Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina
Abstract
1. Introduction
2. Results
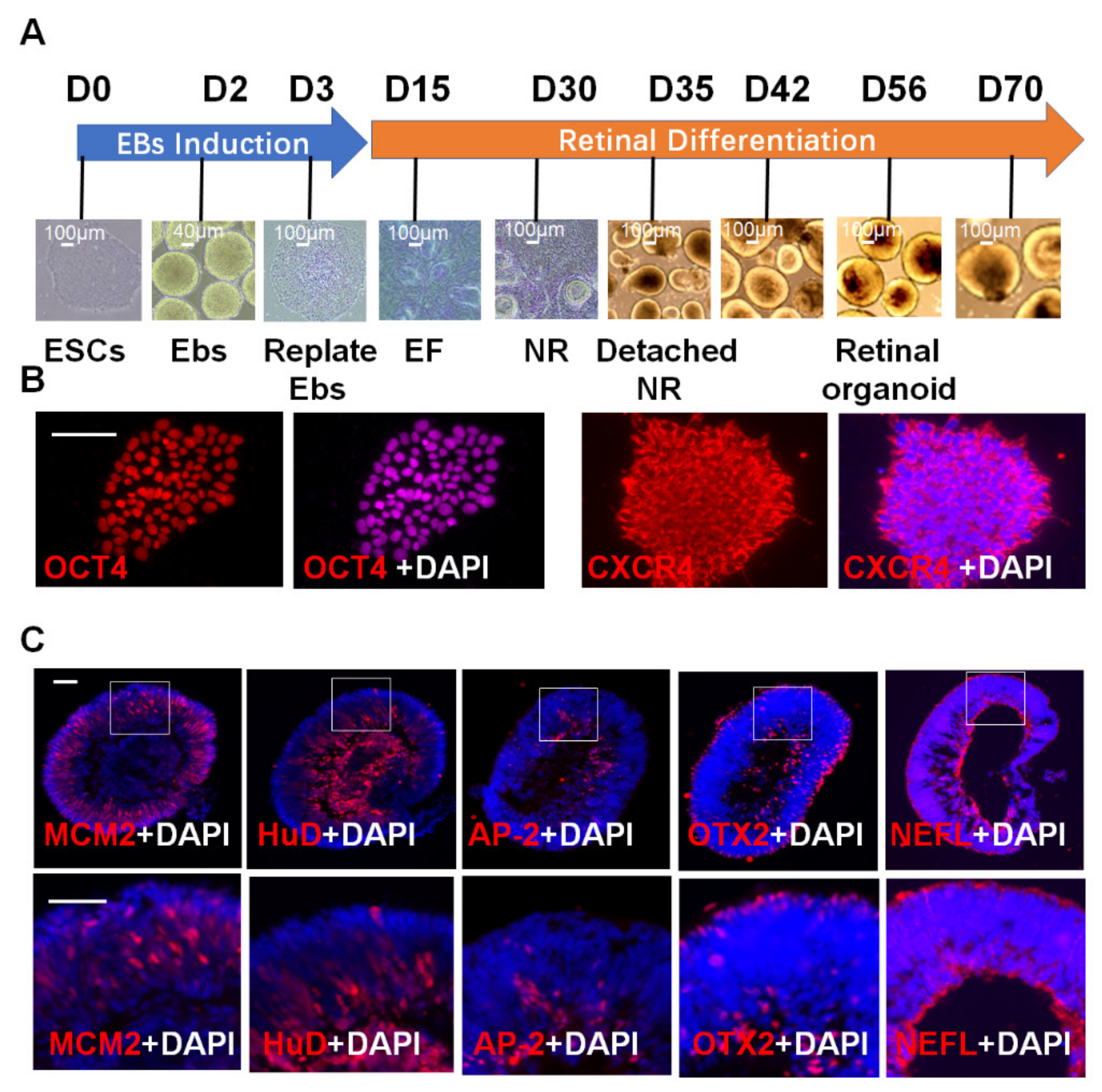
2.1. 3D Retinal Organoids