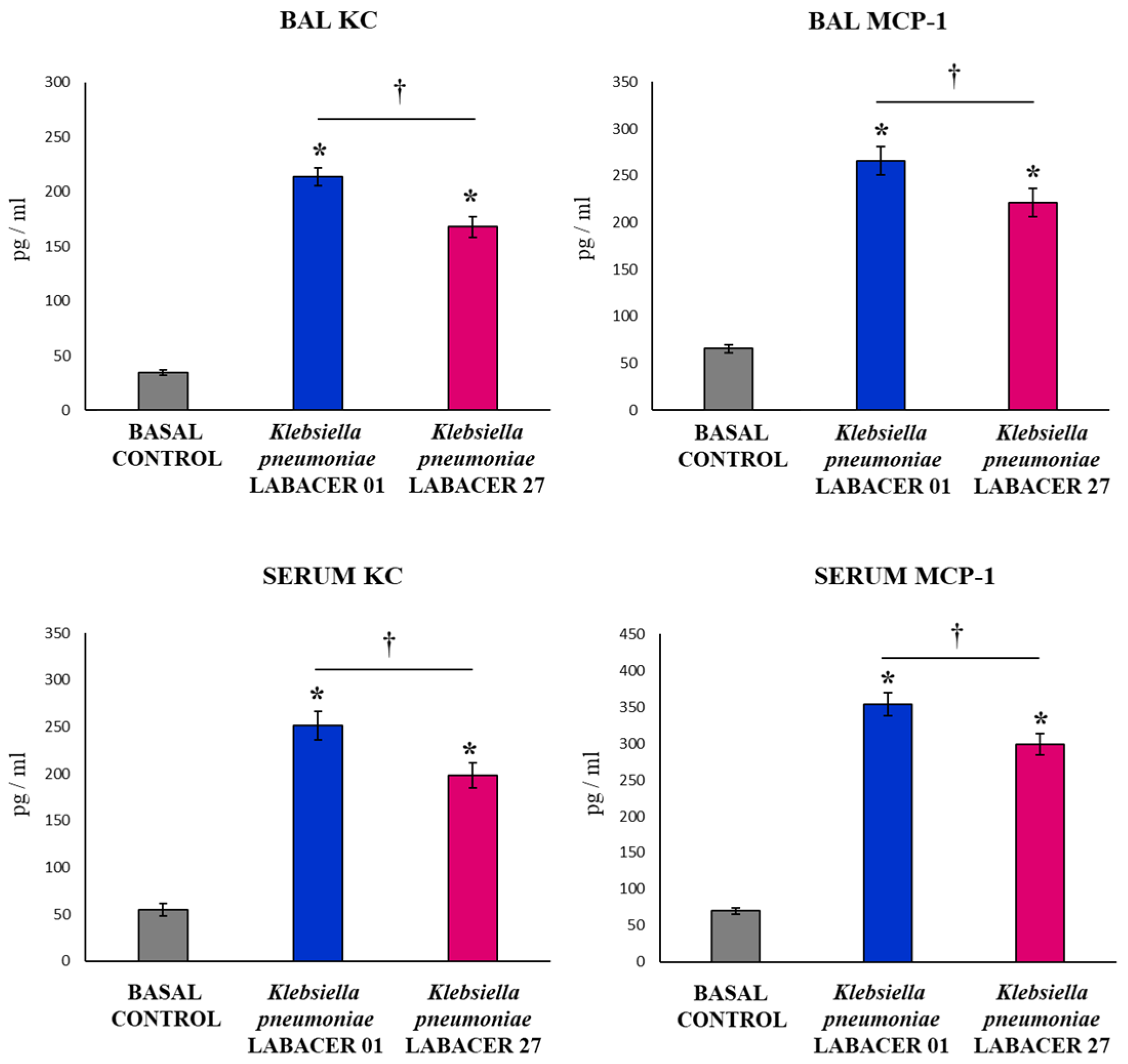
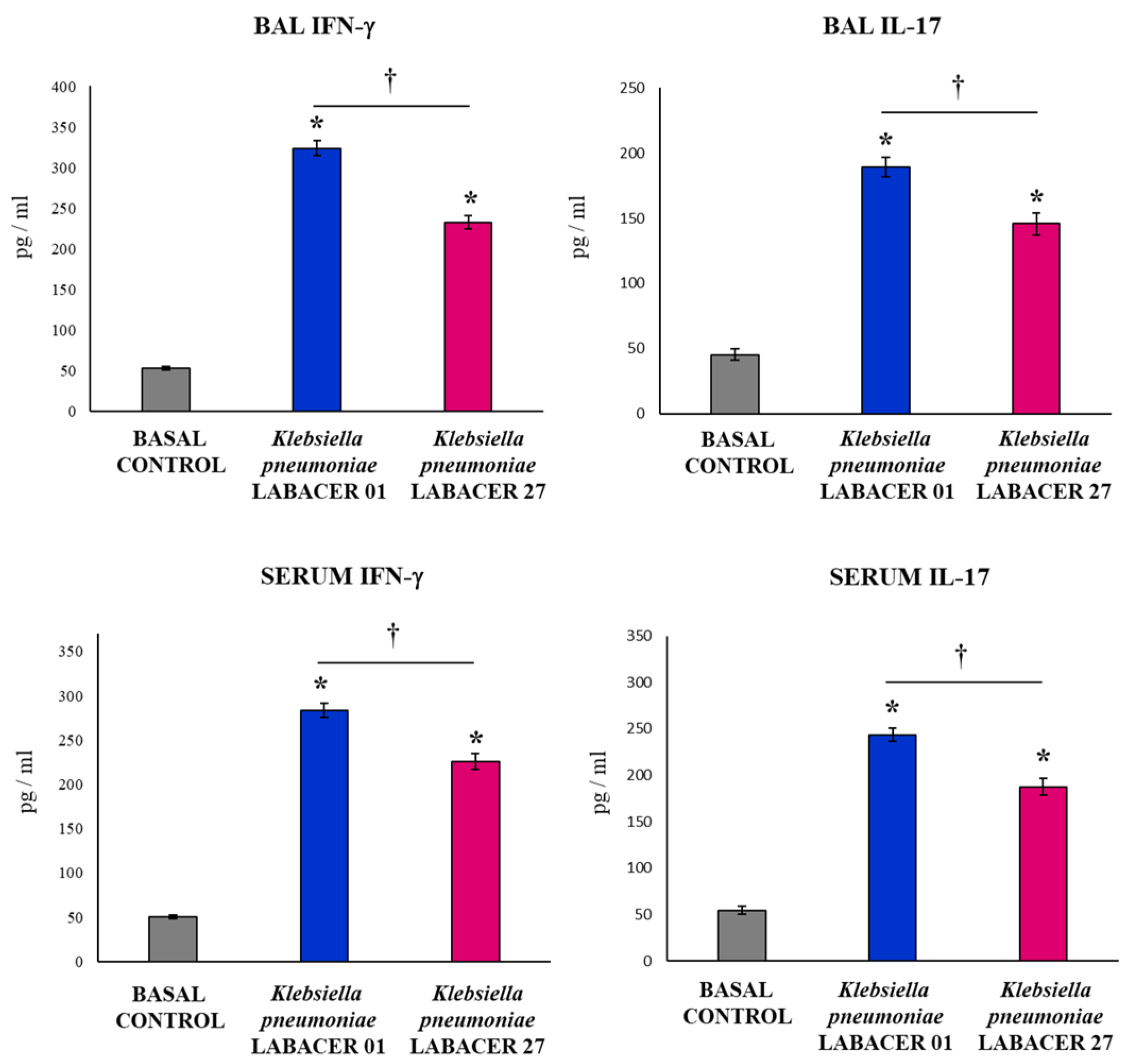
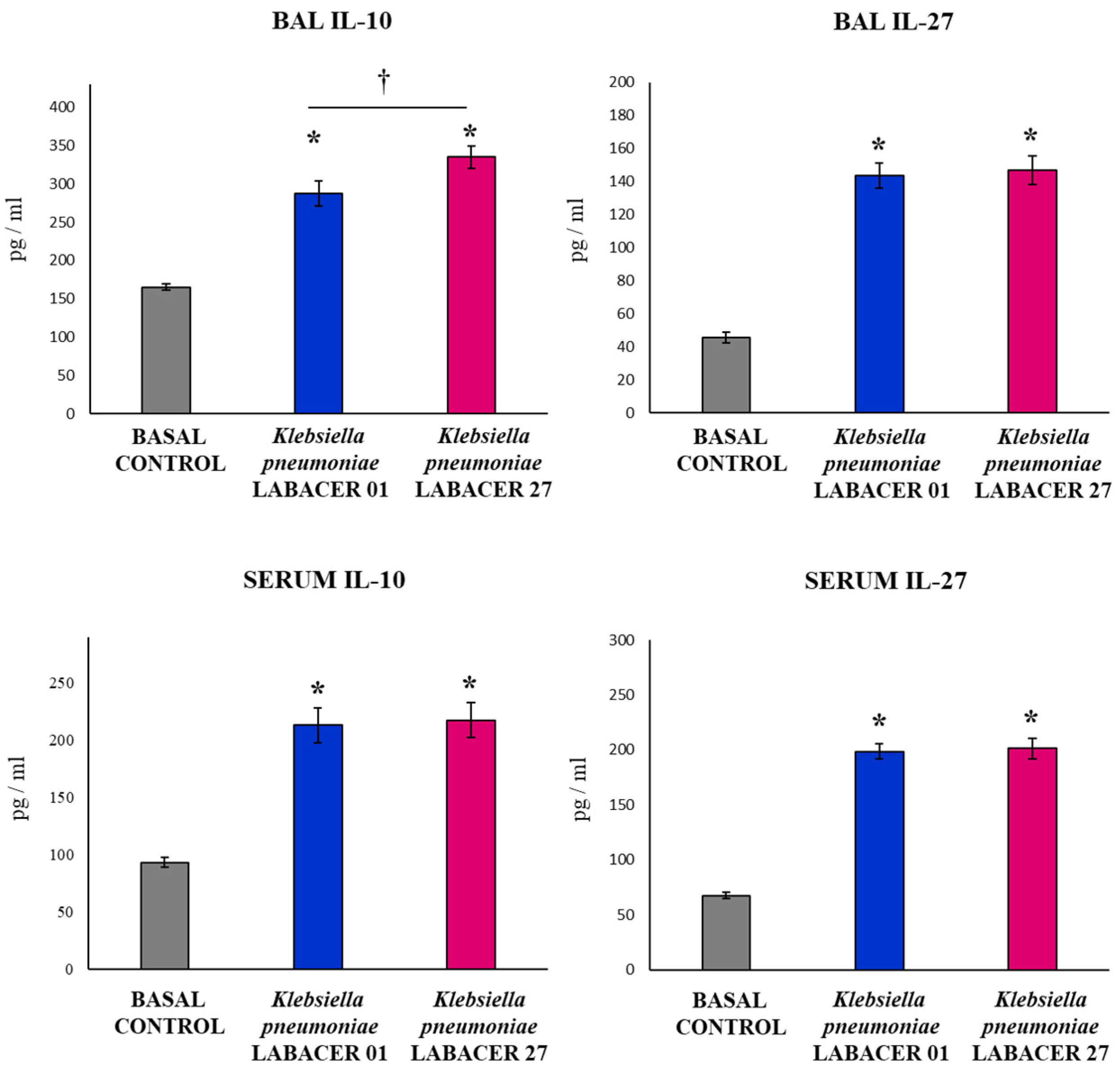
2.2. K. pneumoniae LABACER 01 and LABACER 27 Induce a Marked Respiratory and Systemic Inflammatory Response
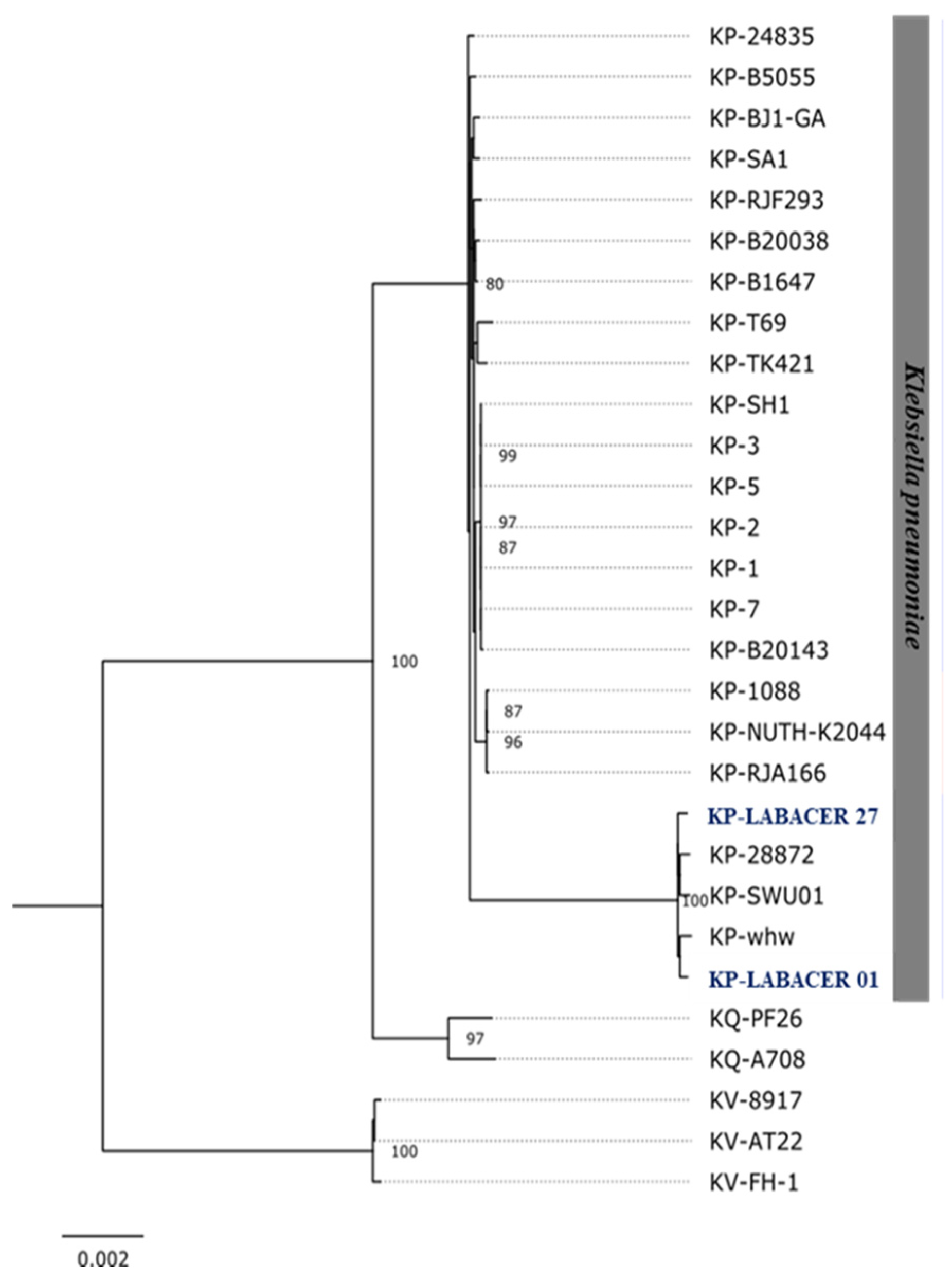
