Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides
Abstract
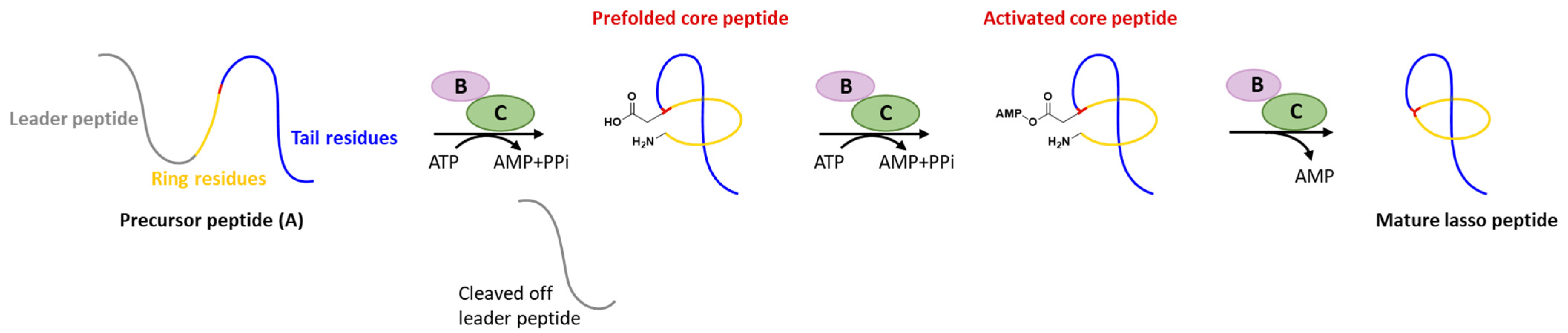
:1. Introduction
2. Disulfuration
3. Phosphorylation
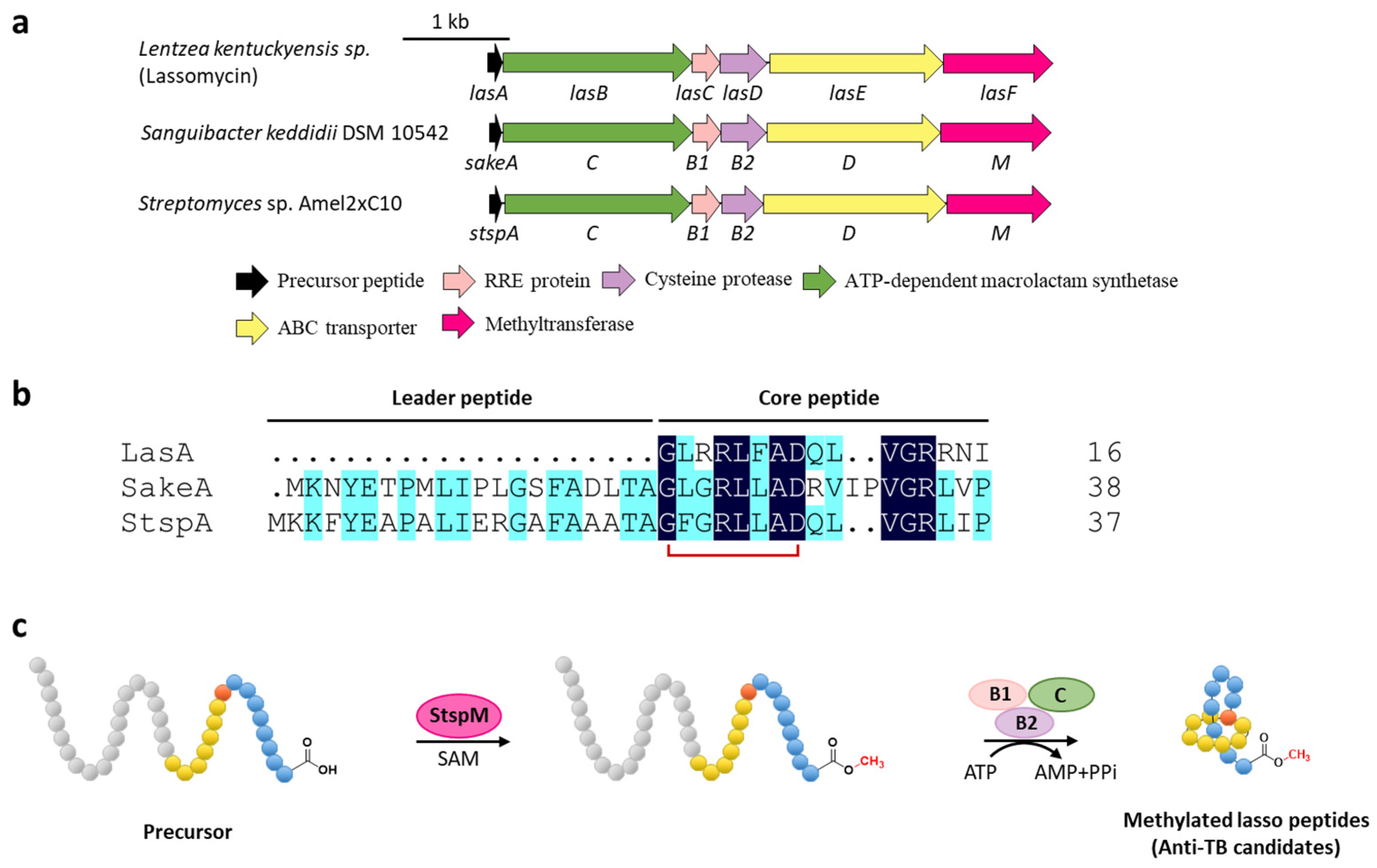
4. Methylation
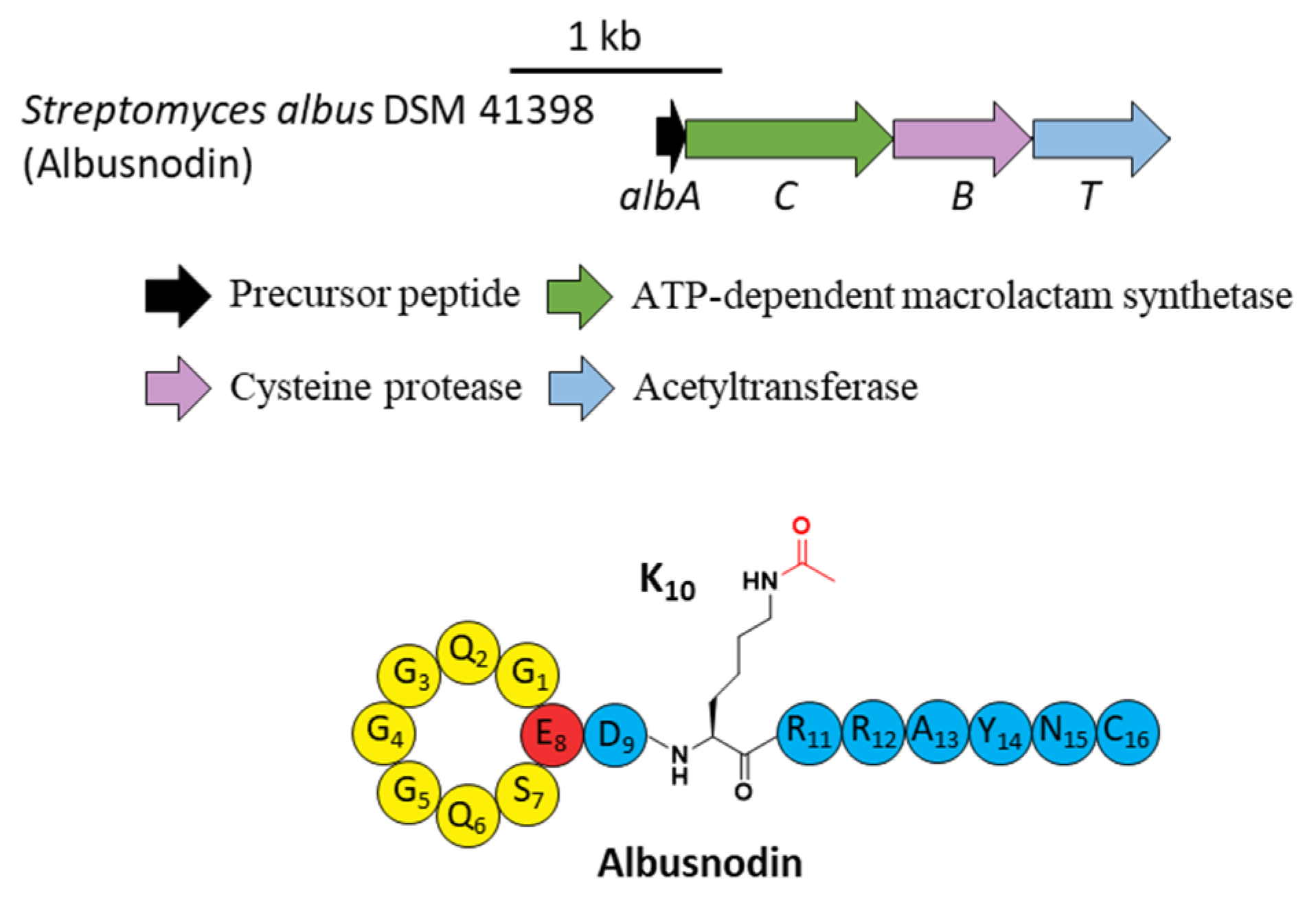
5. Acetylation
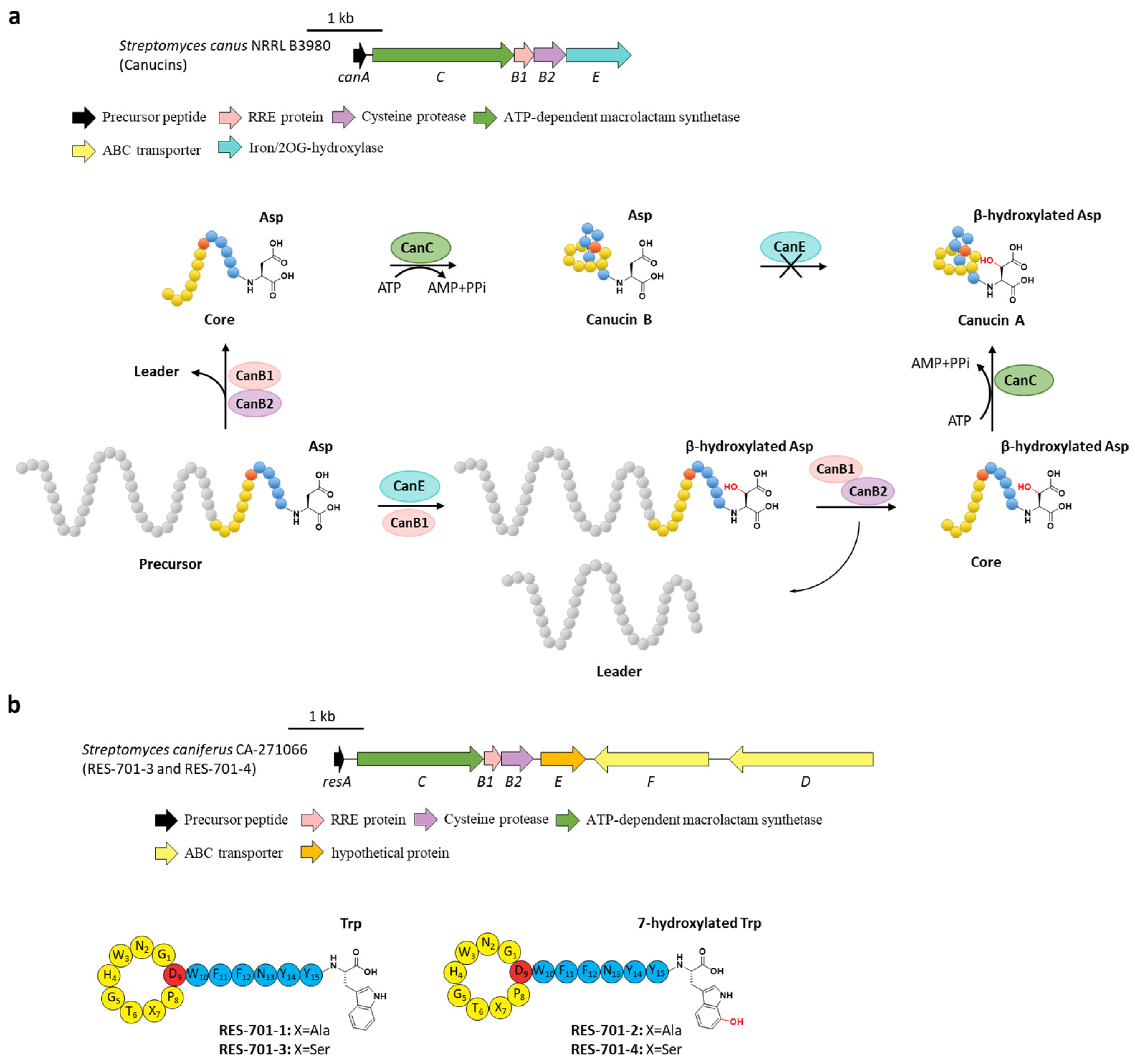
6. Hydroxylation
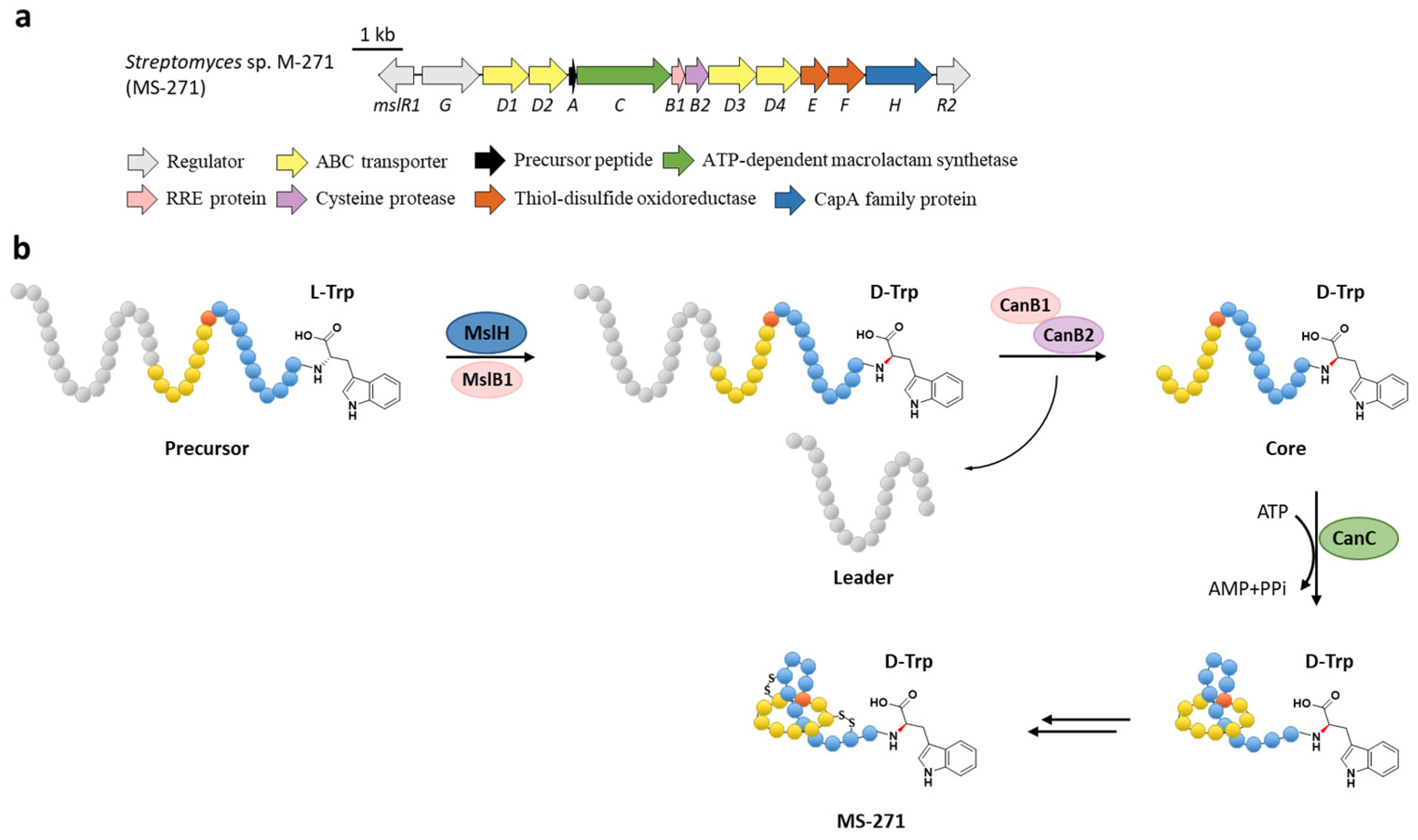
7. Epimerization
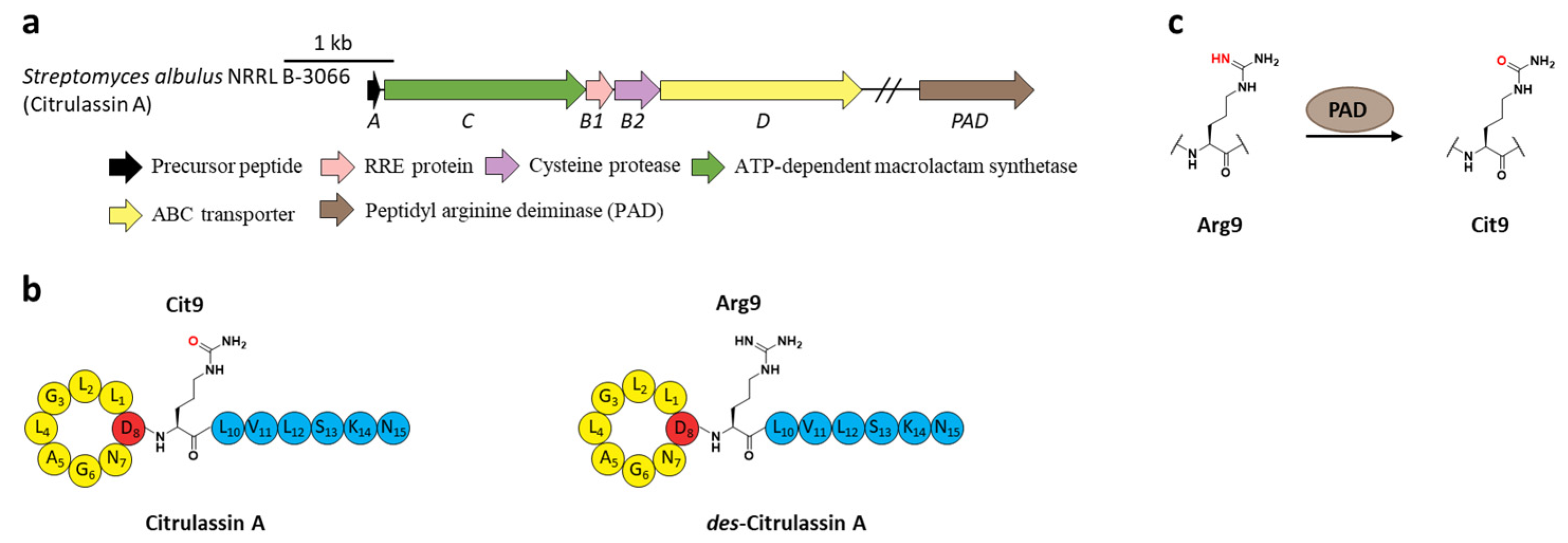
8. Citrullination
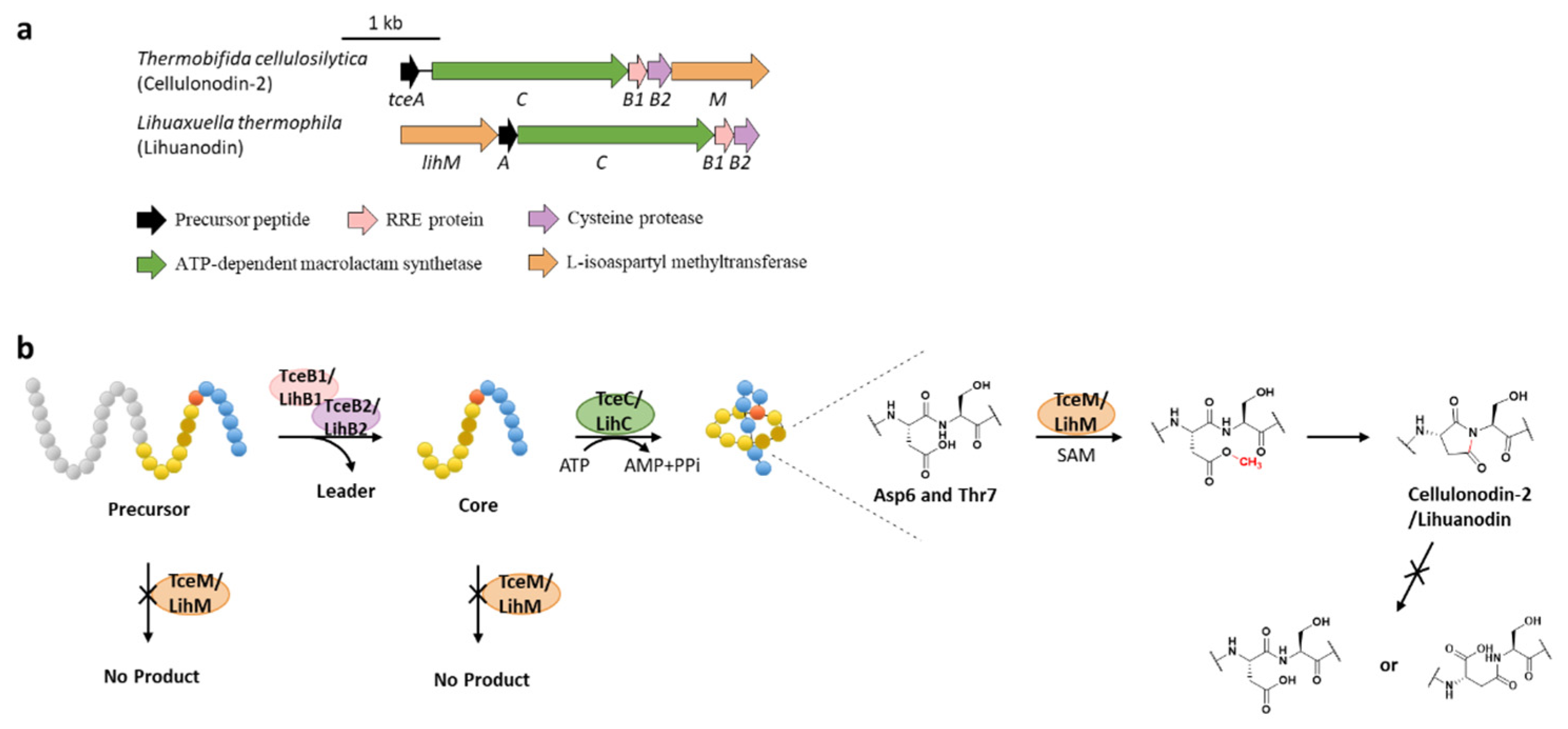
9. Succinimidation
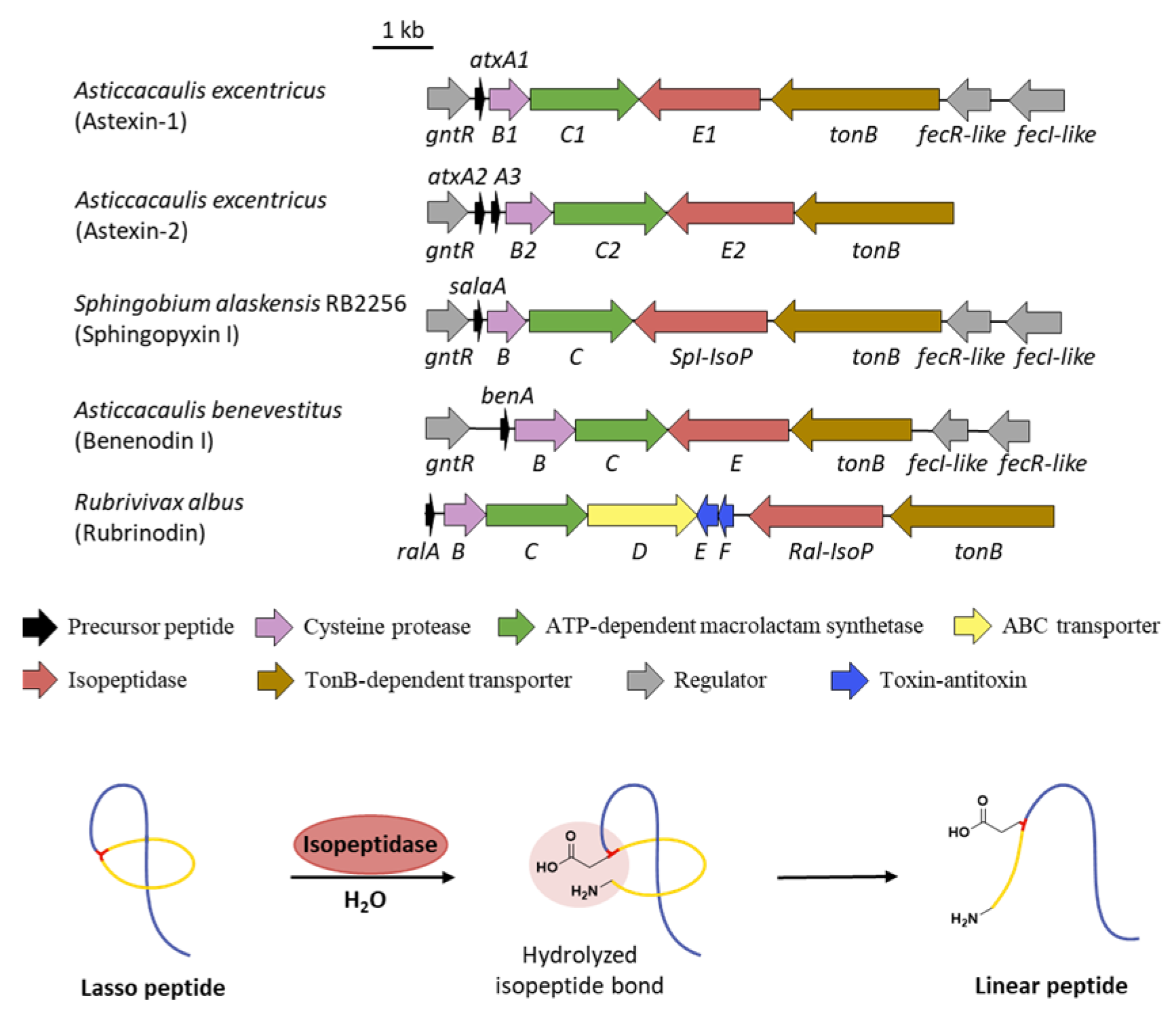
10. Linearization
11. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015, 48, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gómez, H.; Tulla-Puche, J. Lasso peptides: Chemical approaches and structural elucidation. Org. Biomol. Chem. 2018, 16, 5065–5080. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Dit-Foque, K.J.; Xie, X. 2.10—X Lasso peptides: Structure, biosynthesis, activities, and beyond. In Comprehensive Natural Products III, 3rd ed.; Liu, H.-W., Begley, T.P., Eds.; Elsevier: Oxford, UK, 2020; Volume 2, pp. 206–228. [Google Scholar]
- Cheng, C.; Hua, Z.-C. Lasso peptides: Heterologous production and potential medical application. Front. Bioeng. Biotechnol. 2020, 8, 571165. [Google Scholar] [CrossRef]
- Wang, M.; Fage, C.D.; He, Y.; Mi, J.; Yang, Y.; Li, F.; An, X.; Fan, H.; Song, L.; Zhu, S.; et al. Recent advances and perspectives on expanding the chemical diversity of lasso peptides. Front. Bioeng. Biotechnol. 2021, 9, 741364. [Google Scholar] [CrossRef]
- Norris, G.E.; Patchett, M.L. The glycocins: In a class of their own. Curr. Opin. Struct. Biol. 2016, 40, 112–119. [Google Scholar] [CrossRef]
- Narayani, M.; Babu, R.; Chadha, A.; Srivastava, S. Production of bioactive cyclotides: A comprehensive overview. Phytochem. Rev. 2020, 19, 787–825. [Google Scholar] [CrossRef]
- Kaas, Q.; Westermann, J.-C.; Craik, D.J. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef]
- Dorenbos, R.; Stein, T.; Kabel, J.; Bruand, C.; Bolhuis, A.; Bron, S.; Quax, W.J.; van Dijl, J.M. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 2002, 277, 16682–16688. [Google Scholar] [CrossRef] [Green Version]