Generated from H9 hESCs Contain Major Retinal Cell Types Arranged in Proper Layers
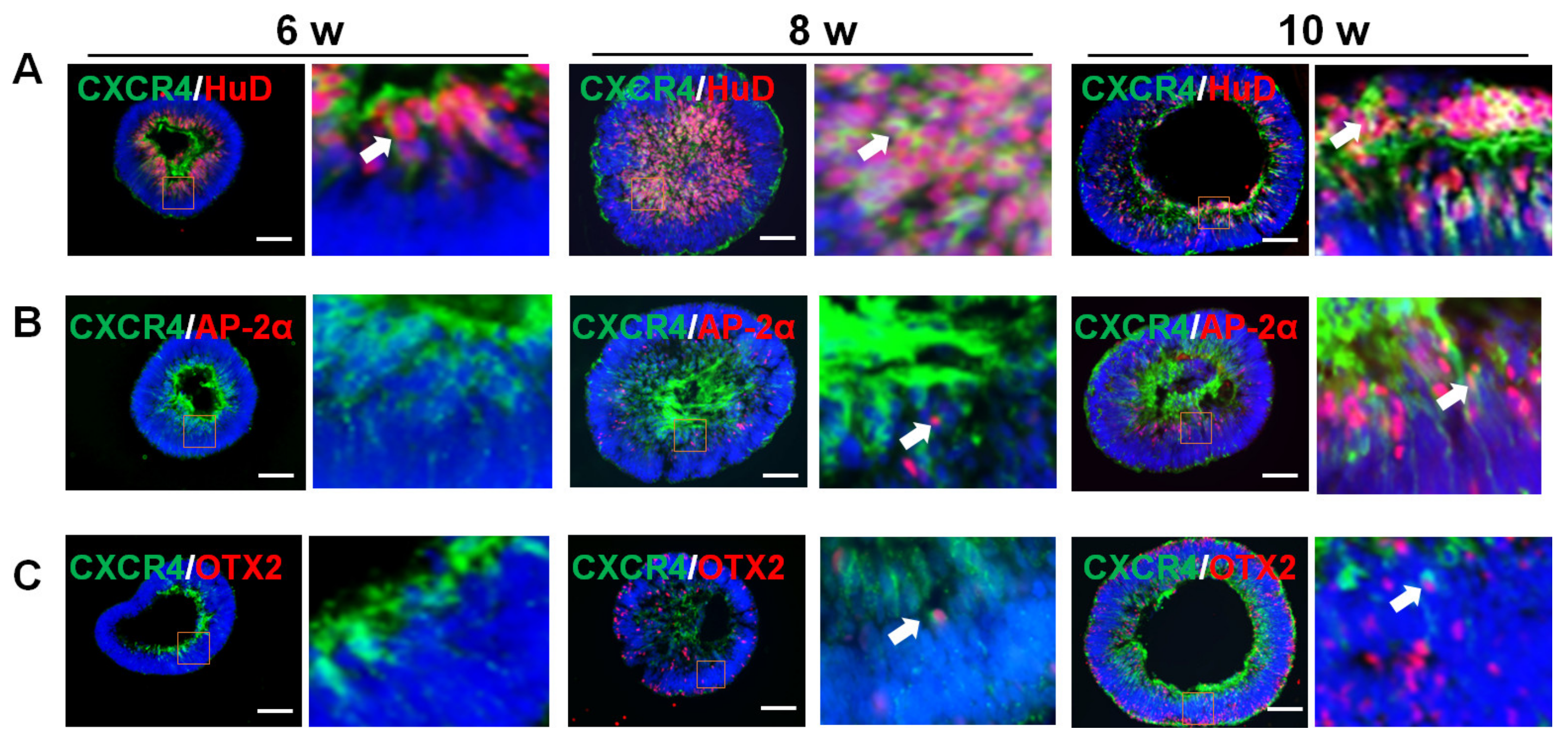
2.2. CXCR4 Location at Various Stages of