The Role of CD4+ T Cells and Microbiota in the Pathogenesis of Asthma
Abstract
1. Introduction
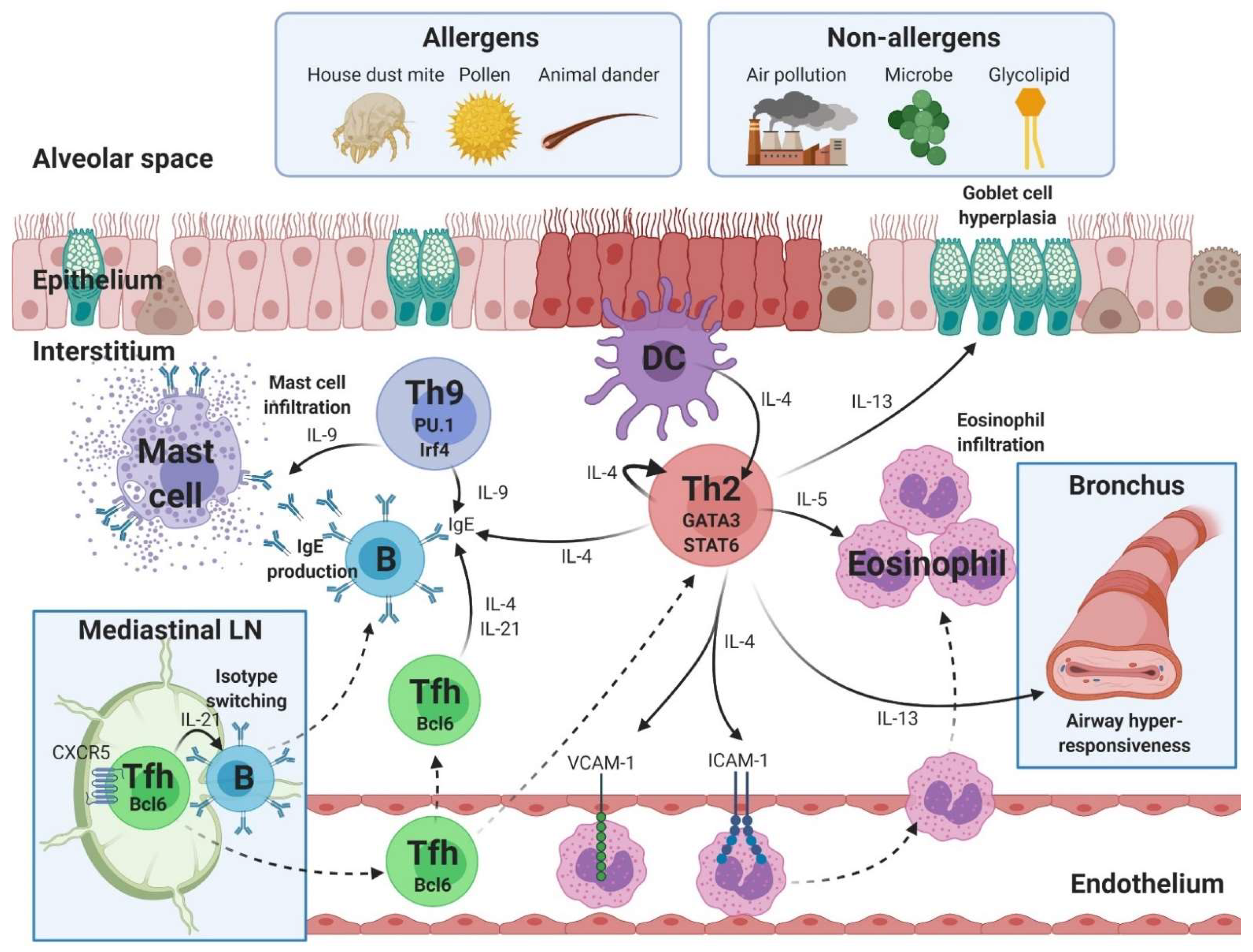
2. Th2-Asthma with Eosinophilic Inflammation
2.1. Th2 Cells
2.2. Th9 Cells
2.3. Tfh Cells
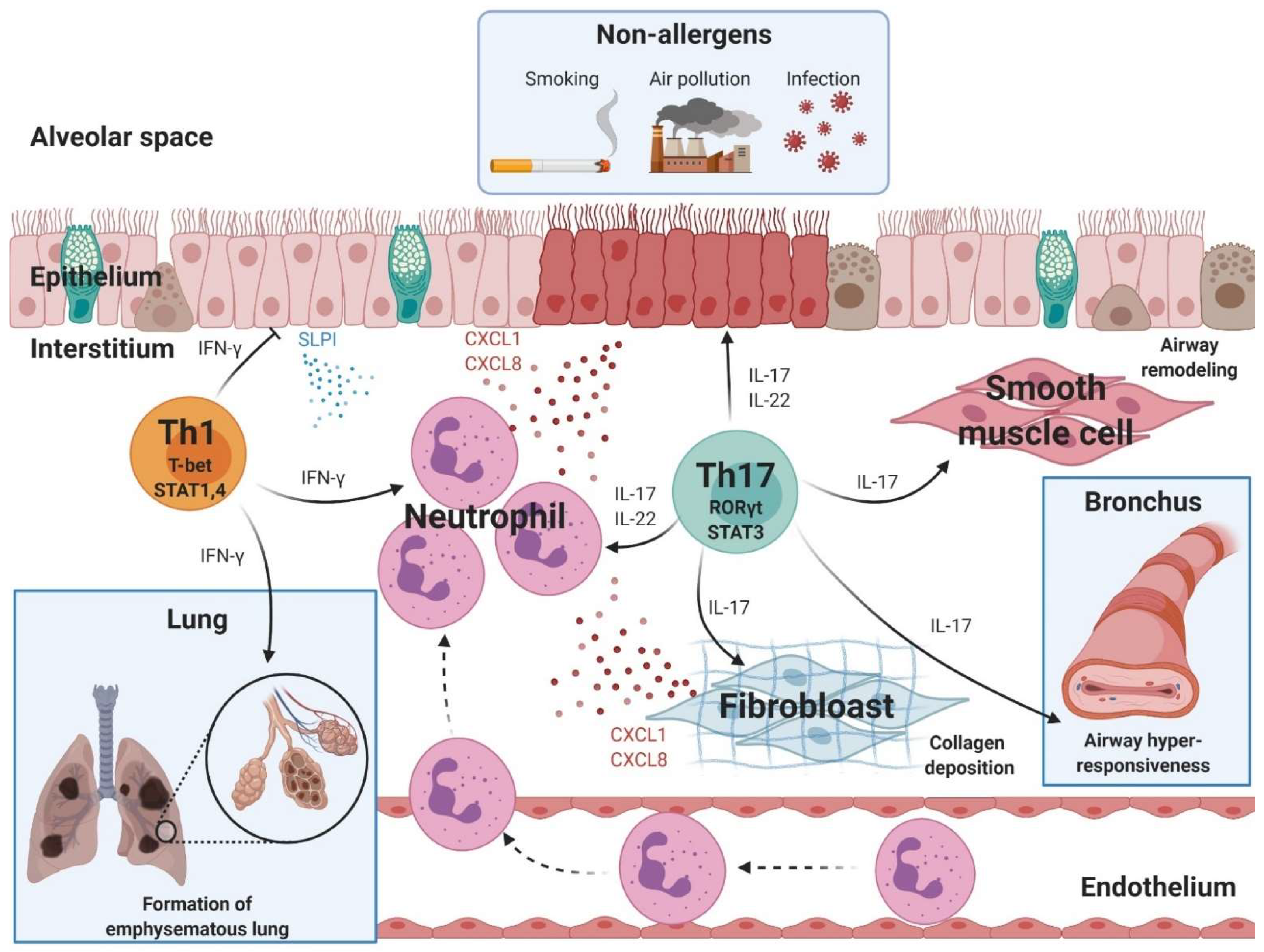
3. Non-Th2 Asthma with Neutrophilic Inflammation
3.1. Th17 Cells
3.2. Th1 Cells
4. Beneficial and Harmful Bacteria in the Pathogenesis of Asthma
4.1. Beneficial Bacteria with Anti-Asthmatic Effects
4.2. Harmful Bacteria with Pro-Asthmatic Effects
5. Dysbiosis-Induced Asthma
5.1. Antibiotics
5.2. Cesarean Section
5.3. Low-Fiber Diet
5.4. Formula Feeding
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hammad, H.; Lambrecht, B.N. The basic immunology of asthma. Cell 2021, 184, 1469–1485. [Google Scholar] [CrossRef] [PubMed]
- Enilari, O.; Sinha, S. The Global Impact of Asthma in Adult Populations. Ann. Glob. Health 2019, 85, 1–7. [Google Scholar] [CrossRef]
- 2021 GINA Main Report Global Initiative for Asthma–GINA. Available online: https://ginasthma.org/gina-reports/ (accessed on 12 August 2021).
- Mina Gaga, E.Z. Oral steroids in asthma: A double-edged sword. Eur. Respir. J. 2019, 54, 1902034. [Google Scholar] [CrossRef]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef]
- Doroudchi, A.; Pathria, M.; Modena, B.D. Asthma biologics: Comparing trial designs, patient cohorts and study results. Ann. Allergy Asthma Immunol. 2020, 124, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Sze, E.; Bhalla, A.; Nair, P. Mechanisms and therapeutic strategies for non-T2 asthma. Allergy Eur. J. Allergy Clin. Immunol. 2020, 75, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Muehling, L.M.; Lawrence, M.G.; Woodfolk, J.A. Pathogenic CD4+ T cells in patients with asthma. J. Allergy Clin. Immunol. 2017, 140, 1523–1540. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Simpson, J.L.; Daly, J.; Baines, K.J.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Hugenholtz, P.; Willner, D.; et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J. 2016, 47, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V.; Nariya, S.; Bhakta, N.R.; Beigelman, A.; Castro, M.; Dyer, A.-M.; Israel, E.; Kraft, M.; Martin, R.J.; et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017, 140, 63–75. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.M.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities—2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, M.E.; Lee, F.E.-H.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2018, 56, 219–233. [Google Scholar] [CrossRef]
- Brusselle, G.G.; Maes, T.; Bracke, K.R. Eosinophils in the Spotlight: Eosinophilic airway inflammation in nonallergic asthma. Nat. Med. 2013, 19, 977–979. [Google Scholar] [CrossRef]
- Dong, C. Cytokine Regulation and Function in T Cells. Annu. Rev. Immunol. 2021, 39, 51–76. [Google Scholar] [CrossRef]
- Knochelmann, H.M.; Dwyer, C.J.; Bailey, S.R.; Amaya, S.M.; Elston, D.M.; Mazza-McCrann, J.M.; Paulos, C.M. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell. Mol. Immunol. 2018, 15, 458–469. [Google Scholar] [CrossRef]
- Saeki, M.; Nishimura, T.; Kitamura, N.; Hiroi, T.; Mori, A.; Kaminuma, O. Potential Mechanisms of T Cell-Mediated and Eosinophil-Independent Bronchial Hyperresponsiveness. Int. J. Mol. Sci. 2019, 20, 2980. [Google Scholar] [CrossRef] [PubMed]