- Gruber, C.W.; Čemažar, M.; Clark, R.J.; Horibe, T.; Renda, R.F.; Anderson, M.A.; Craik, D.J. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 2007, 282, 20435–20446. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Ogasawara, Y.; Nomura, S.; Dairi, T. Biosynthetic gene cluster of a d-tryptophan-containing lasso peptide, MS-271. ChemBioChem 2018, 19, 2045–2048. [Google Scholar] [CrossRef]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Harada, S.; Kodani, S. Isolation and structure determination of a new lasso peptide specialicin based on genome mining. Bioorg. Med. Chem. 2018, 26, 6050–6055. [Google Scholar] [CrossRef]
- Shao, M.; Ma, J.; Li, Q.; Ju, J. Identification of the anti-infective aborycin biosynthetic gene cluster from deep-sea-derived Streptomyces sp. SCSIO ZS0098 enables production in a heterologous host. Mar. Drugs 2019, 17, 127. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Hidalgo, M.; Martin, J.; Genilloud, O. Identification and heterologous expression of the biosynthetic gene cluster encoding the lasso peptide humidimycin, a caspofungin activity potentiator. Antibiotics 2020, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ducasse, R.; Zirah, S.; Blond, A.; Goulard, C.; Lescop, E.; Giraud, C.; Hartke, A.; Guittet, E.; Pernodet, J.L.; et al. Characterization of sviceucin from Streptomyces provides insight into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem. Biol. 2015, 10, 2641–2649. [Google Scholar] [CrossRef]
- Stariha, L.M.; McCafferty, D.G. Discovery of the class I antimicrobial lasso peptide arcumycin. ChemBioChem 2021, 22, 2632–2640. [Google Scholar] [CrossRef]
- Potterat, O.; Wagner, K.; Gemmecker, G.; Mack, J.; Puder, C.; Vettermann, R.; Streicher, R. BI-32169, a bicyclic 19-peptide with strong glucagon receptor antagonist activity from Streptomyces sp. J. Nat. Prod. 2004, 67, 1528–1531. [Google Scholar] [CrossRef]
- Mevaere, J.; Goulard, C.; Schneider, O.; Sekurova, O.N.; Ma, H.; Zirah, S.; Afonso, C.; Rebuffat, S.; Zotchev, S.B.; Li, Y. An orthogonal system for heterologous expression of actinobacterial lasso peptides in Streptomyces hosts. Sci. Rep. 2018, 8, 8232. [Google Scholar] [CrossRef]