Organoid Differentiation
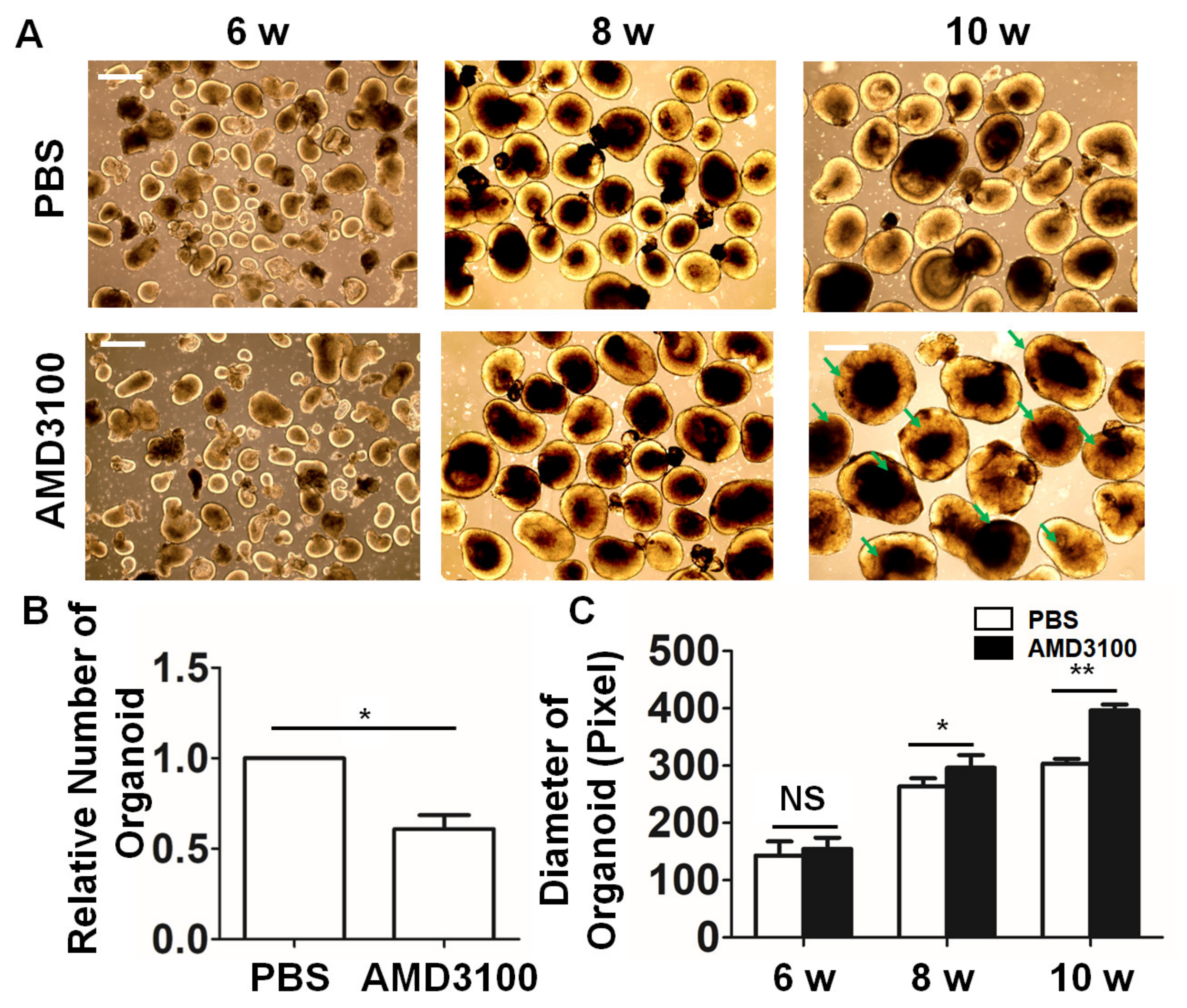
2.3. Blockade of CXCR4 with the Antagonist AMD3100 Affects the Formation of Retinal Organoids