- Seumois, G.; Zapardiel-Gonzalo, J.; White, B.; Singh, D.; Schulten, V.; Dillon, M.; Hinz, D.; Broide, D.H.; Sette, A.; Peters, B.; et al. Transcriptional Profiling of Th2 Cells Identifies Pathogenic Features Associated with Asthma. J. Immunol. 2016, 197, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Junttila, I.S. Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, B.N.; Hammad, H.; Fahy, J.V. The Cytokines of Asthma. Immunity 2019, 50, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the Pathophysiology of Severe Asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef]
- Foster, P.S.; Hogan, S.P.; Ramsay, A.J.; Matthaei, K.I.; Young, I.G. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J. Exp. Med. 1996, 183, 195–201. [Google Scholar] [CrossRef]
- Johnston, L.K.; Hsu, C.-L.; Krier-Burris, R.A.; Chhiba, K.D.; Chien, K.B.; McKenzie, A.; Berdnikovs, S.; Bryce, P.J. IL-33 Precedes IL-5 in Regulating Eosinophil Commitment and Is Required for Eosinophil Homeostasis. J. Immunol. 2016, 197, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.W. Eosinophil Activation Status in Separate Compartments and Association with Asthma. Front. Med. 2017, 4, 75. [Google Scholar] [CrossRef]
- Seibold, M.A. Interleukin-13 Stimulation Reveals the Cellular and Functional Plasticity of the Airway Epithelium. Ann. Am. Thorac. Soc. 2018, 15, S98–S106. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ye, Z. The Potential Role and Regulatory Mechanisms of MUC5AC in Chronic Obstructive Pulmonary Disease. Molecules 2020, 25, 4437. [Google Scholar] [CrossRef]
- Marone, G.; Granata, F.; Pucino, V.; Pecoraro, A.; Heffler, E.; Loffredo, S.; Scadding, G.W.; Varricchi, G. The Intriguing Role of Interleukin 13 in the Pathophysiology of Asthma. Front. Pharmacol. 2019, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Bel, E.H.; Wenzel, S.E.; Thompson, P.J.; Prazma, C.M.; Keene, O.N.; Yancey, S.W.; Ortega, H.G.; Pavord, I.D. Oral Glucocorticoid-Sparing Effect of Mepolizumab in Eosinophilic Asthma. N. Eng. J. Med. 2014, 371, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Ortega, H.G.; Liu, M.C.; Pavord, I.D.; Brusselle, G.G.; FitzGerald, J.M.; Chetta, A.; Humbert, M.; Katz, L.E.; Keene, O.N.; Yancey, S.W.; et al. Mepolizumab Treatment in Patients with Severe Eosinophilic Asthma. N. Eng. J. Med. 2014, 371, 1198–1207. [Google Scholar] [CrossRef]
- Castro, M.; Zangrilli, J.; Wechsler, M.E.; Bateman, E.D.; Brusselle, G.G.; Bardin, P.; Murphy, K.; Maspero, J.F.; O’Brien, C.; Korn, S. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: Results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir. Med. 2015, 3, 355–366. [Google Scholar] [CrossRef]
- Nair, P.; Wenzel, S.; Rabe, K.F.; Bourdin, A.; Lugogo, N.L.; Kuna, P.; Barker, P.; Sproule, S.; Ponnarambil, S.; Goldman, M. Oral Glucocorticoid–Sparing Effect of Benralizumab in Severe Asthma. N. Eng. J. Med. 2017, 376, 2448–2458. [Google Scholar] [CrossRef]
- Castro, M.; Corren, J.; Pavord, I.D.; Maspero, J.; Wenzel, S.; Rabe, K.F.; Busse, W.W.; Ford, L.; Sher, L.; FitzGerald, J.M.; et al. Dupilumab Efficacy and Safety in Moderate-to-Severe Uncontrolled Asthma. N. Eng. J. Med. 2018, 378, 2486–2496. [Google Scholar] [CrossRef]
- Garn, H.; Renz, H. GATA-3-specific DNAzyme A novel approach for stratified asthma therapy. Eur. J. Immunol. 2017, 47, 22–30. [Google Scholar] [CrossRef]
- Krug, N.; Hohlfeld, J.M.; Kirsten, A.-M.; Kornmann, O.; Beeh, K.M.; Kappeler, D.; Korn, S.; Ignatenko, S.; Timmer, W.; Rogon, C.; et al. Allergen-Induced Asthmatic Responses Modified by a GATA3-Specific DNAzyme. N. Eng. J. Med. 2015, 372, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Badolati, I.; Sverremark-Ekström, E.; Heiden, M. van der Th9 cells in allergic diseases: A role for the microbiota? Scand. J. Immunol. 2020, 91, e12857. [Google Scholar] [CrossRef]