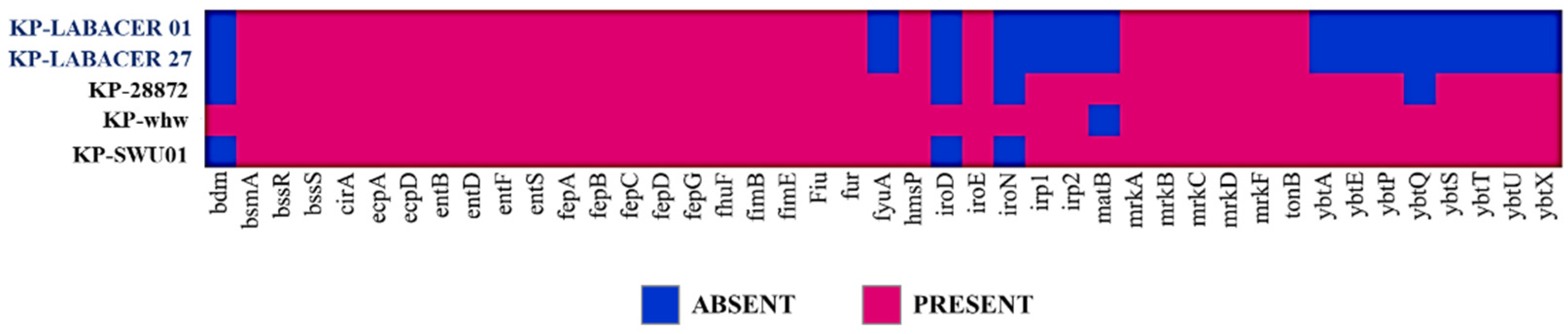
2.3. Genomic Characterization of K. pneumoniae LABACER 01 and LABACER 07
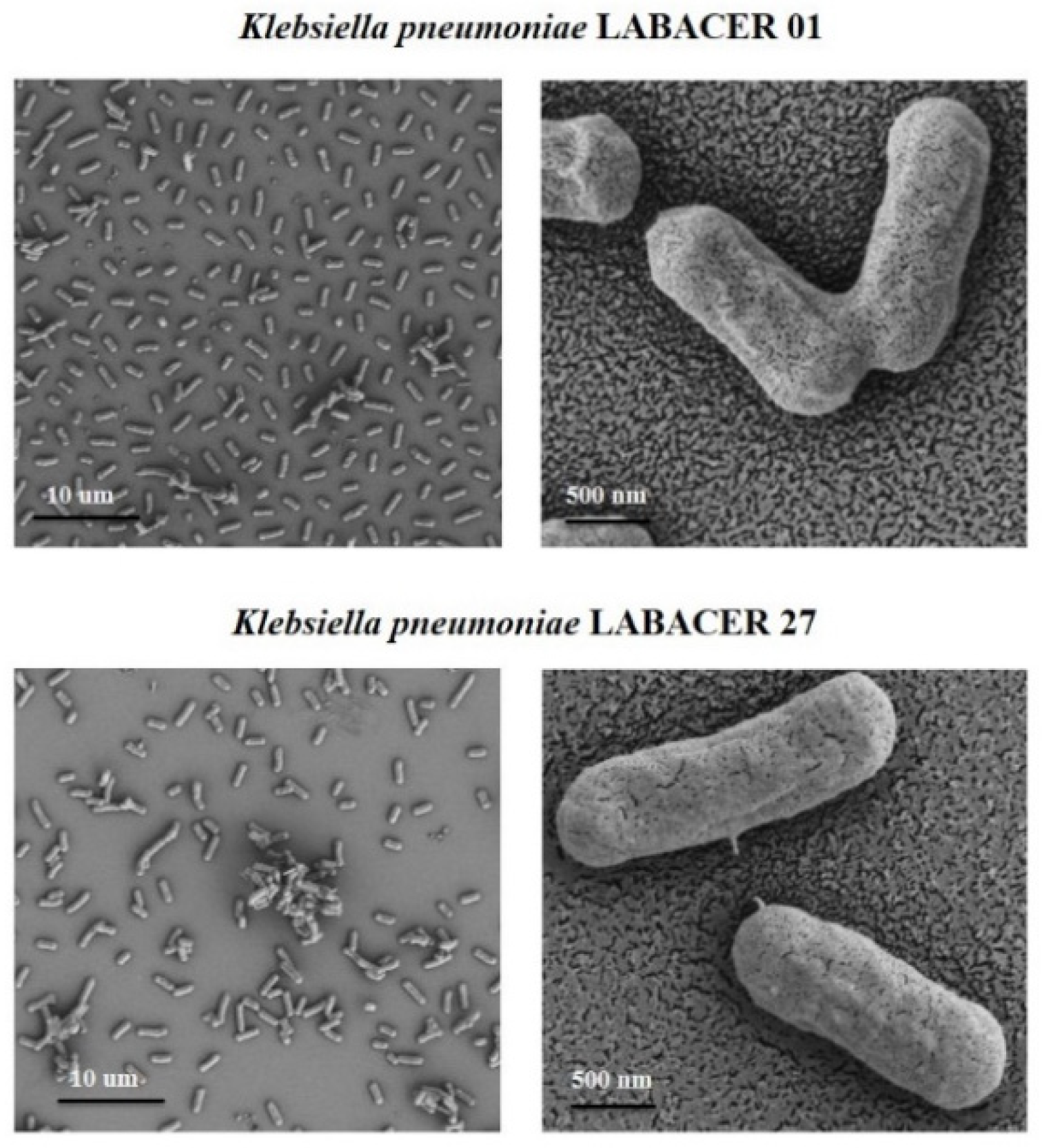
2.4. Morphological Characterization of K. pneumoniae LABACER 01 and LABACER 27
3. Discussion
4. Materials and Methods
4.1. K. pneumoniae LABACER 01 and LABACER 27
4.2. Murine Infection Model
4.3. Respiratory Infection with K. pneumoniae LABACER 01 and LABACER 27
4.4. K. pneumoniae Counts in Lungs
4.5. Lung Damage
4.6. Serum Cytokines and Bronchoalveolar Lavages
4.7. Comparative and Functional Genomics Studies
4.8. Phylogenomic Analysis and Virulence Factor Genes
4.9. Electron Microscopy
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef]
- Antimicrobial Surveillance Program SENTRY, World Health Organization, 2014. Available online: www.who.int (accessed on 3 May 2022).