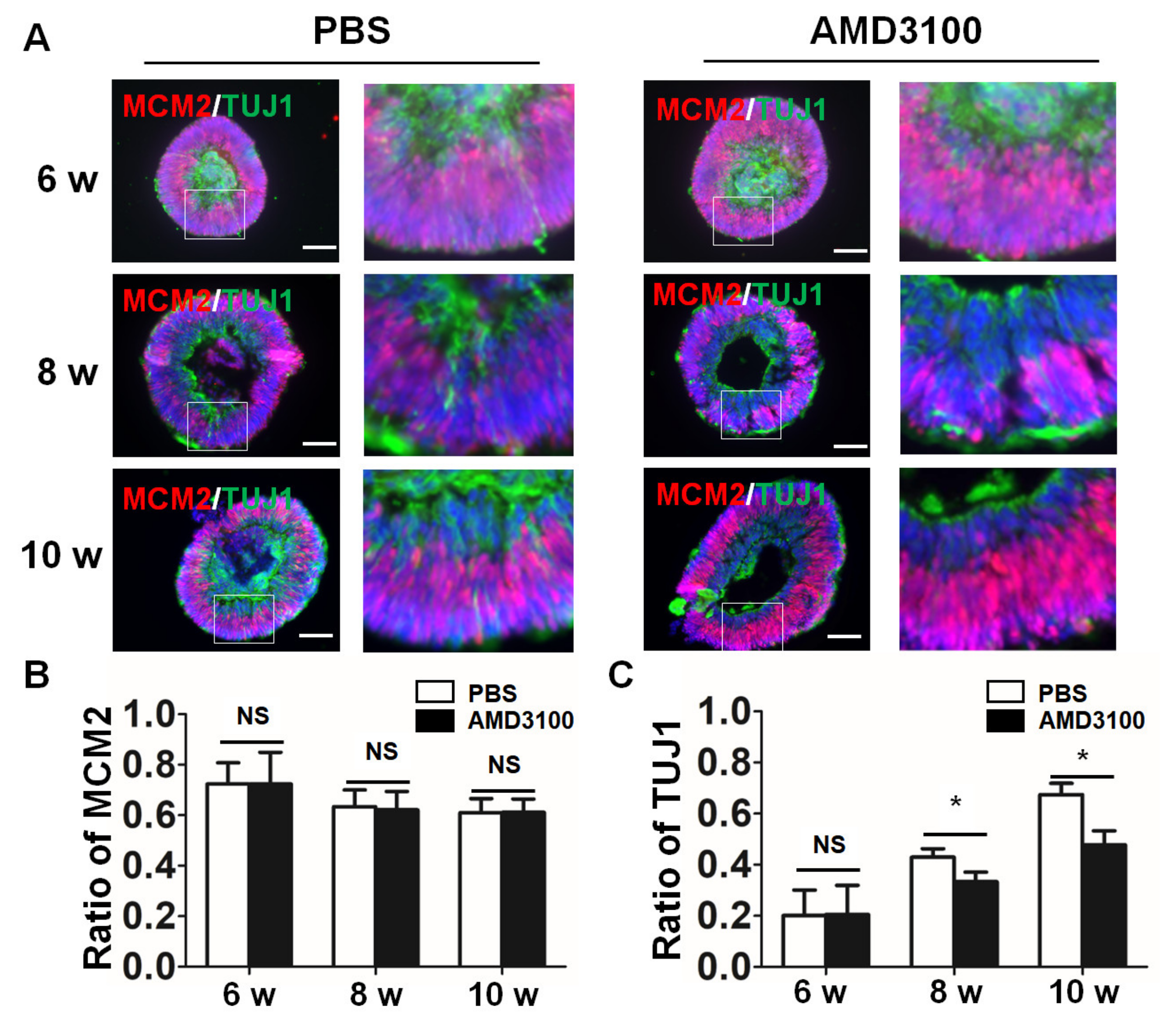
2.4. Blockade of CXCR4 Impairs Retinal Progenitor Cells’ Differentiation in Retinal Organoids
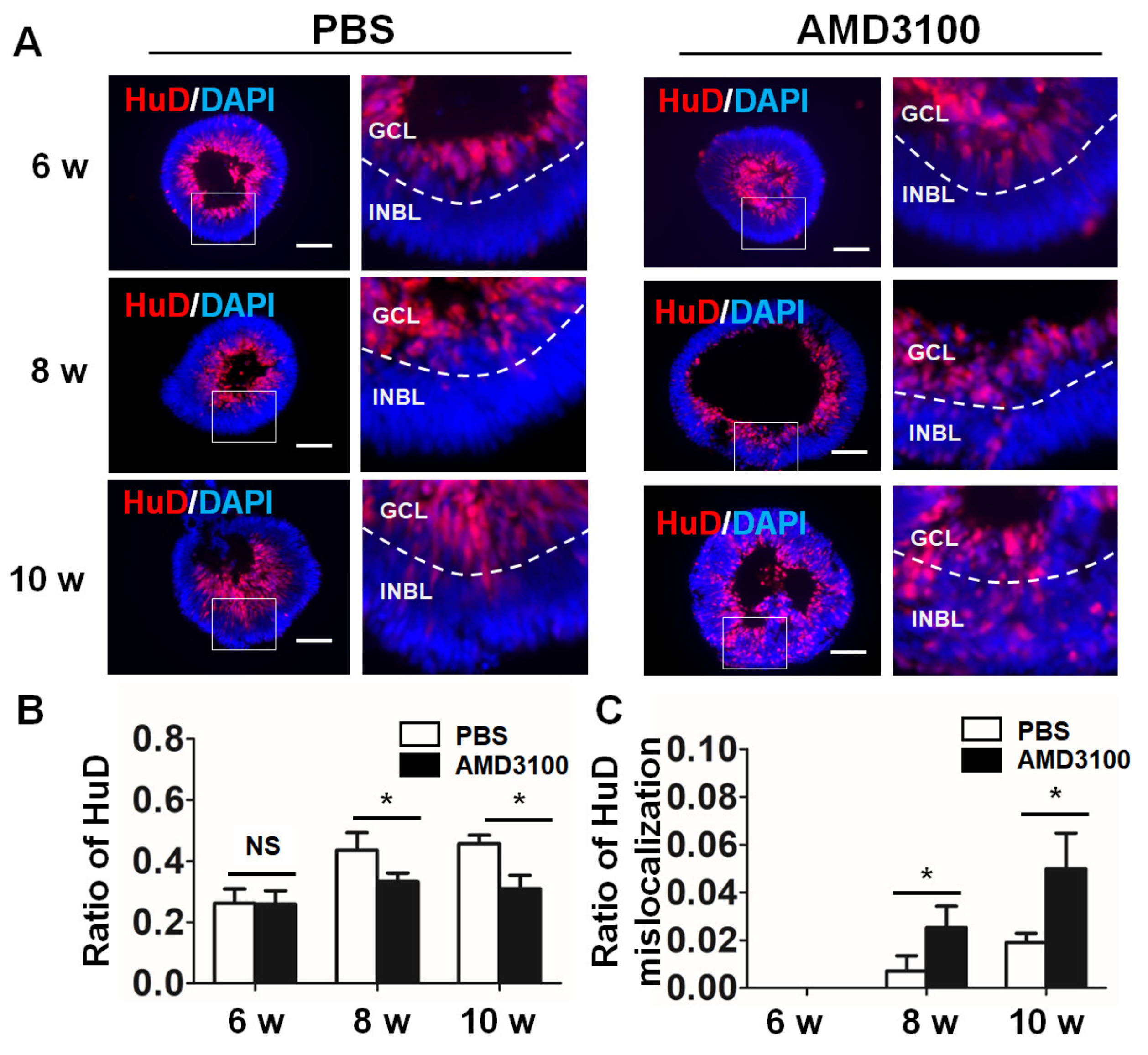