- Neurath, M.F.; Kaplan, M.H. Th9 cells in immunity and immunopathological diseases. Semin. Immunopathol. 2017, 39, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Angkasekwinai, P.; Dong, C. IL-9-producing T cells: Potential players in allergy and cancer. Nat. Rev. Immunol. 2021, 21, 37–48. [Google Scholar] [CrossRef]
- Koch, S.; Sopel, N.; Finotto, S. Th9 and other IL-9-producing cells in allergic asthma. Semin. Immunopathol. 2016, 39, 55–68. [Google Scholar] [CrossRef]
- Petit-Frere, C.; Dugas, B.; Braquet, P.; Mencia-Huerta, J.M. Interleukin-9 potentiates the interleukin-4-induced IgE and IgG1 release from murine B lymphocytes. Immunology 1993, 79, 146. [Google Scholar]
- McLane, M.P.; Haczku, A.; van de Rijn, M.; Weiss, C.; Ferrante, V.; MacDonald, D.; Renauld, J.-C.; Nicolaides, N.C.; Holroyd, K.J.; Levitt, R.C. Interleukin-9 promotes allergen-induced eosinophilic inflammation and airway hyperresponsiveness in transgenic mice. Am. J. Respir. Cell Mol. Biol. 1998, 19, 713–720. [Google Scholar] [CrossRef]
- Jones, C.P.; Gregory, L.G.; Causton, B.; Campbell, G.A.; Lloyd, C.M. Activin A and TGF-β promote TH9 cell–mediated pulmonary allergic pathology. J. Allergy Clin. Immunol. 2012, 129, 1000–1010.e3. [Google Scholar] [CrossRef]
- Temann, U.A.; Ray, P.; Flavell, R.A. Pulmonary overexpression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J. Clin. Investig. 2002, 109, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Louahed, J.; Zhou, Y.; Maloy, W.L.; Rani, P.U.; Weiss, C.; Tomer, Y.; Vink, A.; Renauld, J.; Snick van, J.; Nicolaides, N.C.; et al. Interleukin 9 promotes influx and local maturation of eosinophils. Blood 2001, 97, 1035–1042. [Google Scholar] [CrossRef]
- Vermeer, P.D.; Harson, R.; Einwalter, L.A.; Moninger, T.; Zabner, J. Interleukin-9 induces goblet cell hyperplasia during repair of human airway epithelia. Am. J. Respir. Cell Mol. Biol. 2003, 28, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-C.; Sehra, S.; Goswami, R.; Yao, W.; Yu, Q.; Stritesky, G.L.; Jabeen, R.; McKinley, C.; Ahyi, A.-N.; Han, L.; et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 2010, 11, 527–534. [Google Scholar] [CrossRef] [PubMed]
- McLeod, J.J.A.; Baker, B.; Ryan, J.J. Mast cell production and response to IL-4 and IL-13. Cytokine 2015, 75, 57–61. [Google Scholar] [CrossRef]
- Sehra, S.; Yao, W.; Nguyen, E.T.; Glosson-Byers, N.L.; Akhtar, N.; Zhou, B.; Kaplan, M.H. TH9 cells are required for tissue mast cell accumulation during allergic inflammation. J. Allergy Clin. Immunol. 2015, 136, 433–440.e1. [Google Scholar] [CrossRef]
- Cheng, G.; Arima, M.; Honda, K.; Hirata, H.; Eda, F.; Yoshida, N.; Fukushima, F.; Ishii, Y.; Fukuda, T. Anti-interleukin-9 antibody treatment inhibits airway inflammation and hyperreactivity in mouse asthma model. Am. J. Respir. Crit. Care Med. 2002, 166, 409–416. [Google Scholar] [CrossRef]
- Kim, M.S.; Cho, K.-A.; Cho, Y.J.; Woo, S.-Y. Effects of interleukin-9 blockade on chronic airway inflammation in murine asthma models. Allergy Asthma Immunol. Res. 2013, 5, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.K.; Leigh, R.; McLaurin, K.K.; Kim, K.; Hultquist, M.; Molfino, N.A. A randomized, controlled trial to evaluate the effect of an anti-interleukin-9 monoclonal antibody in adults with uncontrolled asthma. Respir. Res. 2013, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.M.; Harker, J.A. Epigenetic Control of Interleukin-9 in Asthma. N. Eng. J. Med. 2018, 379, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Fan, Y.; Li, J.; Zhang, X.; Lou, X.; Dou, Y.; Shi, X.; Lan, P.; Xiao, Y.; Minze, L.; et al. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J. Exp. Med. 2018, 215, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Sanders, Y.Y.; Thannickal, V.J. BETting on Novel Treatments for Asthma: Bromodomain 4 Inhibitors. Am. J. Respir. Cell Mol. Biol. 2019, 60, 7. [Google Scholar] [CrossRef]
