Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro
Abstract
1. Introduction
2. Results
2.1. Experimental Approach for Analysis of the Effect of PARP1 on Transcription through a Nucleosome
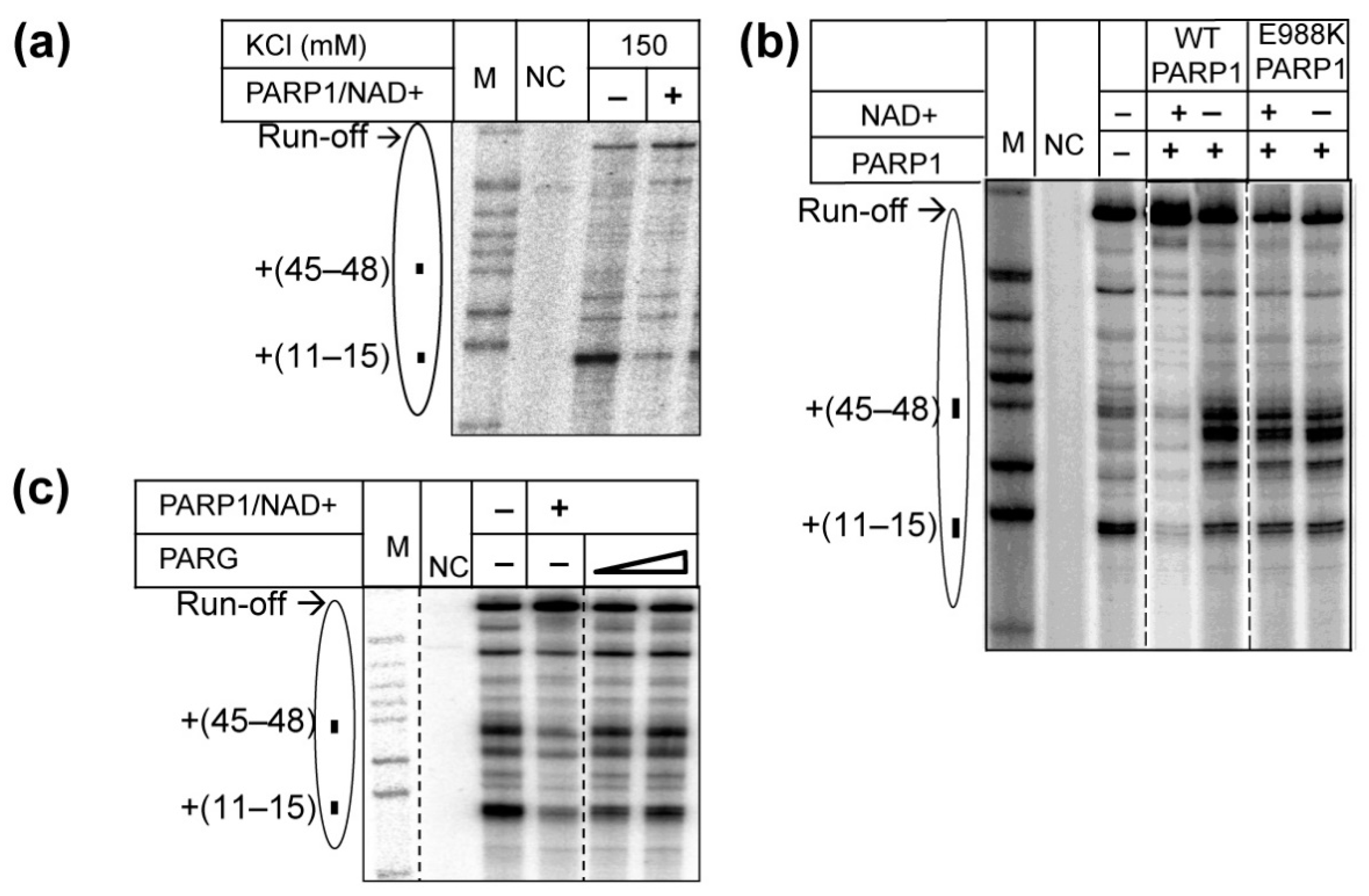
2.2. PARP1 Facilitates Transcription through the Nucleosome by Pol II
2.3. Catalytic Activity of PARP1 Is Required to Facilitate Transcription through the Nucleosome
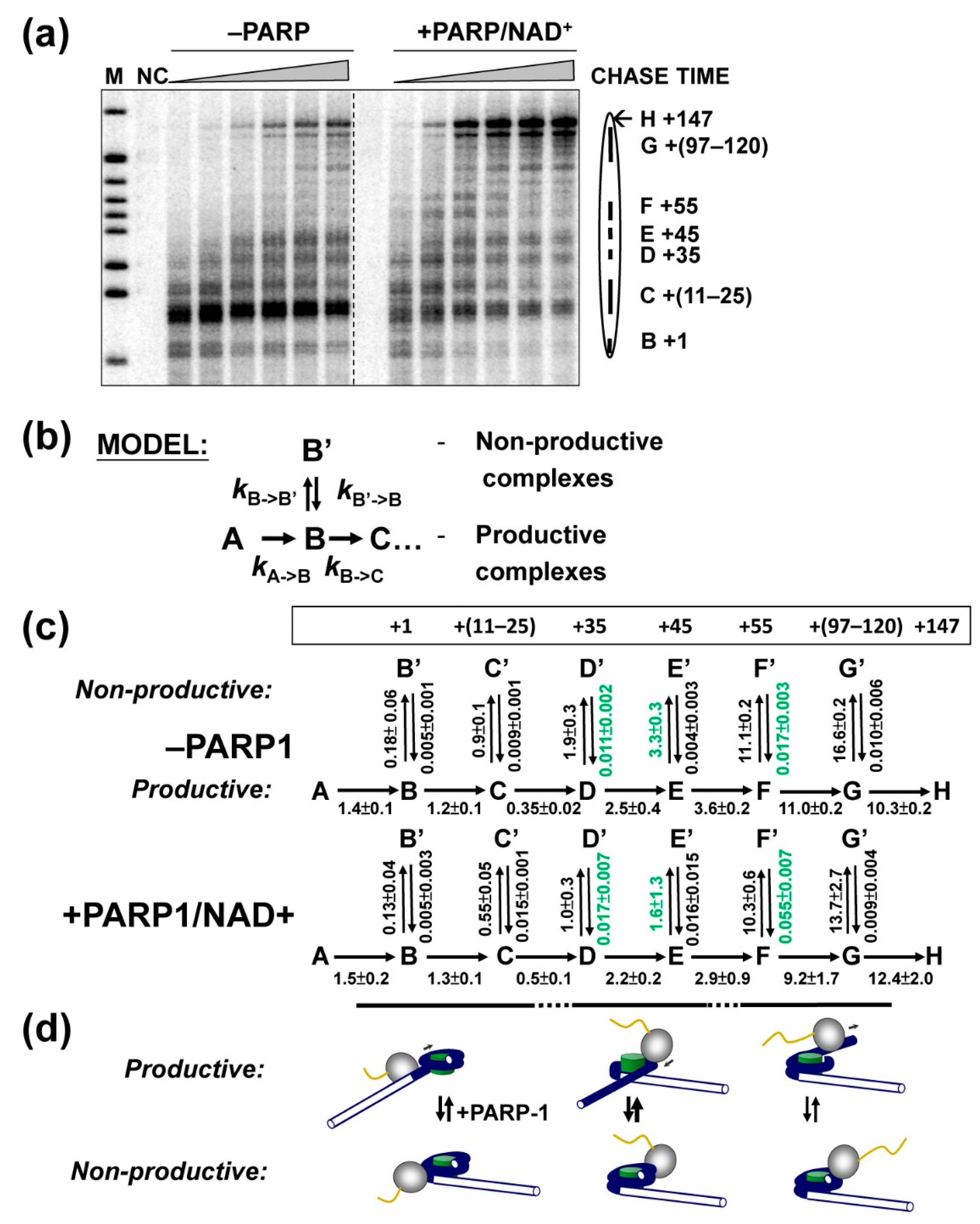
2.4. PARP1-Induced PARylation Facilitates Formation of Productive Elongation Complexes
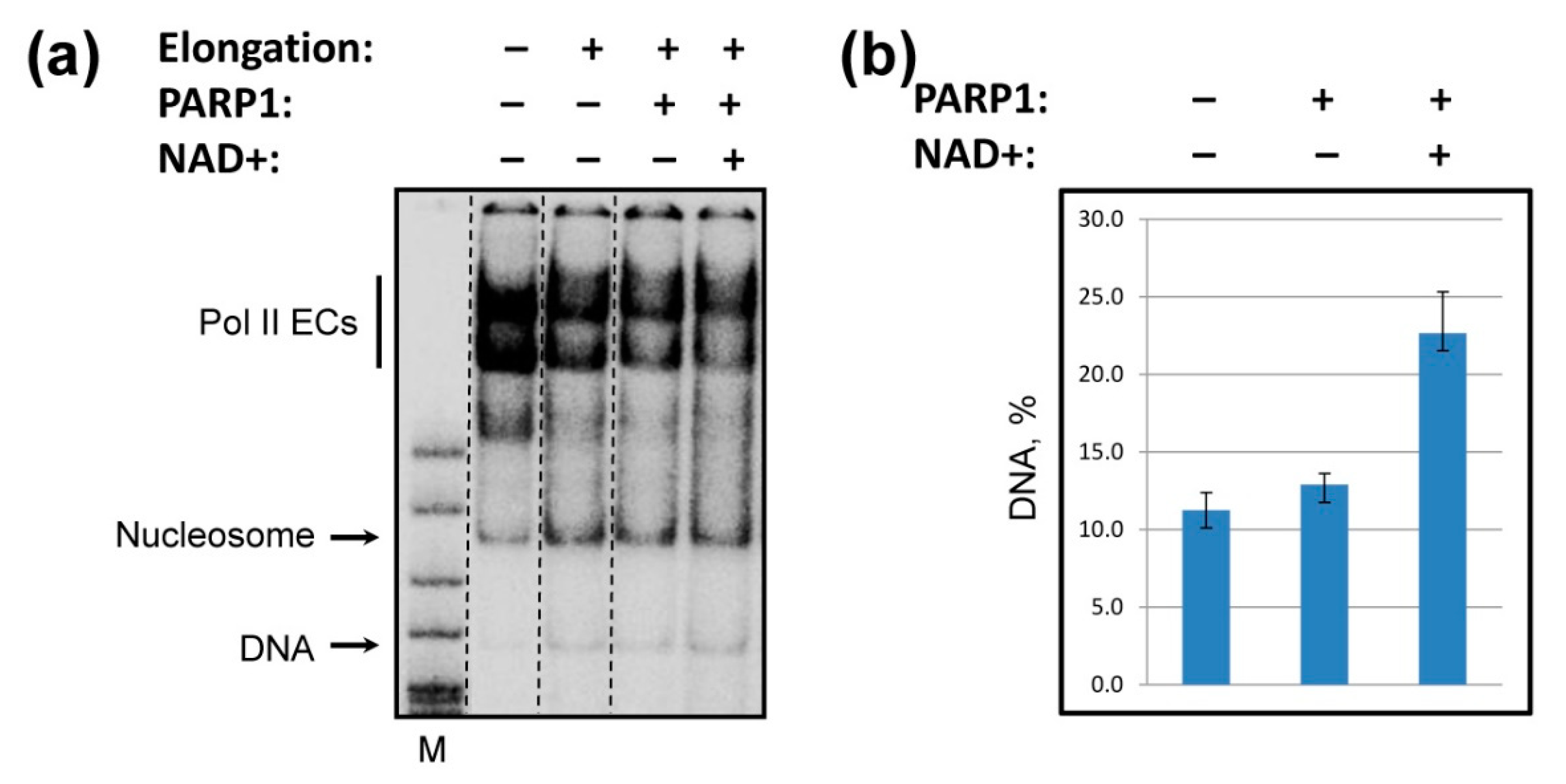
2.5. PARP1 Facilitates Histone Eviction during Transcription
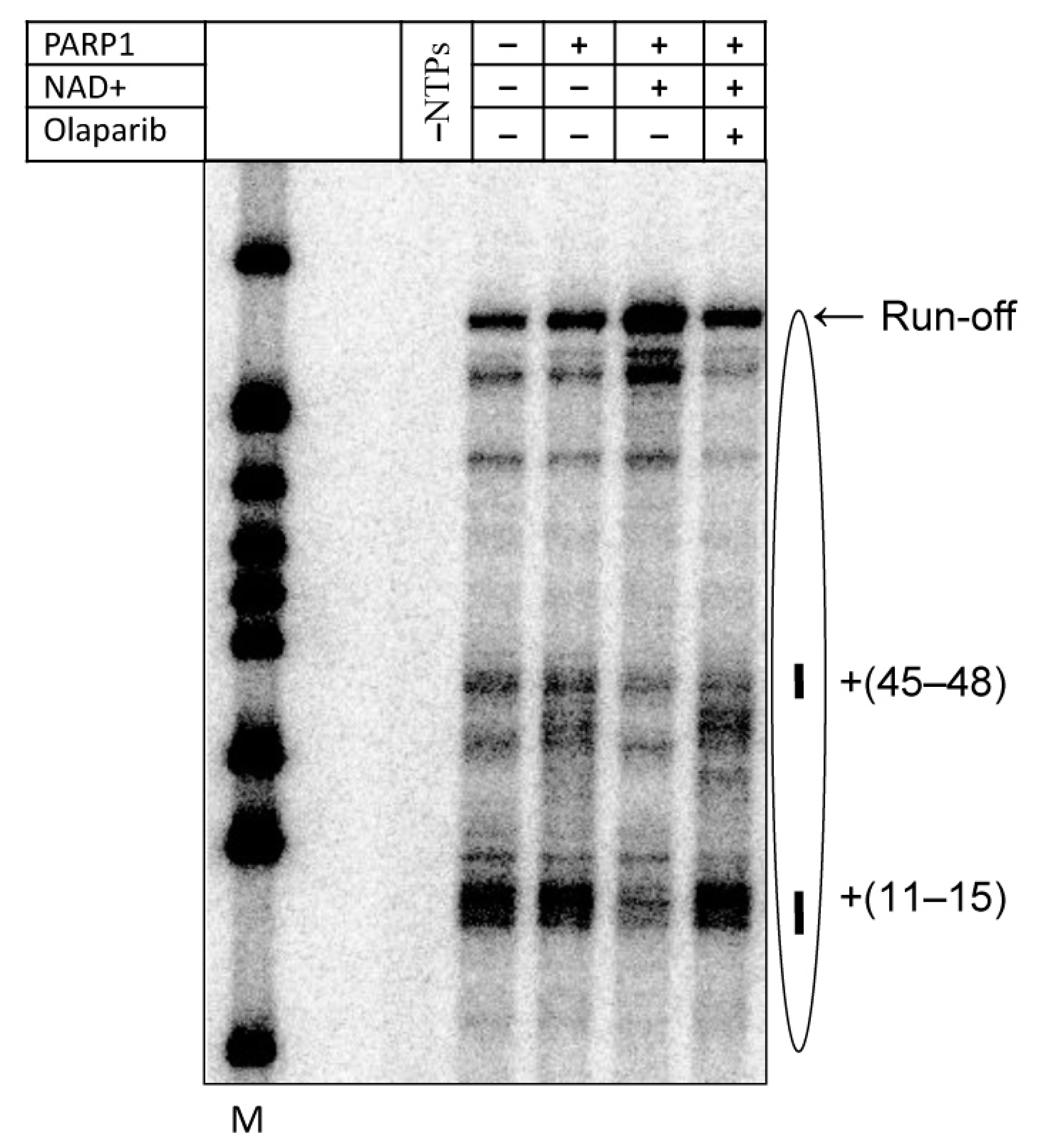
2.6. Anti-Cancer Drug Olaparib Strongly Inhibits PARP1-Dependent Transcription through a Nucleosome
3. Discussion
3.1. Mechanism of PARP1 Action during Transcription through a Nucleosome
3.2. Mechanism of Olaparib Action during Transcription through a Nucleosome
3.3. Implications In Vivo
3.4. Conclusions
4. Materials and Methods
4.1. Purification of Proteins, DNA Templates, and Nucleosomes
4.2. Transcription and Nucleosome Fate Assays
4.3. Time-Courses of yPol II Transcription
4.4. PARP1 Automodification
4.5. Core Histones ADP-Ribosylation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Kutuzov, M.M.; Belousova, E.A.; Kurgina, T.A.; Ukraintsev, A.A.; Vasil’eva, I.A.; Khodyreva, S.N.; Lavrik, O.I. The contribution of PARP1, PARP2 and poly(ADP-ribosyl)ation to base excision repair in the nucleosomal context. Sci. Rep. 2021, 11, 4849. [Google Scholar] [CrossRef] [PubMed]
- Shieh, W.M.; Ame, J.C.; Wilson, M.V.; Wang, Z.Q.; Koh, D.W.; Jacobson, M.K.; Jacobson, E.L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998, 273, 30069–30072. [Google Scholar] [CrossRef]