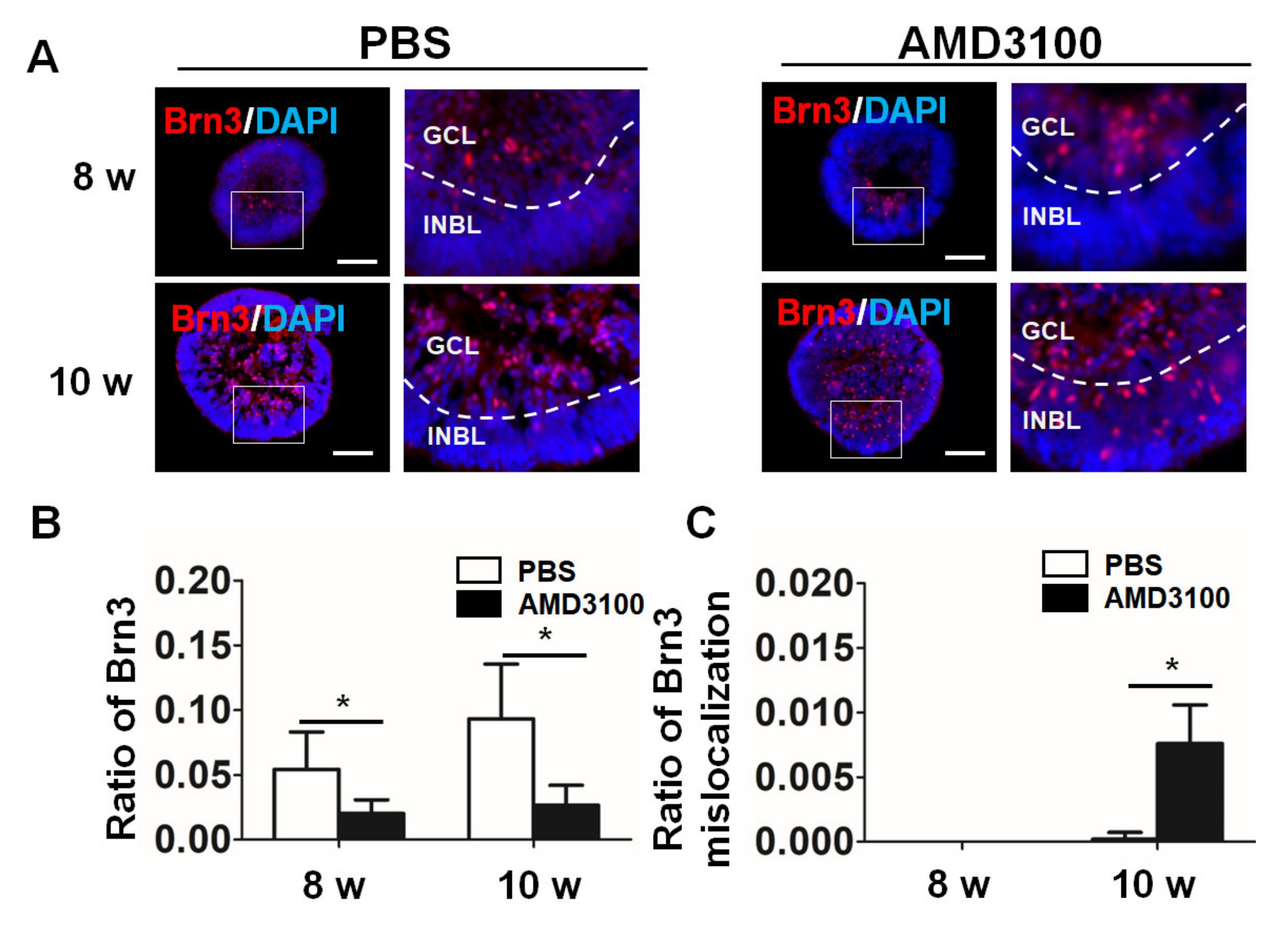
2.5. Blockade of CXCR4 Decreases the Ratio of Ganglion Cells and Results in Ganglion Cell Mislocalization in Retinal Organoids