- Gong, F.; Zheng, T.; Zhou, P. T Follicular Helper Cell Subsets and the Associated Cytokine IL-21 in the Pathogenesis and Therapy of Asthma. Front. Immunol. 2019, 10, 2918. [Google Scholar] [CrossRef]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef]
- Vinuesa, C.G.; Linterman, M.A.; Yu, D.; MacLennan, I.C.M. Follicular Helper T Cells. Annu. Rev. Immunol. 2016, 34, 335–368. [Google Scholar] [CrossRef]
- Corry, D.B.; Kheradmand, F. Induction and regulation of the IgE response. Nature 1999, 402, 18–23. [Google Scholar] [CrossRef]
- Kobayashi, T.; Iijima, K.; Dent, A.L.; Kita, H. Follicular helper T cells mediate IgE antibody response to airborne allergens. J. Allergy Clin. Immunol. 2017, 139, 300–313.e7. [Google Scholar] [CrossRef]
- Noble, A.; Zhao, J. Follicular helper T cells are responsible for IgE responses to Der p 1 following house dust mite sensitization in mice. Clin. Exp. Allergy 2016, 46, 1075–1082. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, C.L.; Wang, N.; Wang, Z.C.; Ma, J.; Zhu, R.F.; Xu, X.Y.; Zhou, P.C.; Yu, D.; Liu, Z. Correlation of allergen-specific T follicular helper cell counts with specific IgE levels and efficacy of allergen immunotherapy. J. Allergy Clin. Immunol. 2018, 142, 321–324.e10. [Google Scholar] [CrossRef]
- Coquet, J.M.; Schuijs, M.J.; Smyth, M.J.; Deswarte, K.; Beyaert, R.; Braun, H.; Boon, L.; Hedestam, G.B.K.; Nutt, S.L.; Hammad, H.; et al. Interleukin-21-Producing CD4+ T Cells Promote Type 2 Immunity to House Dust Mites. Immunity 2015, 43, 318–330. [Google Scholar] [CrossRef]
- Ballesteros-Tato, A.; Randall, T.D.; Lund, F.E.; Spolski, R.; Leonard, W.J.; León, B. T Follicular Helper Cell Plasticity Shapes Pathogenic T Helper 2 Cell-Mediated Immunity to Inhaled House Dust Mite. Immunity 2016, 44, 259–273. [Google Scholar] [CrossRef]
- Uwadiae, F.I.; Pyle, C.J.; Walker, S.A.; Lloyd, C.M.; Harker, J.A. Targeting the ICOS/ICOS-L pathway in a mouse model of established allergic asthma disrupts T follicular helper cell responses and ameliorates disease. Allergy 2019, 74, 650–662. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, Y.; Sun, G. miR-192 suppresses T follicular helper cell differentiation by targeting CXCR5 in childhood asthma. Scand. J. Clin. Lab. Investig. 2018, 78, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Thomson, N.C. Novel approaches to the management of noneosinophilic asthma. Ther. Adv. Respir. Dis. Rev. 2016, 10, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Wener, R.R.L.; Bel, E.H. Severe refractory asthma: An update. Eur. Respir. Rev. 2013, 22, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Laan, M.; Cui, Z.H.; Hoshino, H.; Lötvall, J.; Sjöstrand, M.; Gruenert, D.C.; Skoogh, B.E.; Lindén, A. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J. Immunol. 1999, 162, 2347–2352. [Google Scholar]
- Pène, J.; Chevalier, S.; Preisser, L.; Vénéreau, E.; Guilleux, M.-H.; Ghannam, S.; Molès, J.-P.; Danger, Y.; Ravon, E.; Lesaux, S.; et al. Chronically Inflamed Human Tissues Are Infiltrated by Highly Differentiated Th17 Lymphocytes. J. Immunol. 2008, 180, 7423–7430. [Google Scholar] [CrossRef]