- Guerrero-Garzón, J.F.; Madland, E.; Zehl, M.; Singh, M.; Rezaei, S.; Aachmann, F.L.; Courtade, G.; Urban, E.; Rückert, C.; Busche, T.; et al. Class IV lasso peptides synergistically induce proliferation of cancer cells and sensitize them to doxorubicin. iScience 2020, 23, 101785. [Google Scholar] [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Cao, L.; Link, A.J. Pandonodin: A proteobacterial lasso peptide with an exceptionally long c-terminal tail. ACS Chem. Biol. 2019, 14, 2783–2792. [Google Scholar] [CrossRef]
- Liu, T.; Ma, X.; Yu, J.; Yang, W.; Wang, G.; Wang, Z.; Ge, Y.; Song, J.; Han, H.; Zhang, W.; et al. Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications. Chem. Sci. 2021, 12, 12353–12364. [Google Scholar] [CrossRef]
- Zhu, S.; Hegemann, J.D.; Fage, C.D.; Zimmermann, M.; Xie, X.; Linne, U.; Marahiel, M.A. Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem. 2016, 291, 13662–13678. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Fage, C.D.; Hegemann, J.D.; Yan, D.; Marahiel, M.A. Dual substrate-controlled kinase activity leads to polyphosphorylated lasso peptides. FEBS Lett. 2016, 590, 3323–3334. [Google Scholar] [CrossRef]
- Zyubko, T.; Serebryakova, M.; Andreeva, J.; Metelev, M.; Lippens, G.; Dubiley, S.; Severinov, K. Efficient in vivo synthesis of lasso peptide pseudomycoidin proceeds in the absence of both the leader and the leader peptidase. Chem. Sci. 2019, 10, 9699–9707. [Google Scholar] [CrossRef] [Green Version]
- Gavrish, E.; Sit, C.S.; Cao, S.G.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the atp-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Lear, S.; Munshi, T.; Hudson, A.S.; Hatton, C.; Clardy, J.; Mosely, J.A.; Bull, T.J.; Sit, C.S.; Cobb, S.L. Total chemical synthesis of lassomycin and lassomycin-amide. Org. Biomol. Chem. 2016, 14, 4534–4541. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.W.R.; Cook, G.M.; Leung, I.K.H.; Brimble, M.A. An efficient chemical synthesis of lassomycin enabled by an on-resin lactamisation-off-resin methanolysis strategy and preparation of chemical variants. Aust. J. Chem. 2017, 70, 172–183. [Google Scholar] [CrossRef]
- Su, Y.; Han, M.; Meng, X.; Feng, Y.; Luo, S.; Yu, C.; Zheng, G.; Zhu, S. Discovery and characterization of a novel C-terminal peptide carboxyl methyltransferase in a lassomycin-like lasso peptide biosynthetic pathway. Appl. Microbiol. Biotechnol. 2019, 103, 2649–2664. [Google Scholar] [CrossRef]