- Eisemann, T.; Pascal, J.M. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell Mol. Life Sci. 2020, 77, 19–33. [Google Scholar] [CrossRef]
- van Beek, L.; McClay, E.; Patel, S.; Schimpl, M.; Spagnolo, L.; Maia de Oliveira, T. PARP Power: A Structural Perspective on PARP1, PARP2, and PARP3 in DNA Damage Repair and Nucleosome Remodelling. Int. J. Mol. Sci. 2021, 22, 5112. [Google Scholar] [CrossRef]
- Eustermann, S.; Wu, W.F.; Langelier, M.F.; Yang, J.C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Zandarashvili, L.; Aguiar, P.M.; Black, B.E.; Pascal, J.M. NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 2018, 9, 844. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zamudio, R.; Ha, H.C. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol. Cell Biol. 2012, 32, 2490–2502. [Google Scholar] [CrossRef]
- Tanuma, S.; Kawashima, K.; Endo, H. Comparison of ADP-ribosylation of chromosomal proteins between intact and broken cells. Biochem. Biophys. Res. Commun. 1985, 127, 896–902. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Feofanov, A.V.; Studitsky, V.M. PARP-1-Associated Pathological Processes: Inhibition by Natural Polyphenols. Int. J. Mol. Sci. 2021, 22, 1441. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, J.; Jung, K.; Luger, K. Inhibitors of PARP: Number crunching and structure gazing. Proc. Natl. Acad. Sci. USA 2022, 119, e2121979119. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, C.C.; Moore, K.N. Olaparib: An oral PARP-1 and PARP-2 inhibitor with promising activity in ovarian cancer. Future Oncol. 2015, 11, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Tulin, A.V. Poly-ADP-ribose polymerase: Machinery for nuclear processes. Mol. Asp. Med. 2013, 34, 1124–1137. [Google Scholar] [CrossRef]