- Bellini, A.; Marini, M.A.; Bianchetti, L.; Barczyk, M.; Schmidt, M.; Mattoli, S. Interleukin (IL)-4, IL-13, and IL-17A differentially affect the profibrotic and proinflammatory functions of fibrocytes from asthmatic patients. Mucosal Immunol. 2011, 5, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, D.C.; Boswell, M.G.; Reiss, S.; Zhou, W.; Goleniewska, K.; Toki, S.; Harintho, M.T.; Lukacs, N.W.; Kolls, J.K.; Peebles, R.S. IL-17A inhibits airway reactivity induced by respiratory syncytial virus infection during allergic airway inflammation. Thorax 2013, 68, 717–723. [Google Scholar] [CrossRef]
- Camargo, L.D.N.; Righetti, R.F.; Aristóteles, L.R.D.C.R.B.; Dos Santos, T.M.; de Souza, F.C.R.; Fukuzaki, S.; Cruz, M.M.; Alonso-Vale, M.I.C.; Saraiva-Romanholo, B.M.; Prado, C.M.; et al. Effects of Anti-IL-17 on Inflammation, Remodeling, and Oxidative Stress in an Experimental Model of Asthma Exacerbated by LPS. Front. Immunol. 2018, 8, 1835. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Al-Alwan, L.; Risse, P.A.; Roussel, L.; Rousseau, S.; Halayko, A.J.; Martin, J.G.; Hamid, Q.; Eidelman, D.H. TH17 cytokines induce human airway smooth muscle cell migration. J. Allergy Clin. Immunol. 2011, 127, 1046–1053.e2. [Google Scholar] [CrossRef]
- Kudo, M.; Melton, A.C.; Chen, C.; Engler, M.B.; Huang, K.E.; Ren, X.; Wang, Y.; Bernstein, X.; Li, J.T.; Atabai, K.; et al. IL-17A produced by αβ T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat. Med. 2012, 18, 547–554. [Google Scholar] [CrossRef]
- Chang, Y.; Al-Alwan, L.; Risse, P.-A.; Halayko, A.J.; Martin, J.G.; Baglole, C.J.; Eidelman, D.H.; Hamid, Q. Th17-associated cytokines promote human airway smooth muscle cell proliferation. FASEB J. 2012, 26, 5152–5160. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, R.K.; Bajbouj, K.; Al Heialy, S.; Mahboub, B.; Ansari, A.W.; Hachim, I.Y.; Rawat, S.; Salameh, L.; Hachim, M.Y.; Olivenstein, R.; et al. IL-17 Induced Autophagy Regulates Mitochondrial Dysfunction and Fibrosis in Severe Asthmatic Bronchial Fibroblasts. Front. Immunol. 2020, 11, 1002. [Google Scholar] [CrossRef]
- Chiba, Y.; Tanoue, G.; Suto, R.; Suto, W.; Hanazaki, M.; Katayama, H.; Sakai, H. Interleukin-17A directly acts on bronchial smooth muscle cells and augments the contractility. Pharmacol. Rep. 2016, 69, 377–385. [Google Scholar] [CrossRef]
- Willis, C.R.; Siegel, L.; Leith, A.; Mohn, D.; Escobar, S.; Wannberg, S.; Misura, K.; Rickel, E.; Rottman, J.B.; Comeau, M.R.; et al. IL-17RA signaling in airway inflammation and bronchial hyperreactivity in allergic asthma. Am. J. Respir. Cell Mol. Biol. 2015, 53, 810–821. [Google Scholar] [CrossRef]
- Nanzer, A.M.; Chambers, E.S.; Ryanna, K.; Richards, D.F.; Black, C.; Timms, P.M.; Martineau, A.R.; Griffiths, C.J.; Corrigan, C.J.; Hawrylowicz, C.M. Enhanced production of IL-17A in patients with severe asthma is inhibited by 1α,25-dihydroxyvitamin D3 in a glucocorticoid-independent fashion. J. Allergy Clin. Immunol. 2013, 132, 3037. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Nanzer, A.M.; Pfeffer, P.E.; Richards, D.F.; Timms, P.M.; Martineau, A.R.; Griffiths, C.J.; Corrigan, C.J.; Hawrylowicz, C.M. Distinct endotypes of steroid-resistant asthma characterized by IL-17Ahigh and IFN-γhigh immunophenotypes: Potential benefits of calcitriol. J. Allergy Clin. Immunol. 2015, 136, 628–637.e4. [Google Scholar] [CrossRef]
- Safety, Tolerability, and Efficacy of AIN457 in Patients with Uncontrolled Asthma–Study Results–ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/results/NCT01478360?cond=AIN457&draw=2#part (accessed on 12 August 2021).