- Singh, S.; Chang, A.; Goff, R.D.; Bingman, C.A.; Gruschow, S.; Sherman, D.H.; Phillips, G.N.; Thorson, J.S. Structural characterization of the mitomycin 7-O-methyltransferase. Proteins 2011, 79, 2181–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwatsuki, M.; Tomoda, H.; Uchida, R.; Gouda, H.; Hirono, S.; Omura, S. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J. Am. Chem. Soc. 2006, 128, 7486–7491. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Uchida, R.; Takakusagi, Y.; Matsumoto, A.; Jiang, C.L.; Takahashi, Y.; Arai, M.; Kobayashi, S.; Matsumoto, M.; Inokoshi, J.; et al. Lariatins, novel anti-mycobacterial peptides with a lasso structure, produced by Rhodococcus jostii K01-B0171. J. Antibiot. 2007, 60, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Koizumi, Y.; Gouda, H.; Hirono, S.; Tomoda, H.; Omura, S. Lys17 in the ‘lasso’ peptide lariatin A is responsible for anti-mycobacterial activity. Bioorg. Med. Chem. Lett. 2009, 19, 2888–2890. [Google Scholar] [CrossRef] [PubMed]
- Inokoshi, J.; Koyama, N.; Miyake, M.; Shimizu, Y.; Tomoda, H. Structure-activity analysis of Gram-positive Bacterium-producing lasso peptides with anti-mycobacterial activity. Sci. Rep. 2016, 6, 30375. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Cheung-Lee, W.L.; Elashal, H.E.; Raj, M.; Link, A.J. Albusnodin: An acetylated lasso peptide from Streptomyces albus. Chem. Commun. 2018, 54, 1339–1342. [Google Scholar] [CrossRef]
- Son, S.; Jang, M.; Lee, B.; Hong, Y.-S.; Ko, S.-K.; Jang, J.-H.; Ahn, J.S. Ulleungdin, a lasso peptide with cancer cell migration inhibitory activity discovered by the genome mining approach. J. Nat. Prod. 2018, 81, 2205–2211. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Y.H.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef]
- Zhang, C.; Seyedsayamdost, M.R. CanE, an iron/2-oxoglutarate-dependent lasso peptide hydroxylase from Streptomyces canus. ACS Chem. Biol. 2020, 15, 890–894. [Google Scholar] [CrossRef]
- Krebs, C.; Fujimori, D.G.; Walsh, C.T.; Bollinger, J.M. Non-heme Fe(IV)-oxo intermediates. Acc. Chem. Res. 2007, 40, 484–492. [Google Scholar] [CrossRef] [Green Version]