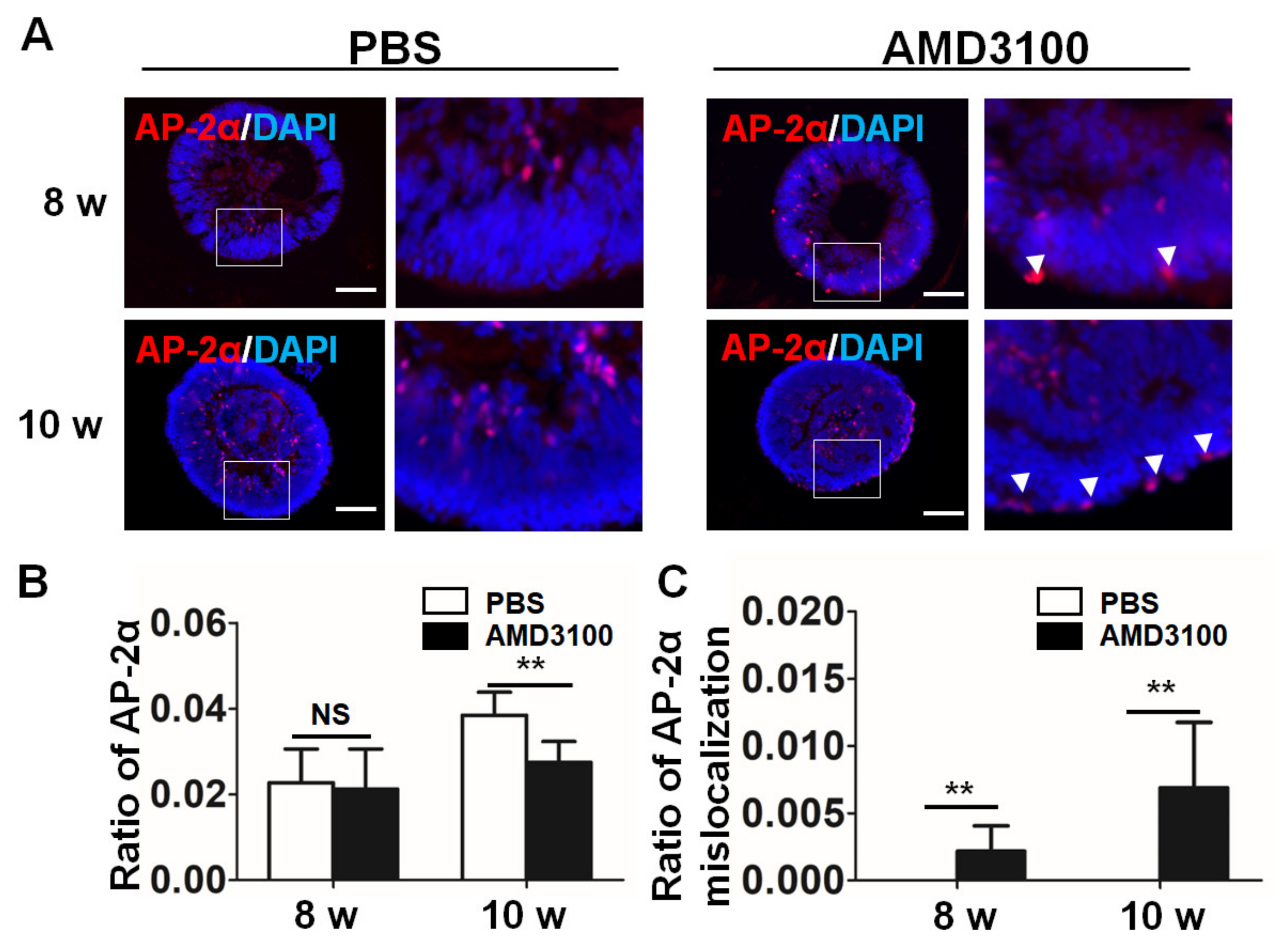
2.6. Blockade of CXCR4 Leads to a Decrease in Amacrine Cells and Their Mislocalization in Retinal Organoids