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (hypermucoviscous)Klebsiella pneumoniae. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef]
- Prokesch, B.C.; TeKippe, M.; Kim, J.; Raj, P.D.; TeKippe, E.M.; Greenberg, E.D. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 2016, 16, e190–e195. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio 2015, 6, e00630-15. [Google Scholar] [CrossRef]
- Patel, P.K.; Russo, T.A.; Karchmer, A.W. Hypervirulent Klebsiella pneumoniae. Open Forum Infect. Dis. 2014, 1, ofu028. [Google Scholar] [CrossRef]
- Kitchel, B.; Rasheed, J.K.; Patel, J.B.; Srinivasan, A.; Navon-Venezia, S.; Carmeli, Y.; Brolund, A.; Giske, C.G. Molecular Epidemiology of KPC-Producing Klebsiella pneumoniae Isolates in the United States: Clonal Expansion of Multilocus Sequence Type 258. Antimicrob. Agents Chemother. 2009, 53, 3365–3370. [Google Scholar] [CrossRef]
- Gomez, S.; Pasteran, F.; Faccone, D.; Tijet, N.; Rapoport, M.; Lucero, C.; Lastovetska, O.; Albornoz, E.; Galas, M.; Melano, R.; et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin. Microbiol. Infect. 2011, 17, 1520–1524. [Google Scholar] [CrossRef][Green Version]
- Cejas, D.; Elena, A.; Nuñez, D.G.; Platero, P.S.; De Paulis, A.; Magariños, F.; Alfonso, C.; Berger, M.A.; Fernández-Canigia, L.; Gutkind, G.; et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019, 18, 238–242. [Google Scholar] [CrossRef]
- Vargas, J.M.; Moreno Mochi, M.P.; Nuñez, J.M.; Cáceres, M.; Mochi, S.; Del Campo Moreno, R.; Jure, M.A. Virulence factors and clinical patterns of multi-ple-clone hypermucoviscous KPC-2 producing Klebsiella pneumoniae. Heliyon 2019, 5, e01829. [Google Scholar] [CrossRef] [PubMed]
- Jure, M.A.; Castillo, M.E.; Musa, H.E.; López, C.G.; Cáceres, M.A.; Mochi, S.D.; Bousquet, A.A.; Genel, N.A.; Arlet, G.A.; Decré, D.C. Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucumán, Argentina. J. Glob. Antimicrob. Resist. 2019, 19, 183–187. [Google Scholar] [CrossRef]
- Jure, M.A.; Albarracin, L.; Vargas, J.M.; Maidana, S.D.; Zamar, J.C.; Kitazawa, H.; Villena, J. Draft genome sequences of two hypermucoviscous carbapenem-resistant ST25 Klebsiella pneumoniae strains causing respiratory and systemic infections. J. Glob. Antimicrob. Resist. 2021, 26, 174–176. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Shen, W.; Wang, S.M.; Wang, G.; Zhou, Y. Whole-genome sequence of a carbapenem-resistant hypermucoviscous Klebsiella pneumoniae isolate SWU01 with capsular serotype K47 belonging to ST11 from a patient in China. J. Glob. Antimicrob. Resist. 2017, 11, 87–89. [Google Scholar] [CrossRef]
- Fu, L.; Tang, L.; Wang, S.; Liu, Q.; Liu, Y.; Zhang, Z.; Zhang, L.; Li, Y.; Chen, W.; Wang, G.; et al. Co-location of the blaKPC-2, blaCTX-M-65, rmtB and virulence relevant factors in an IncFII plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb. Pathog. 2018, 124, 301–304. [Google Scholar] [CrossRef]
- Lan, P.; Zhao, D.; Gu, J.; Shi, Q.; Yan, R.; Jiang, Y.; Zhou, J.; Yu, Y. Genome-Based Analysis of a Sequence Type 1049 Hypervirulent Klebsiella pneumoniae Causing Bacteremic Neck Abscess. Front. Microbiol. 2021, 11, 617651. [Google Scholar] [CrossRef]
- Wysocka, M.; Zamudio, R.; Oggioni, M.R.; Gołębiewska, J.; Dudziak, A.; Krawczyk, B. The New Klebsiella pneumoniae ST152 Variants with Hypermucoviscous Phenotype Isolated from Renal Transplant Recipients with Asymptomatic Bacteriuria—Genetic Characteristics by WGS. Genes 2020, 11, 1189. [Google Scholar] [CrossRef]
- Budnick, J.A.; Bina, X.R.; Bina, J.E. Complete Genome Sequence of Klebsiella pneumoniae Strain ATCC 43816. Microbiol. Resour. Announc. 2021, 10, e01441-20. [Google Scholar] [CrossRef]
- Hsieh, P.-F.; Lu, Y.-R.; Lin, T.-L.; Lai, L.-Y.; Wang, J.-T. Klebsiella pneumoniae Type VI Secretion System Contributes to Bacterial Competition, Cell Invasion, Type-1 Fimbriae Expression, and In Vivo Colonization. J. Infect. Dis. 2018, 219, 637–647. [Google Scholar] [CrossRef]