- Maksimov, M.O.; Pan, S.J.; Link, A.J. Lasso peptides: Structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 2012, 29, 996–1006. [Google Scholar] [CrossRef]
- Burkhart, B.J.; Hudson, G.A.; Dunbar, K.L.; Mitchell, D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 2015, 11, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Fage, C.D.; Hegemann, J.D.; Mielcarek, A.; Yan, D.; Linne, U.; Marahiel, M.A. The B1 protein guides the biosynthesis of a lasso peptide. Sci. Rep. 2016, 6, 35604. [Google Scholar] [CrossRef] [Green Version]
- Cheung, W.L.; Chen, M.Y.; Maksimov, M.O.; Link, A.J. Lasso peptide biosynthetic protein larB1 binds both leader and core peptide regions of the precursor protein LarA. ACS Cent. Sci. 2016, 2, 702–709. [Google Scholar] [CrossRef]
- Yamasaki, M.; Yano, K.; Yoshida, M.; Matsuda, Y.; Yamaguchi, K. RES-701-1, a novel and selective endothelin type B receptor antagonist produced by Streptomyces sp. RE-701. II. Determination of the primary sequence. J. Antibiot. 1994, 47, 276–280. [Google Scholar] [CrossRef] [Green Version]
- Yano, K.; Yamasaki, M.; Yoshida, M.; Matsuda, Y.; Yamaguchi, K. Res-701-2, a novel and selective endothelin type-B receptor antagonist produced by Streptomyces Sp. 2. Determination of the primary structure. J. Antibiot. 1995, 48, 1368–1370. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, T.; Ochiai, K.; Tanaka, T.; Tsukuda, E.; Chiba, S.; Yano, K.; Yamasaki, M.; Yoshida, M.; Matsuda, Y. Res-701-2, Res-701-3 and Res-701-4, novel and selective endothelin type-B receptor antagonists produced by Streptomyces Sp. 1. Taxonomy of producing strains, fermentation, isolation, and biochemical-properties. J. Antibiot. 1995, 48, 1213–1220. [Google Scholar] [CrossRef] [Green Version]
- Oves-Costales, D.; Sánchez-Hidalgo, M.; Martín, J.; Genilloud, O. Identification, cloning and heterologous expression of the gene cluster directing RES-701-3, -4 lasso peptides biosynthesis from a marine Streptomyces strain. Mar. Drugs 2020, 18, 238. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Shigematsu, M.; Sato, S.; Kato, H.; Dairi, T. Involvement of peptide epimerization in poly-γ-glutamic acid biosynthesis. Org. Lett. 2019, 21, 3972–3975. [Google Scholar] [CrossRef]