- Kraus, W.L. PARPs and ADP-Ribosylation: 50 Years… and Counting. Mol. Cell 2015, 58, 902–910. [Google Scholar] [CrossRef]
- Kraus, W.L. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008, 20, 294–302. [Google Scholar] [CrossRef]
- Petesch, S.J.; Lis, J.T. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012, 28, 285–294. [Google Scholar] [CrossRef]
- Ji, Y.; Tulin, A.V. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 2010, 20, 512–518. [Google Scholar] [CrossRef]
- Gibson, B.A.; Zhang, Y.; Jiang, H.; Hussey, K.M.; Shrimp, J.H.; Lin, H.; Schwede, F.; Yu, Y.; Kraus, W.L. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 2016, 353, 45–50. [Google Scholar] [CrossRef]
- Gadad, S.S.; Camacho, C.V.; Malladi, V.; Hutti, C.R.; Nagari, A.; Kraus, W.L. PARP-1 Regulates Estrogen-Dependent Gene Expression in Estrogen Receptor alpha-Positive Breast Cancer Cells. Mol. Cancer Res. MCR 2021, 19, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.H.; Castellano, G.; Bonet, J.; Le Dily, F.; Font-Mateu, J.; Ballare, C.; Nacht, A.S.; Soronellas, D.; Oliva, B.; Beato, M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012, 26, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Kraus, W.L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Guastafierro, T.; Cecchinelli, B.; Zampieri, M.; Reale, A.; Riggio, G.; Sthandier, O.; Zupi, G.; Calabrese, L.; Caiafa, P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J. Biol. Chem. 2008, 283, 21873–21880. [Google Scholar] [CrossRef]
- Petesch, S.J.; Lis, J.T. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol. Cell 2012, 45, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Petesch, S.J.; Lis, J.T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 2008, 134, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Mauro, S.; Gevry, N.; Lis, J.T.; Kraus, W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004, 119, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Gaykalova, D.A.; Pestov, N.A.; Golovastov, V.V.; Vassylyev, D.G.; Artsimovitch, I.; Studitsky, V.M. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009, 16, 1272–1278. [Google Scholar] [CrossRef]
- Bondarenko, V.A.; Steele, L.M.; Ujvari, A.; Gaykalova, D.A.; Kulaeva, O.I.; Polikanov, Y.S.; Luse, D.S.; Studitsky, V.M. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 2006, 24, 469–479. [Google Scholar] [CrossRef]
- Kulaeva, O.I.; Gaykalova, D.A.; Studitsky, V.M. Transcription through chromatin by RNA polymerase II: Histone displacement and exchange. Mutat. Res. 2007, 618, 116–129. [Google Scholar] [CrossRef]
- Kulaeva, O.I.; Hsieh, F.K.; Studitsky, V.M. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl. Acad. Sci. USA 2010, 107, 11325–11330. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Studitsky, V.M. Mechanism of histone survival during transcription by RNA polymerase II. Transcription 2010, 1, 85–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, H.W.; Valieva, M.E.; Safina, A.; Chereji, R.V.; Wang, J.; Kulaeva, O.I.; Morozov, A.V.; Kirpichnikov, M.P.; Feofanov, A.V.; Gurova, K.V.; et al. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci. Adv. 2018, 4, eaav2131. [Google Scholar] [CrossRef]
- Chang, H.W.; Kulaeva, O.I.; Shaytan, A.K.; Kibanov, M.; Kuznedelov, K.; Severinov, K.V.; Kirpichnikov, M.P.; Clark, D.J.; Studitsky, V.M. Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 2014, 42, 1619–1627. [Google Scholar] [CrossRef]