- Amarnani, A.; Rosenthal, K.S.; Mercado, J.M.; Brodell, R.T. Concurrent treatment of chronic psoriasis and asthma with ustekinumab. J. Dermatol. Treatm. 2013, 25, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Webb, L.M.; Oyesola, O.O.; Früh, S.P.; Kamynina, E.; Still, K.M.; Patel, R.K.; Peng, S.A.; Cubitt, R.L.; Grimson, A.; Grenier, J.K.; et al. The Notch signaling pathway promotes basophil responses during helminth-induced type 2 inflammation. J. Exp. Med. 2019, 216, 1268–1279. [Google Scholar] [CrossRef]
- Garn, H.; Potaczek, D.P.; Pfefferle, P.I. The Hygiene Hypothesis and New Perspectives—Current Challenges Meeting an Old Postulate. Front. Immunol. 2021, 12, 847. [Google Scholar] [CrossRef]
- Cui, J.; Pazdziorko, S.; Miyashiro, J.S.; Thakker, P.; Pelker, J.W.; Declercq, C.; Jiao, A.; Gunn, J.; Mason, L.; Leonard, J.P.; et al. TH1-mediated airway hyperresponsiveness independent of neutrophilic inflammation. J. Allergy Clin. Immunol. 2005, 115, 309–315. [Google Scholar] [CrossRef]
- Raundhal, M.; Morse, C.; Khare, A.; Oriss, T.B.; Milosevic, J.; Trudeau, J.; Huff, R.; Pilewski, J.; Holguin, F.; Kolls, J.; et al. High IFN-γ and low SLPI mark severe asthma in mice and humans. J. Clin. Investig. 2015, 125, 3037–3050. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Oh, S.-Y.; Jeon, S.G.; Park, H.-W.; Lee, S.-Y.; Chun, E.-Y.; Bang, B.; Lee, H.-S.; Oh, M.-H.; Kim, Y.-S.; et al. Airway Exposure Levels of Lipopolysaccharide Determine Type 1 versus Type 2 Experimental Asthma. J. Immunol. 2007, 178, 5375–5382. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, T.; Zhu, Z.; Homer, R.J.; Riese, R.J.; Chapman, H.A.; Shapiro, S.D.; Elias, J.A. Interferon γ induction of pulmonary emphysema in the adult murine lung. J. Exp. Med. 2000, 192, 1587–1599. [Google Scholar] [CrossRef]
- Loverdos, K.; Bellos, G.; Kokolatou, L.; Vasileiadis, I.; Giamarellos, E.; Pecchiari, M.; Koulouris, N.; Koutsoukou, A.; Rovina, N. Lung Microbiome in Asthma: Current Perspectives. J. Clin. Med. 2019, 8, 1967. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut–Lung Axis. Int. J. Mol. Sci. 2019, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Paramithiotis, S.; Shin, H.-S. Kimchi and Other Widely Consumed Traditional Fermented Foods of Korea: A Review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef]
- Redondo-Useros, N.; Gheorghe, A.; Díaz-Prieto, L.E.; Villavisencio, B.; Marcos, A.; Nova, E. Associations of Probiotic Fermented Milk (PFM) and Yogurt Consumption with Bifidobacterium and Lactobacillus Components of the Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 651. [Google Scholar] [CrossRef]
- Spacova, I.; Petrova, M.I.; Fremau, A.; Pollaris, L.; Vanoirbeek, J.; Ceuppens, J.L.; Seys, S.; Lebeer, S. Intranasal administration of probiotic Lactobacillus rhamnosus GG prevents birch pollen-induced allergic asthma in a murine model. Allergy 2019, 74, 100–110. [Google Scholar] [CrossRef]
- Li, L.; Fang, Z.; Liu, X.; Hu, W.; Lu, W.; Lee, Y.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS ONE 2020, 15, e0231865. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Jan, R.-L.; Lin, Y.-L.; Chen, H.-H.; Wang, J.-Y. Randomized placebo-controlled trial of lactobacillus on asthmatic children with allergic rhinitis. Pediatr. Pulmonol. 2010, 45, 1111–1120. [Google Scholar] [CrossRef]
- Raftis, E.J.; Delday, M.I.; Cowie, P.; McCluskey, S.M.; Singh, M.D.; Ettorre, A.; Mulder, I.E. Bifidobacterium breve MRx0004 protects against airway inflammation in a severe asthma model by suppressing both neutrophil and eosinophil lung infiltration. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Sun, S.; Luo, L.; Liang, W.; Yin, Q.; Guo, J.; Rush, A.M.; Lv, Z.; Liang, Q.; Fischbach, M.A.; Sonnenburg, J.L.; et al. Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc. Natl. Acad. Sci. USA 2020, 117, 27509–27515. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, M.M.; Indolfi, C.; Capasso, M.; Maiello, N.; Decimo, F.; Ciprandi, G. Bifidobacterium mixture (B longum BB536, B infantis M-63, B breve M-16V) treatment in children with seasonal allergic rhinitis and intermittent asthma. Ital. J. Pediatrics 2017, 43, 3405. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; Angelis, M. De The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Michalovich, D.; Rodriguez-Perez, N.; Smolinska, S.; Pirozynski, M.; Mayhew, D.; Uddin, S.; Van Horn, S.; Sokolowska, M.; Altunbulakli, C.; Eljaszewicz, A.; et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kuczma, M.P.; Szurek, E.A.; Cebula, A.; Ngo, V.L.; Pietrzak, M.; Kraj, P.; Denning, T.L.; Ignatowicz, L. Self and microbiota-derived epitopes induce CD4+ T cell anergy and conversion into CD4+Foxp3+ regulatory cells. Mucosal Immunol. 2020, 14, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.; Tokman, H.B.; Uysal, H.K.; Demiryas, S.; Karakullukcu, A.; Saribas, S.; Cokugras, H.; Kocazeybek, B.S. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol. Et. Immunopathol. 2019, 47, 365–371. [Google Scholar] [CrossRef]