- Fodah, R.A.; Scott, J.B.; Tam, H.-H.; Yan, P.; Pfeffer, T.L.; Bundschuh, R.; Warawa, J.M. Correlation of Klebsiella pneumoniae Comparative Genetic Analyses with Virulence Profiles in a Murine Respiratory Disease Model. PLoS ONE 2014, 9, e107394. [Google Scholar] [CrossRef]
- Clarke, T.B. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 2014, 82, 4596–4606. [Google Scholar] [CrossRef]
- Xiong, H.; Carter, R.A.; Leiner, I.M.; Tang, Y.-W.; Chen, L.; Kreiswirth, B.N.; Pamer, E.G. Distinct Contributions of Neutrophils and CCR2+ Monocytes to Pulmonary Clearance of Different Klebsiella pneumoniae Strains. Infect. Immun. 2015, 83, 3418–3427. [Google Scholar] [CrossRef]
- Zhang, G.; Jiang, C.; Xie, N.; Xu, Y.; Liu, L.; Liu, N. Treatment with andrographolide sulfonate provides additional benefits to imipenem in a mouse model of Klebsiella pneumoniae pneumonia. Biomed. Pharmacother. 2019, 117, 109065. [Google Scholar] [CrossRef]
- Bruchmann, S.; Feltwell, T.; Parkhill, J.; Short, F.L. Identifying virulence determinants of multidrug-resistant Klebsiella pneumoniae in Galleria mellonella. Pathog. Dis. 2021, 79, ftab009. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-M.; Li, L.-H.; Yan, J.-J.; Tsao, N.; Liao, T.-L.; Tsai, H.-C.; Fung, C.-P.; Chen, H.-J.; Liu, Y.-M.; Wang, J.-T.; et al. Genome Sequencing and Comparative Analysis of Klebsiella pneumoniae NTUH-K2044, a Strain Causing Liver Abscess and Meningitis. J. Bacteriol. 2009, 191, 4492–4501. [Google Scholar] [CrossRef]
- Wang, X.; Xie, Y.; Li, G.; Liu, J.; Li, X.; Tian, L.; Sun, J.; Ou, H.-Y.; Qu, H. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence 2018, 9, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Meijer, M.T.; de Vos, A.F.; Scicluna, B.P.; Roelofs, J.J.; Fayçal, C.A.; Orend, G.; Uhel, F.; van der Poll, T. Tenascin-C Deficiency Is Associated With Reduced Bacterial Outgrowth During Klebsiella pneumoniae-Evoked Pneumosepsis in Mice. Front. Immunol. 2021, 12, 600979. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, D.K.; Allen, I.C. Using Klebsiella pneumoniae to Model Acute Lung Inflammation in Mice. In Mouse Models of Innate Immunity; Humana Press: New York, NY, USA, 2019; Volume 1960, pp. 169–180. [Google Scholar] [CrossRef]
- Ramirez-Moral, I.; Blok, D.C.; Bernink, J.H.; Garcia-Laorden, M.I.; Florquin, S.; Boon, L.; Veer, C.V.; Mack, M.; Saluzzo, S.; Knapp, S.; et al. Interleukin-33 improves local immunity during Gram-negative pneumonia by a combined effect on neutrophils and inflammatory monocytes. J. Pathol. 2020, 253, 374–383. [Google Scholar] [CrossRef]
- Lasko, M.J.; Abdelraouf, K.; Nicolau, D.P. Comparative in vivo activity of human-simulated plasma and epithelial lining fluid exposures of WCK 5222 (cefepime/zidebactam) against KPC- and OXA-48-like-producing Klebsiella pneumoniae in the neutropenic murine pneumonia model. J. Antimicrob. Chemother. 2021, 76, 2310–2316. [Google Scholar] [CrossRef]
- Tan, S.; Gao, J.; Li, Q.; Guo, T.; Dong, X.; Bai, X.; Yang, J.; Hao, S.; He, F. Synergistic effect of chlorogenic acid and levofloxacin against Klebsiella pneumonia infection in vitro and in vivo. Sci. Rep. 2020, 10, 20013. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Z.; Yuan, Y.; Jing, H.; Zou, J.; Zhang, X.; Zeng, X.; Zhang, W.; Zou, Q.; Zhang, J. Innate Immune Effectors Play Essential Roles in Acute Respiratory Infection Caused by Klebsiella pneumoniae. J. Immunol. Res. 2020, 2020, 5291714. [Google Scholar] [CrossRef] [PubMed]
- Olonisakin, T.F.; Li, H.; Xiong, Z.; Kochman, E.J.K.; Yu, M.; Qu, Y.; Hulver, M.; Kolls, J.K.; Croix, C.S.; Doi, Y.; et al. CD36 Provides Host Protection Against Klebsiella pneumoniae Intrapulmonary Infection by Enhancing Lipopolysaccharide Responsiveness and Macrophage Phagocytosis. J. Infect. Dis. 2016, 214, 1865–1875. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, F.R.; Tomokiyo, M.; Moyano, R.O.; Quilodrán-Vega, S.; Yamamuro, H.; Kanmani, P.; Melnikov, V.; Kurata, S.; Kitazawa, H.; Villena, J. The Respiratory Commensal Bacterium Dolosigranulum pigrum 040417 Improves the Innate Immune Response to Streptococcus pneumoniae. Microorganisms 2021, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Moyano, R.O.; Tonetti, F.R.; Tomokiyo, M.; Kanmani, P.; Vizoso-Pinto, M.G.; Kim, H.; Quilodrán-Vega, S.; Melnikov, V.; Alvarez, S.; Takahashi, H.; et al. The Ability of Respiratory Commensal Bacteria to Beneficially Modulate the Lung Innate Immune Response Is a Strain Dependent Characteristic. Microorganisms 2020, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Clua, P.; Kanmani, P.; Zelaya, H.; Tada, A.; Kober, A.K.M.H.; Salva, S.; Alvarez, S.; Kitazawa, H.; Villena, J. Peptidoglycan from Immunobiotic Lactobacillus rhamnosus Improves Resistance of Infant Mice to Respiratory Syncytial Viral Infection and Secondary Pneumococcal Pneumonia. Front. Immunol. 2017, 8, 948. [Google Scholar] [CrossRef]
- Clua, P.; Tomokiyo, M.; Tonetti, F.R.; Islam, A.; Castillo, V.G.; Marcial, G.; Salva, S.; Alvarez, S.; Takahashi, H.; Kurata, S.; et al. The Role of Alveolar Macrophages in the Improved Protection against Respiratory Syncytial Virus and Pneumococcal Superinfection Induced by the Peptidoglycan of Lactobacillus rhamnosus CRL1505. Cells 2020, 9, 1653. [Google Scholar] [CrossRef]
- Hsieh, P.; Lin, T.; Lee, C.; Tsai, S.; Wang, J. Serum-Induced Iron-Acquisition Systems and TonB Contribute to Virulence in Klebsiella pneumoniae Causing Primary Pyogenic Liver Abscess. J. Infect. Dis. 2008, 197, 1717–1727. [Google Scholar] [CrossRef]
- Khater, F.; Balestrino, D.; Charbonnel, N.; Dufayard, J.F.; Brisse, S.; Forestier, C. In Silico Analysis of Usher Encoding Genes in Klebsiella pneumoniae and Characterization of Their Role in Adhesion and Colonization. PLoS ONE 2015, 10, e0116215. [Google Scholar] [CrossRef]
- Bachman, M.A.; Miller, V.L.; Weiser, J.N. Mucosal Lipocalin 2 Has Pro-Inflammatory and Iron-Sequestering Effects in Response to Bacterial Enterobactin. PLoS Pathog. 2009, 5, e1000622. [Google Scholar] [CrossRef]
- El Fertas-Aissani, R.; Messai, Y.; Alouache, S.; Bakour, R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 2013, 61, 209–216. [Google Scholar] [CrossRef]
- Bachman, M.A.; Lenio, S.; Schmidt, L.; Oyler, J.E.; Weiser, J.N. Interaction of Lipocalin 2, Transferrin, and Siderophores Determines the Replicative Niche of Klebsiella pneumoniae during Pneumonia. mBio 2012, 3, e00224-11. [Google Scholar] [CrossRef] [PubMed]
- Holden, V.I.; Lenio, S.; Kuick, R.; Ramakrishnan, S.K.; Shah, Y.M.; Bachman, M.A. Bacterial Siderophores That Evade or Overwhelm Lipocalin 2 Induce Hypoxia Inducible Factor 1α and Proinflammatory Cytokine Secretion in Cultured Respiratory Epithelial Cells. Infect. Immun. 2014, 82, 3826–3836. [Google Scholar] [CrossRef] [PubMed]
- Holden, V.; Breen, P.; Houle, S.; Dozois, C.M.; Bachman, M.A. Klebsiella pneumoniae Siderophores Induce Inflammation, Bacterial Dissemination, and HIF-1α Stabilization during Pneumonia. mBio 2016, 7, e01397-16. [Google Scholar] [CrossRef] [PubMed]
- Lavender, H.; Jagnow, J.J.; Clegg, S. Klebsiella pneumoniae type 3 fimbria-mediated immunity to infection in the murine model of respiratory disease. Int. J. Med. Microbiol. 2005, 295, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef]
- Li, G.; Sun, S.; Zhao, Z.Y.; Sun, Y. The pathogenicity of rmpA or aerobactin-positive Klebsiella pneumoniae in infected mice. J. Int. Med. Res. 2019, 47, 4344–4352. [Google Scholar] [CrossRef]
- Jung, H.-J.; Sorbara, M.T.; Pamer, E.G. TAM mediates adaptation of carbapenem-resistant Klebsiella pneumoniae to antimicrobial stress during host colonization and infection. PLoS Pathog. 2021, 17, e1009309. [Google Scholar] [CrossRef]
- Jung, H.-J.; Littmann, E.R.; Seok, R.; Leiner, I.M.; Taur, Y.; Peled, J.; Van Den Brink, M.; Ling, L.; Chen, L.; Kreiswirth, B.N.; et al. Genome-Wide Screening for Enteric Colonization Factors in Carbapenem-Resistant ST258 Klebsiella pneumoniae. mBio 2019, 10, e02663-18. [Google Scholar] [CrossRef]
- Mizunoe, Y.; Nakabeppu, Y.; Sekiguchi, M.; Kawabata, S.; Moriya, T.; Amako, K. Cloning and sequence of the gene encoding the major structural component of mannose-resistant fimbriae of Serratia marcescens. J. Bacteriol. 1988, 170, 3567–3574. [Google Scholar] [CrossRef]