- Feng, Z.; Ogasawara, Y.; Dairi, T. Identification of the peptide epimerase MslH responsible for d-amino acid introduction at the C-terminus of ribosomal peptides. Chem. Sci. 2020, 12, 2567–2574. [Google Scholar] [CrossRef]
- Freeman, M.F.; Helf, M.J.; Bhushan, A.; Morinaka, B.I.; Piel, J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat. Chem. 2017, 9, 387–395. [Google Scholar] [CrossRef]
- Benjdia, A.; Guillot, A.; Ruffié, P.; Leprince, J.; Berteau, O. Post-translational modification of ribosomally synthesized peptides by a radical SAM epimerase in Bacillus subtilis. Nat. Chem. 2017, 9, 698–707. [Google Scholar] [CrossRef]
- Yang, X.; van der Donk, W.A. Post-translational introduction of d-alanine into ribosomally synthesized peptides by the dehydroalanine reductase NpnJ. J. Am. Chem. Soc. 2015, 137, 12426–12429. [Google Scholar] [CrossRef]
- Huo, L.; van der Donk, W.A. Discovery and characterization of bicereucin, an unusual d-amino acid-containing mixed two-component lantibiotic. J. Am. Chem. Soc. 2016, 138, 5254–5257. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, F.; Cheng, Z.; Bashiri, G.; Wang, J.; Hong, J.; Wang, Y.; Xu, L.; Chen, X.; Huang, S.-X.; et al. Functional genome mining reveals a class V lanthipeptide containing a d-amino acid introduced by an F420H2-dependent reductase. Angew. Chem. Int. Ed. 2020, 59, 18029–18035. [Google Scholar] [CrossRef]
- Harris, L.A.; Saint-Vincent, P.M.B.; Guo, X.; Hudson, G.A.; DiCaprio, A.J.; Zhu, L.; Mitchell, D.A. Reactivity-based screening for citrulline-containing natural products reveals a family of bacterial peptidyl arginine deiminases. ACS Chem. Biol. 2020, 15, 3167–3175. [Google Scholar] [CrossRef]
- Kamble, N.U.; Majee, M. Protein l-isoaspartyl methyltransferase (PIMT) in plants: Regulations and functions. Biochem. J. 2020, 477, 4453–4471. [Google Scholar] [CrossRef]
- Cao, L.; Beiser, M.; Koos, J.D.; Orlova, M.; Elashal, H.E.; Schroder, H.V.; Link, A.J. Cellulonodin-2 and lihuanodin: Lasso peptides with an aspartimide post-translational modification. J. Am. Chem. Soc. 2021, 143, 11690–11702. [Google Scholar] [CrossRef]