- Falcone, M.; Tiseo, G.; Menichetti, F. Community-acquired Pneumonia Owing to Multidrug-Resistant Pathogens: A Step toward an Early Identification. Ann. Am. Thorac. Soc. 2021, 18, 211. [Google Scholar] [CrossRef]
- Lee, H.S.; Plechot, K.; Gohil, S.; Le, J. Clostridium difficile: Diagnosis and the Consequence of Over Diagnosis. Infect. Dis. Ther. 2021, 10, 687–697. [Google Scholar] [CrossRef]
- Van Nimwegen, F.A.; Penders, J.; Stobberingh, E.E.; Postma, D.S.; Koppelman, G.H.; Kerkhof, M.; Reijmerink, N.E.; Dompeling, E.; Van Den Brandt, P.A.; Ferreira, I.; et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 2011, 128, 948–955. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Arrieta, M.-C.; Dimitriu, P.A.; Cheng, J.; Thorson, L.; Lefebvre, D.L.; Azad, M.B.; Subbarao, P.; Mandhane, P.; Becker, A.; et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. 2016, 130, 2199–2207. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Chan, Y.L.; Tsai, M.H.; Wang, C.J.; Chiang, M.H.; Chiu, C.C. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ. J. 2019, 12, 100021. [Google Scholar] [CrossRef]
- Stentzel, S.; Teufelberger, A.; Nordengrün, M.; Kolata, J.; Schmidt, F.; van Crombruggen, K.; Michalik, S.; Kumpfmüller, J.; Tischer, S.; Schweder, T.; et al. Staphylococcal serine protease like proteins are pacemakers of allergic airway reactions to Staphylococcus aureus. J. Allergy Clin. Immunol. 2017, 139, 492–500.e8. [Google Scholar] [CrossRef] [PubMed]
- Krysko, O.; Teufelberger, A.; Nevel, S.V.; Krysko, D.V.; Bachert, C. Protease/antiprotease network in allergy: The role of Staphylococcus aureus protease-like proteins. Allergy 2019, 74, 2077–2086. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.F.; Peng, R.D.; McCormack, M.C.; Matsui, E.C. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J. Allergy Clin. Immunol. 2015, 135, 811–813.e5. [Google Scholar] [CrossRef]
- Zhang, Q.; Illing, R.; Hui, C.K.; Downey, K.; Carr, D.; Stearn, M.; Alshafi, K.; Menzies-Gow, A.; Zhong, N.; Fan Chung, K. Bacteria in sputum of stable severe asthma and increased airway wall thickness. Respir. Res. 2012, 13, 1–8. [Google Scholar] [CrossRef]
- Tuli, J.F.; Ramezanpour, M.; Cooksley, C.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Association between mucosal barrier disruption by Pseudomonas aeruginosa exoproteins and asthma in patients with chronic rhinosinusitis. Allergy 2021, 76, 1–11. [Google Scholar] [CrossRef]
- Nakamoto, K.; Watanabe, M.; Sada, M.; Inui, T.; Nakamura, M.; Honda, K.; Ishii, H.; Takizawa, H. IL-6 and IL-8 production induced by Pseudomonas aeruginosa flagellin in human bronchial epithelial cells. Eur. Respir. J. 2017, 50, PA989. [Google Scholar] [CrossRef]
- Green, B.J.; Wiriyachaiporn, S.; Grainge, C.; Rogers, G.B.; Kehagia, V.; Lau, L.; Carroll, M.P.; Bruce, K.D.; Howarth, P.H. Potentially Pathogenic Airway Bacteria and Neutrophilic Inflammation in Treatment Resistant Severe Asthma. PLoS ONE 2014, 9, e100645. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Tiffany, C.R.; Bäumler, A.J. Dysbiosis: From fiction to function. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G602–G608. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Örtqvist, A.K.; Lundholm, C.; Kieler, H.; Ludvigsson, J.F.; Fall, T.; Ye, W.; Almqvist, C. Antibiotics in fetal and early life and subsequent childhood asthma: Nationwide population based study with sibling analysis. BMJ 2014, 349, g6979. [Google Scholar] [CrossRef]
- Ni, J.; Friedman, H.; Boyd, B.C.; McGurn, A.; Babinski, P.; Markossian, T.; Dugas, L.R. Early antibiotic exposure and development of asthma and allergic rhinitis in childhood. BMC Pediatrics 2019, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Patrick, D.M.; Sbihi, H.; Dai, D.L.Y.; Al Mamun, A.; Rasali, D.; Rose, C.; Marra, F.; Boutin, R.C.T.; Petersen, C.; Stiemsma, L.T.; et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: Evidence from population-based and prospective cohort studies. Lancet Respir. Med. 2020, 8, 1094–1105. [Google Scholar] [CrossRef]
- Zhang, M.; Litonjua, A.A.; Mueller, N.T. Maternal antibiotic use and child asthma: Is the association causal? Eur. Respir. J. 2018, 52, 1801007. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Adami, A.J.; Bracken, S.J.; Guernsey, L.A.; Rafti, E.; Maas, K.R.; Graf, J.; Matson, A.P.; Thrall, R.S.; Schramm, C.M. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatric. Res. 2018, 84, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Eunju, O.; Lee, J.Y.; Lee, M.; Han, D.; Ko, H.J.; Sprent, J.; Surh, C.D.; Kim, K.S. Food antigens drive spontaneous IgE elevation in the absence of commensal microbiota. Sci. Adv. 2019, 5, eaaw1507. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, H.K. The role of skin and orogenital microbiota in protective immunity and chronic immune-mediated inflammatory disease. Front. Immunol. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Kim, G.; Bae, J.; Kim, M.J.; Kwon, H.; Park, G.; Kim, S.J.; Choe, Y.H.; Kim, J.; Park, S.H.; Choe, B.H.; et al. Delayed Establishment of Gut Microbiota in Infants Delivered by Cesarean Section. Front. Microbiol. 2020, 11, 2099. [Google Scholar] [CrossRef] [PubMed]