- Hsieh, F.K.; Kulaeva, O.I.; Patel, S.S.; Dyer, P.N.; Luger, K.; Reinberg, D.; Studitsky, V.M. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 2013, 110, 7654–7659. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Pestov, N.A.; Hsieh, F.K.; Studitsky, V.M. Experimental analysis of the mechanism of chromatin remodeling by RNA polymerase II. Methods Enzym. 2012, 512, 293–314. [Google Scholar] [CrossRef]
- Jungmichel, S.; Rosenthal, F.; Altmeyer, M.; Lukas, J.; Hottiger, M.O.; Nielsen, M.L. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 2013, 52, 272–285. [Google Scholar] [CrossRef]
- Sultanov, D.C.; Gerasimova, N.S.; Kudryashova, K.S.; Maluchenko, N.V.; Kotova, E.Y.; Langelier, M.F.; Pascal, J.M.; Kirpichnikov, M.P.; Feofanov, A.V.; Studitsky, V.M. Unfolding of core nucleosomes by PARP-1 revealed by spFRET microscopy. AIMS Genet. 2017, 4, 21–31. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Hepler, M.R.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757. [Google Scholar] [CrossRef]
- Clark, N.J.; Kramer, M.; Muthurajan, U.M.; Luger, K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J. Biol. Chem. 2012, 287, 32430–32439. [Google Scholar] [CrossRef]
- Ruf, A.; Rolli, V.; de Murcia, G.; Schulz, G.E. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 1998, 278, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wacker, D.A.; Ruhl, D.D.; Balagamwala, E.H.; Hope, K.M.; Zhang, T.; Kraus, W.L. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell Biol. 2007, 27, 7475–7485. [Google Scholar] [CrossRef] [PubMed]
- Gaykalova, D.A.; Kulaeva, O.I.; Volokh, O.; Shaytan, A.K.; Hsieh, F.K.; Kirpichnikov, M.P.; Sokolova, O.S.; Studitsky, V.M. Structural analysis of nucleosomal barrier to transcription. Proc. Natl. Acad. Sci. USA 2015, 112, E5787–E5795. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Hsieh, F.K.; Patel, S.S.; Studitsky, V.M. Time-resolved analysis of transcription through chromatin. Methods 2019, 159–160, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.; Levin, M.K.; Patel, S.S. Experimental and computational analysis of DNA unwinding and polymerization kinetics. Methods Mol. Biol. 2010, 587, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Izban, M.G.; Luse, D.S. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991, 5, 683–696. [Google Scholar] [CrossRef]
- Kireeva, M.L.; Hancock, B.; Cremona, G.H.; Walter, W.; Studitsky, V.M.; Kashlev, M. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 2005, 18, 97–108. [Google Scholar] [CrossRef]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef]
- Shen, Y.; Aoyagi-Scharber, M.; Wang, B. Trapping Poly(ADP-Ribose) Polymerase. J. Pharmacol. Exp. Ther. 2015, 353, 446–457. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 2127. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Huang, S.Y.; Renaud, A.; Zhang, Y.; Ji, J.; Takeda, S.; Morris, J.; Teicher, B.; Doroshow, J.H.; Pommier, Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.A.; Cesaroni, M.; Denny, M.F.; Lupey, L.N.; Tempera, I. Global Transcriptome Analysis Reveals That Poly(ADP-Ribose) Polymerase 1 Regulates Gene Expression through EZH2. Mol. Cell Biol. 2015, 35, 3934–3944. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Gamble, M.J.; Frizzell, K.M.; Berrocal, J.G.; Kininis, M.; Kraus, W.L. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 2008, 319, 819–821. [Google Scholar] [CrossRef] [PubMed]
- Nalabothula, N.; Al-jumaily, T.; Eteleeb, A.M.; Flight, R.M.; Xiaorong, S.; Moseley, H.; Rouchka, E.C.; Fondufe-Mittendorf, Y.N. Genome-Wide Profiling of PARP1 Reveals an Interplay with Gene Regulatory Regions and DNA Methylation. PLoS ONE 2015, 10, e0135410. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kraus, W.L. Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Mol. Cell 2017, 65, 589–603.e9. [Google Scholar] [CrossRef] [PubMed]