- Acedo, J.Z.; Bothwell, I.R.; An, L.; Trouth, A.; Frazier, C.; van der Donk, W.A. O-Methyltransferase-mediated incorporation of a beta-amino acid in lanthipeptides. J. Am. Chem. Soc. 2019, 141, 16790–16801. [Google Scholar] [CrossRef]
- Maksimov, M.O.; Pelczer, I.; Link, A.J. Precursor-centric genome-mining approach for lasso peptide discovery. Proc. Natl. Acad. Sci. USA 2012, 109, 15223–15228. [Google Scholar] [CrossRef] [Green Version]
- Maksimov, M.O.; Link, A.J. Discovery and characterization of an isopeptidase that linearizes lasso peptides. J. Am. Chem. Soc. 2013, 135, 12038–12047. [Google Scholar] [CrossRef] [PubMed]
- Fage, C.D.; Hegemann, J.D.; Nebel, A.J.; Steinbach, R.M.; Zhu, S.; Linne, U.; Harms, K.; Bange, G.; Marahiel, M.A. Structure and mechanism of the sphingopyxin I lasso peptide isopeptidase. Angew. Chem. Int. Ed. 2016, 55, 12717–12721. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, M.O.; Koos, J.D.; Zong, C.; Lisko, B.; Link, A.J. Elucidating the specificity determinants of the AtxE2 lasso peptide isopeptidase. J. Biol. Chem. 2015, 290, 30806–30812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chekan, J.R.; Koos, J.D.; Zong, C.; Maksimov, M.O.; Link, A.J.; Nair, S.K. Structure of the lasso peptide isopeptidase identifies a topology for processing threaded substrates. J. Am. Chem. Soc. 2016, 138, 16452–16458. [Google Scholar] [CrossRef] [Green Version]
- Zong, C.H.; Wu, M.J.; Qin, J.Z.; Link, A.J. Lasso peptide benenodin-1 is a thermally actuated [1]rotaxane switch. J. Am. Chem. Soc. 2017, 139, 10403–10409. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Caulosegnins I-III: A highly diverse group of lasso peptides derived from a single biosynthetic gene cluster. J. Am. Chem. Soc. 2013, 135, 210–222. [Google Scholar] [CrossRef]
- Kuroha, M.; Hemmi, H.; Ohnishi-Kameyama, M.; Kodani, S. Isolation and structure determination of a new lasso peptide subterisin from Sphingomonas subterranea. Tetrahedron Lett. 2017, 58, 3429–3432. [Google Scholar] [CrossRef]
- Fuwa, H.; Hemmi, H.; Kaweewan, I.; Kozaki, I.; Honda, H.; Kodani, S. Heterologous production of new lasso peptide koreensin based on genome mining. J. Antibiot. 2021, 74, 42–50. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Xiu, H.; Wang, M.; Fage, C.D.; He, Y.; Niu, X.; Han, M.; Li, F.; An, X.; Fan, H.; Song, L.; et al. Discovery and characterization of rubrinodin provide clues into the evolution of lasso peptides. Biochemistry 2022, 61, 595–607. [Google Scholar] [CrossRef]