- Jingushi, S.; Mitsuyama, M.; Moriya, T.; Amako, K. Antigenic analysis of Serratia marcescens fimbriae with monoclonal antibodies. Infect. Immun. 1987, 55, 1600–1606. [Google Scholar] [CrossRef]
- Verger, D.; Bullitt, E.; Hultgren, S.J.; Waksman, G. Crystal Structure of the P Pilus Rod Subunit PapA. PLoS Pathog. 2007, 3, e73. [Google Scholar] [CrossRef] [PubMed]
- Korea, C.-G.; Badouraly, R.; Prevost, M.; Ghigo, J.-M.; Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010, 12, 1957–1977. [Google Scholar] [CrossRef] [PubMed]
- Larsonneur, F.; Martin, F.A.; Mallet, A.; Martinez-Gil, M.; Semetey, V.; Ghigo, J.-M.; Beloin, C. Functional analysis of E scherichia coli Yad fimbriae reveals their potential role in environmental persistence. Environ. Microbiol. 2016, 18, 5228–5248. [Google Scholar] [CrossRef] [PubMed]
- Lacher, D.W.; Steinsland, H.; Blank, T.E.; Donnenberg, M.S.; Whittam, T.S. Molecular Evolution of Typical Enteropathogenic Escherichia coli: Clonal Analysis by Multilocus Sequence Typing and Virulence Gene Allelic Profiling. J. Bacteriol. 2007, 189, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Caetano, B.A.; Mourão, D.B.; Abreu, P.A.E.; Monaris, D.; Vasconcellos, H.L.; Luz, D.; Galhardo, C.S.; Menezes, M.A.; Lima, F.A.; Elias, W.P.; et al. Macrophage Inflammatory Response Mediated by Intimin and Bundle-Forming Pilus from Enteropathogenic Escherichia coli. Proceedings 2020, 66, 21. [Google Scholar] [CrossRef]
- Bachman, M.A.; Breen, P.; Deornellas, V.; Mu, Q.; Zhao, L.; Wu, W.; Cavalcoli, J.D.; Mobley, H. Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection. mBio 2015, 6, e00775-15. [Google Scholar] [CrossRef]
- Villena, J.; Chiba, E.; Tomosada, Y.; Salva, S.; Marranzino, G.; Kitazawa, H.; Alvarez, S. Orally administered Lactobacillus rhamnosus modulates the respiratory immune response triggered by the viral pathogen-associated molecular pattern poly(I:C). BMC Immunol. 2012, 13, 53. [Google Scholar] [CrossRef]
- Albarracin, L.; Garcia-Castillo, V.; Masumizu, Y.; Indo, Y.; Islam, A.; Suda, Y.; Garcia-Cancino, A.; Aso, H.; Takahashi, H.; Kitazawa, H.; et al. Efficient Selection of New Immunobiotic Strains with Antiviral Effects in Local and Distal Mucosal Sites by Using Porcine Intestinal Epitheliocytes. Front. Immunol. 2020, 11, 543. [Google Scholar] [CrossRef]
- Garcia-Castillo, V.; Tomokiyo, M.; Tonetti, F.R.; Islam, A.; Takahashi, H.; Kitazawa, H.; Villena, J. Alveolar Macrophages Are Key Players in the Modulation of the Respiratory Antiviral Immunity Induced by Orally Administered Lacticaseibacillus rhamnosus CRL1505. Front. Immunol. 2020, 11, 568636. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive web: User-Friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albarracin, L.; Ortiz Moyano, R.; Vargas, J.M.; Andrade, B.G.N.; Cortez Zamar, J.; Dentice Maidana, S.; Fukuyama, K.; Kurata, S.; Jure, M.Á.; Kitazawa, H.; et al. Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina. Int. J. Mol. Sci. 2022, 23, 7361. https://doi.org/10.3390/ijms23137361
Albarracin L, Ortiz Moyano R, Vargas JM, Andrade BGN, Cortez Zamar J, Dentice Maidana S, Fukuyama K, Kurata S, Jure MÁ, Kitazawa H, et al. Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina. International Journal of Molecular Sciences. 2022; 23(13):7361. https://doi.org/10.3390/ijms23137361
Chicago/Turabian StyleAlbarracin, Leonardo, Ramiro Ortiz Moyano, Juan Martin Vargas, Bruno G. N. Andrade, Juan Cortez Zamar, Stefania Dentice Maidana, Kohtaro Fukuyama, Shoichiro Kurata, María Ángela Jure, Haruki Kitazawa, and et al. 2022. "Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina" International Journal of Molecular Sciences 23, no. 13: 7361. https://doi.org/10.3390/ijms23137361
APA StyleAlbarracin, L., Ortiz Moyano, R., Vargas, J. M., Andrade, B. G. N., Cortez Zamar, J., Dentice Maidana, S., Fukuyama, K., Kurata, S., Jure, M. Á., Kitazawa, H., & Villena, J. (2022). Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina. International Journal of Molecular Sciences, 23(13), 7361. https://doi.org/10.3390/ijms23137361