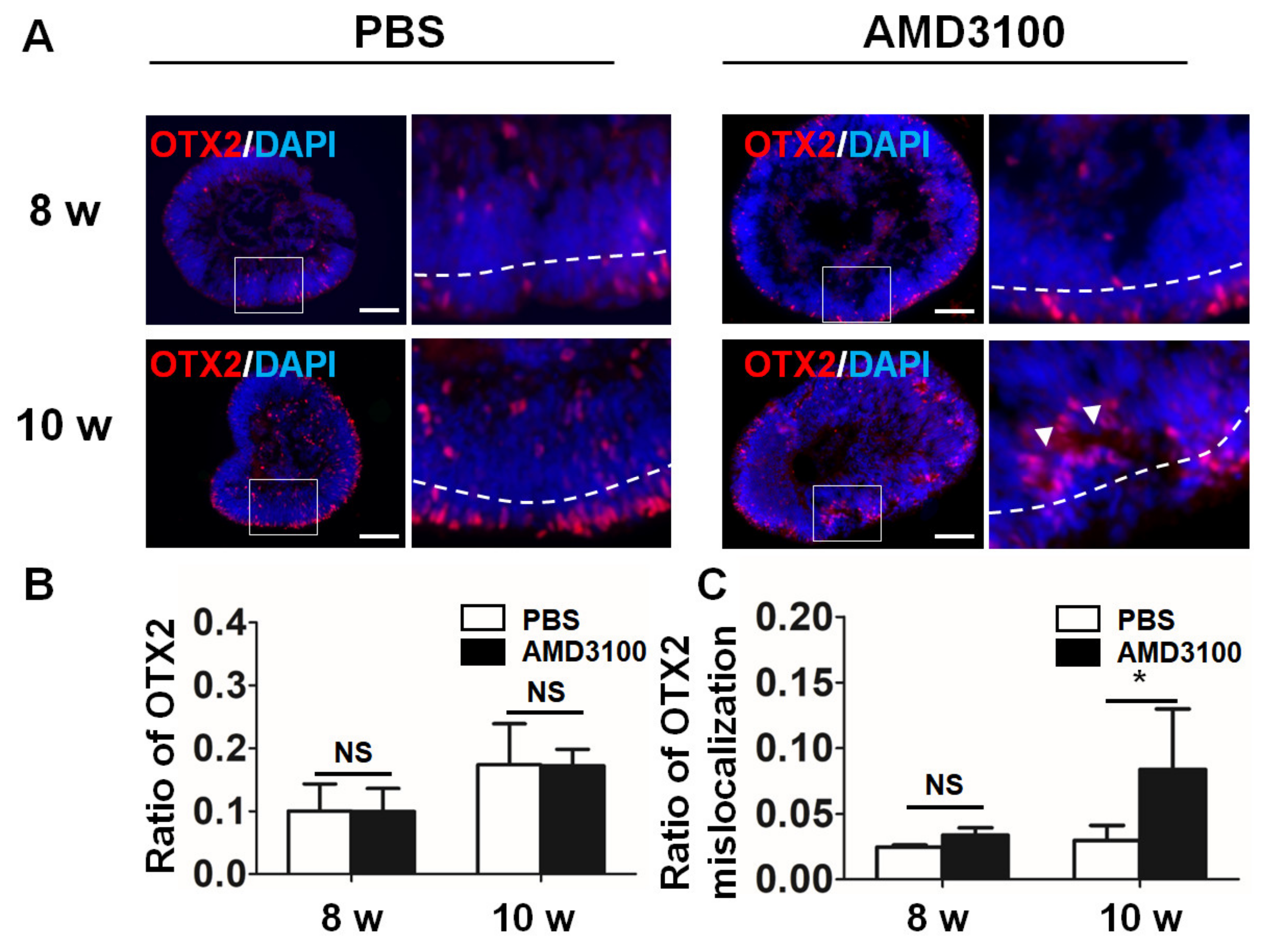
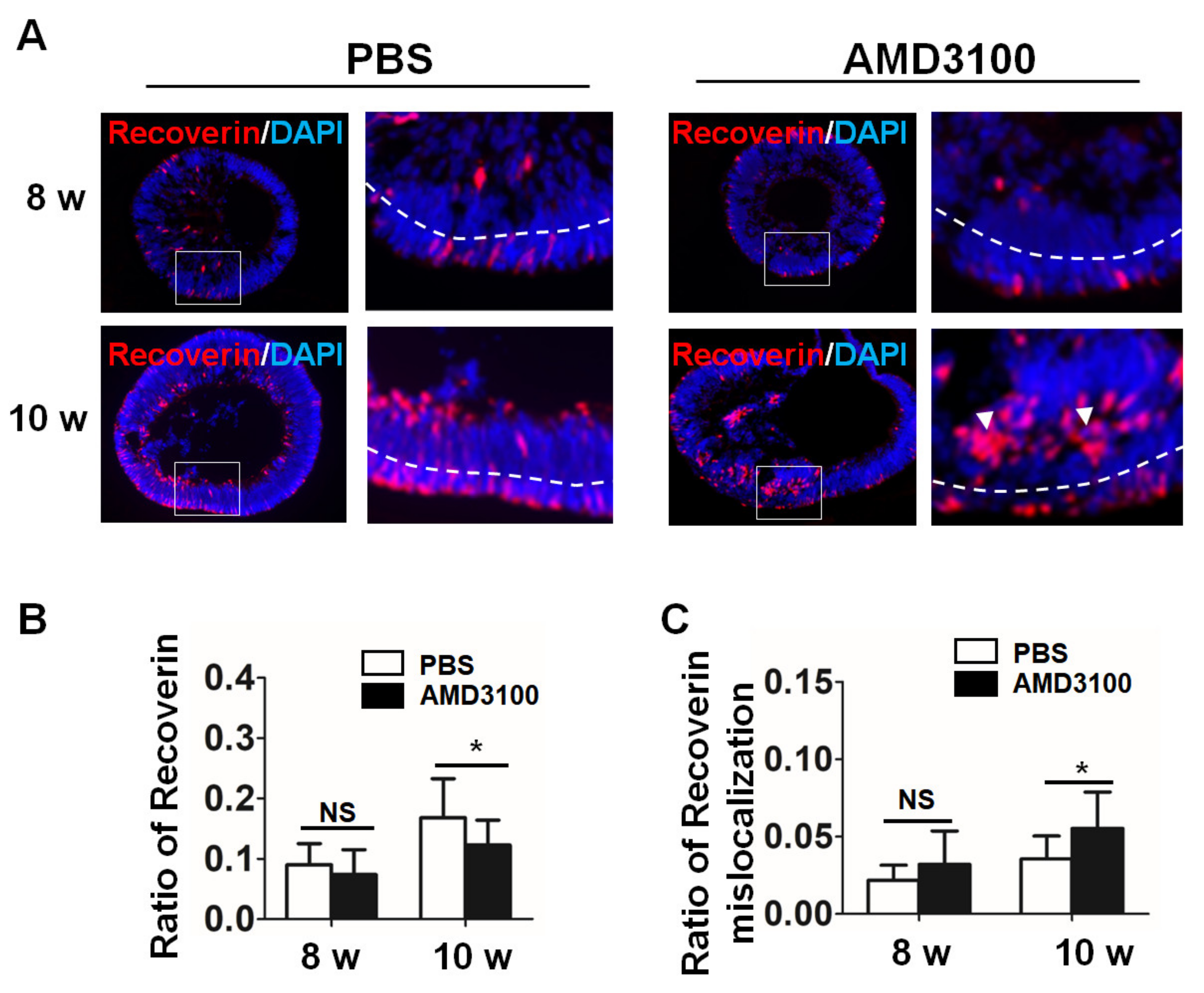
2.7. Blockade of CXCR4 Results in Mislocalization of Photoreceptor Precursors