- Metelev, M.; Arseniev, A.; Bushin, L.B.; Kuznedelov, K.; Artamonova, T.O.; Kondratenko, R.; Khodorkovskii, M.; Seyedsayamdost, M.R.; Severinov, K. Acinetodin and klebsidin, RNA polymerase targeting lasso peptides produced by human isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem. Biol. 2017, 12, 814–824. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Parry, M.E.; Jaramillo Cartagena, A.; Darst, S.A.; Link, A.J. Discovery and structure of the antimicrobial lasso peptide citrocin. J. Biol. Chem. 2019, 294, 6822–6830. [Google Scholar] [CrossRef]
- Lassogen, Inc. Available online: https://www.lassogen.com/ (accessed on 25 June 2022).
- Lassogen, CipherBio. Available online: https://www.cipherbio.com/data-viz/organization/Lassogen/products (accessed on 25 June 2022).
- Funk, M.A.; van der Donk, W.A. Ribosomal natural products, tailored to fit. Acc. Chem. Res. 2017, 50, 1577–1586. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, S.; Martínez-Gómez, A.I.; Rodríguez-Vico, F.; Clemente-Jiménez, J.M.; Las Heras-Vázquez, F.J. d-amino acids related to nutrition and industrial applications. Chem. Biodivers. 2010, 7, 1531–1548. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Niu, W.; Pang, L.; Bian, X.; Zhang, Y.; Zhong, G. Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. Int. J. Mol. Sci. 2022, 23, 7231. https://doi.org/10.3390/ijms23137231
Duan Y, Niu W, Pang L, Bian X, Zhang Y, Zhong G. Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. International Journal of Molecular Sciences. 2022; 23(13):7231. https://doi.org/10.3390/ijms23137231
Chicago/Turabian StyleDuan, Yuwei, Weijing Niu, Linlin Pang, Xiaoying Bian, Youming Zhang, and Guannan Zhong. 2022. "Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides" International Journal of Molecular Sciences 23, no. 13: 7231. https://doi.org/10.3390/ijms23137231
APA StyleDuan, Y., Niu, W., Pang, L., Bian, X., Zhang, Y., & Zhong, G. (2022). Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. International Journal of Molecular Sciences, 23(13), 7231. https://doi.org/10.3390/ijms23137231