- Kireeva, M.L.; Walter, W.; Tchernajenko, V.; Bondarenko, V.; Kashlev, M.; Studitsky, V.M. Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell 2002, 9, 541–552. [Google Scholar] [CrossRef]
- Ujvari, A.; Hsieh, F.K.; Luse, S.W.; Studitsky, V.M.; Luse, D.S. Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J. Biol. Chem. 2008, 283, 32236–32243. [Google Scholar] [CrossRef] [PubMed]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotova, E.Y.; Hsieh, F.-K.; Chang, H.-W.; Maluchenko, N.V.; Langelier, M.-F.; Pascal, J.M.; Luse, D.S.; Feofanov, A.V.; Studitsky, V.M. Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro. Int. J. Mol. Sci. 2022, 23, 7107. https://doi.org/10.3390/ijms23137107
Kotova EY, Hsieh F-K, Chang H-W, Maluchenko NV, Langelier M-F, Pascal JM, Luse DS, Feofanov AV, Studitsky VM. Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro. International Journal of Molecular Sciences. 2022; 23(13):7107. https://doi.org/10.3390/ijms23137107
Chicago/Turabian StyleKotova, Elena Y., Fu-Kai Hsieh, Han-Wen Chang, Natalia V. Maluchenko, Marie-France Langelier, John M. Pascal, Donal S. Luse, Alexey V. Feofanov, and Vasily M. Studitsky. 2022. "Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro" International Journal of Molecular Sciences 23, no. 13: 7107. https://doi.org/10.3390/ijms23137107
APA StyleKotova, E. Y., Hsieh, F.-K., Chang, H.-W., Maluchenko, N. V., Langelier, M.-F., Pascal, J. M., Luse, D. S., Feofanov, A. V., & Studitsky, V. M. (2022). Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro. International Journal of Molecular Sciences, 23(13), 7107. https://doi.org/10.3390/ijms23137107