- Zachariassen, L.F.; Krych, L.; Rasmussen, S.H.; Nielsen, D.S.; Kot, W.; Holm, T.L.; Hansen, A.K.; Hansen, C.H.F. Cesarean Section Induces Microbiota-Regulated Immune Disturbances in C57BL/6 Mice. J. Immunol. 2019, 202, 142–150. [Google Scholar] [CrossRef]
- Stokholm, J.; Thorsen, J.; Blaser, M.J.; Rasmussen, M.A.; Hjelmsø, M.; Shah, S.; Christensen, E.D.; Chawes, B.L.; Bønnelykke, K.; Brix, S.; et al. Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Lan, P.; He, Y.; Li, C.; Ma, X. Effect of the Modifications on the Physicochemical and Biological Properties of β-Glucan—A Critical Review. Molecules 2020, 25, 57. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Sikder, M.A.A.; Curren, B.F.; Werder, R.B.; Simpson, J.; Cuív, P.; Dennis, P.G.; Everard, M.L.; Phipps, S. The influence of the microbiome on early-life severe viral lower respiratory infections and asthma-Food for thought? Front. Immunol. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; MacIa, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.Y.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Saeed, M.A.; Gribben, K.C.; Alam, M.; Lyden, E.R.; Hanson, C.K.; LeVan, T.D. Association of Dietary Fiber on Asthma, Respiratory Symptoms, and Inflammation in the Adult National Health and Nutrition Examination Survey Population. Ann. Am. Thorac. Soc. 2020, 17, 1062–1068. [Google Scholar] [CrossRef]
- Mosconi, E.; Rekima, A.; Seitz-Polski, B.; Kanda, A.; Fleury, S.; Tissandie, E.; Monteiro, R.; Dombrowicz, D.D.; Julia, V.; Glaichenhaus, N.; et al. Breast milk immune complexes are potent inducers of oral tolerance in neonates and prevent asthma development. Mucosal Immunol. 2010, 3, 461–474. [Google Scholar] [CrossRef]
- Hsu, P.S.; Nanan, R. Does breast milk nurture T lymphocytes in their cradle? Front. Pediat. 2018, 6, 268. [Google Scholar] [CrossRef] [PubMed]
- Hasselbalch, H.; Jeppesen, D.L.; Engelmann, M.D.M.; Michaelsen, K.F.; Nielsen, M.B. Decreased thymus size in formula-fed infants compared with breastfed infants. Acta Paediatr. Int. J. Paediatr. 1996, 85, 1029–1032. [Google Scholar] [CrossRef]
- Nakajima, A.; Kaga, N.; Nakanishi, Y.; Ohno, H.; Miyamoto, J.; Kimura, I.; Hori, S.; Sasaki, T.; Hiramatsu, K.; Okumura, K.; et al. Maternal High Fiber Diet during Pregnancy and Lactation Influences Regulatory T Cell Differentiation in Offspring in Mice. J. Immunol. 2017, 199, 3516–3524. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.; Acharjee, A.; Pearce, H.; Quraishi, M.N.; Powell, R.; Rossiter, A.; Beggs, A.; Ewer, A.; Moss, P.; Toldi, G. Breastfeeding promotes early neonatal regulatory T-cell expansion and immune tolerance of non-inherited maternal antigens. Allergy Eur. J. Allergy Clin. Immunol. 2021, 76, 2447–2460. [Google Scholar] [CrossRef]
- Dong, G.-H.; Qian, Z.M.; Liu, M.-M.; Wang, D.; Ren, W.-H.; Bawa, S.; Fu, J.; Wang, J.; Lewis, R.; Zelicoff, A.; et al. Breastfeeding as a modifier of the respiratory effects of air pollution in children. Epidemiology 2013, 24, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Mendes, E.; Acetturi, B.G.; Thomas, A.M.; Martins, F.D.S.; Crisma, A.R.; Murata, G.; Braga, T.T.; Camâra, N.O.; Franco, A.L.D.S.; Setubal, J.C.; et al. Prophylactic Supplementation of Bifidobacterium longum 51A Protects Mice from Ovariectomy-Induced Exacerbated Allergic Airway Inflammation and Airway Hyperresponsiveness. Front. Microbiol. 2017, 8, 1732. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Lin, F.-H.; Lee, Y.-T.; Ku, M.-S.; Lue, K.-H. Effect of Lactobacillus rhamnosus GG immunopathologic changes in chronic mouse asthma model. J. Microbiol. Immunol. Infect. 2019, 52, 911–919. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, J.; Lee, H.K. The Role of CD4+ T Cells and Microbiota in the Pathogenesis of Asthma. Int. J. Mol. Sci. 2021, 22, 11822. https://doi.org/10.3390/ijms222111822
Jeong J, Lee HK. The Role of CD4+ T Cells and Microbiota in the Pathogenesis of Asthma. International Journal of Molecular Sciences. 2021; 22(21):11822. https://doi.org/10.3390/ijms222111822
Chicago/Turabian StyleJeong, Jiung, and Heung Kyu Lee. 2021. "The Role of CD4+ T Cells and Microbiota in the Pathogenesis of Asthma" International Journal of Molecular Sciences 22, no. 21: 11822. https://doi.org/10.3390/ijms222111822
APA StyleJeong, J., & Lee, H. K. (2021). The Role of CD4+ T Cells and Microbiota in the Pathogenesis of Asthma. International Journal of Molecular Sciences, 22(21), 11822. https://doi.org/10.3390/ijms222111822