and Decreases Their Ratio
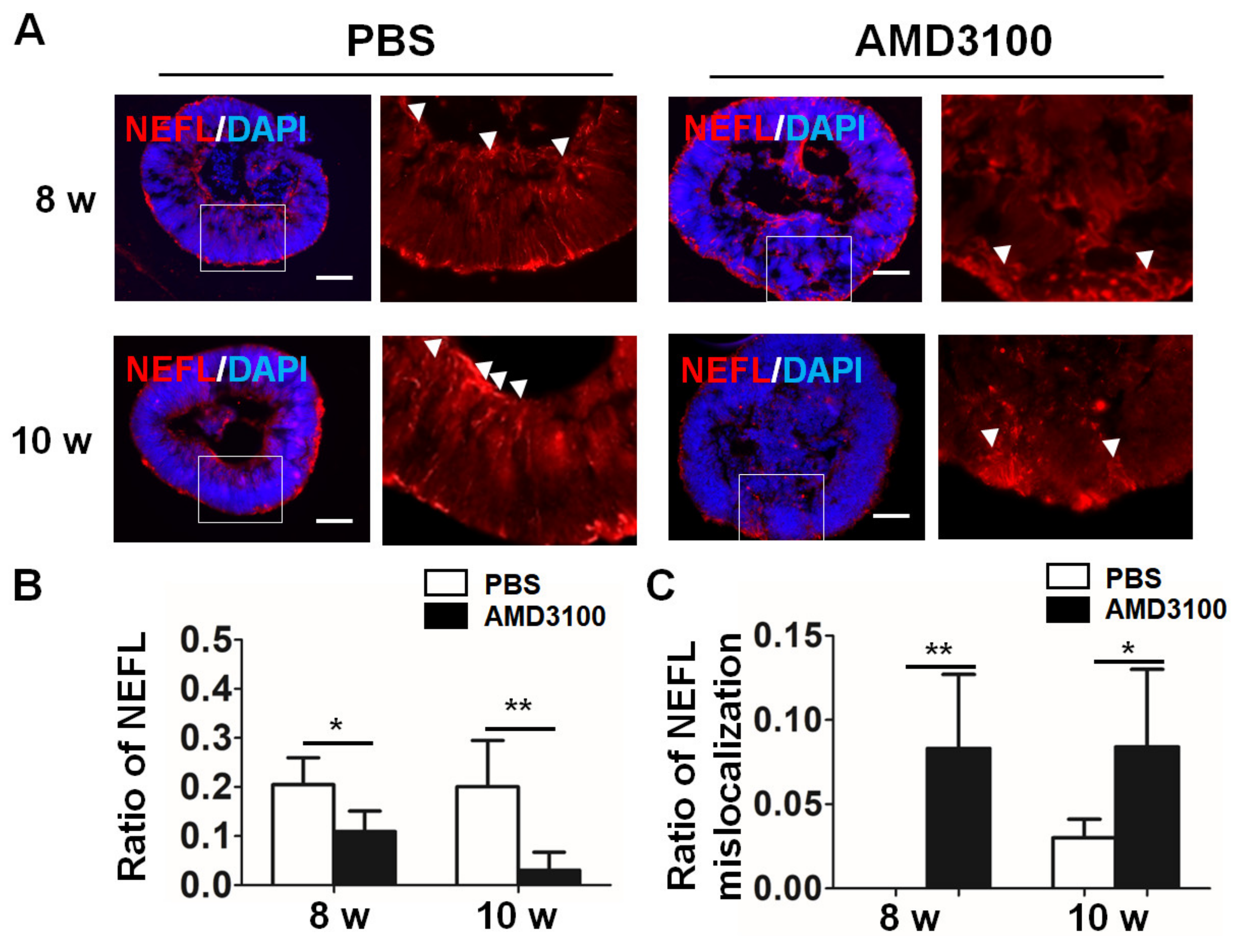
2.8. Blockade of CXCR4 Leads to Abnormal Outgrowth of Neuronal Axons in Retinal Organoids
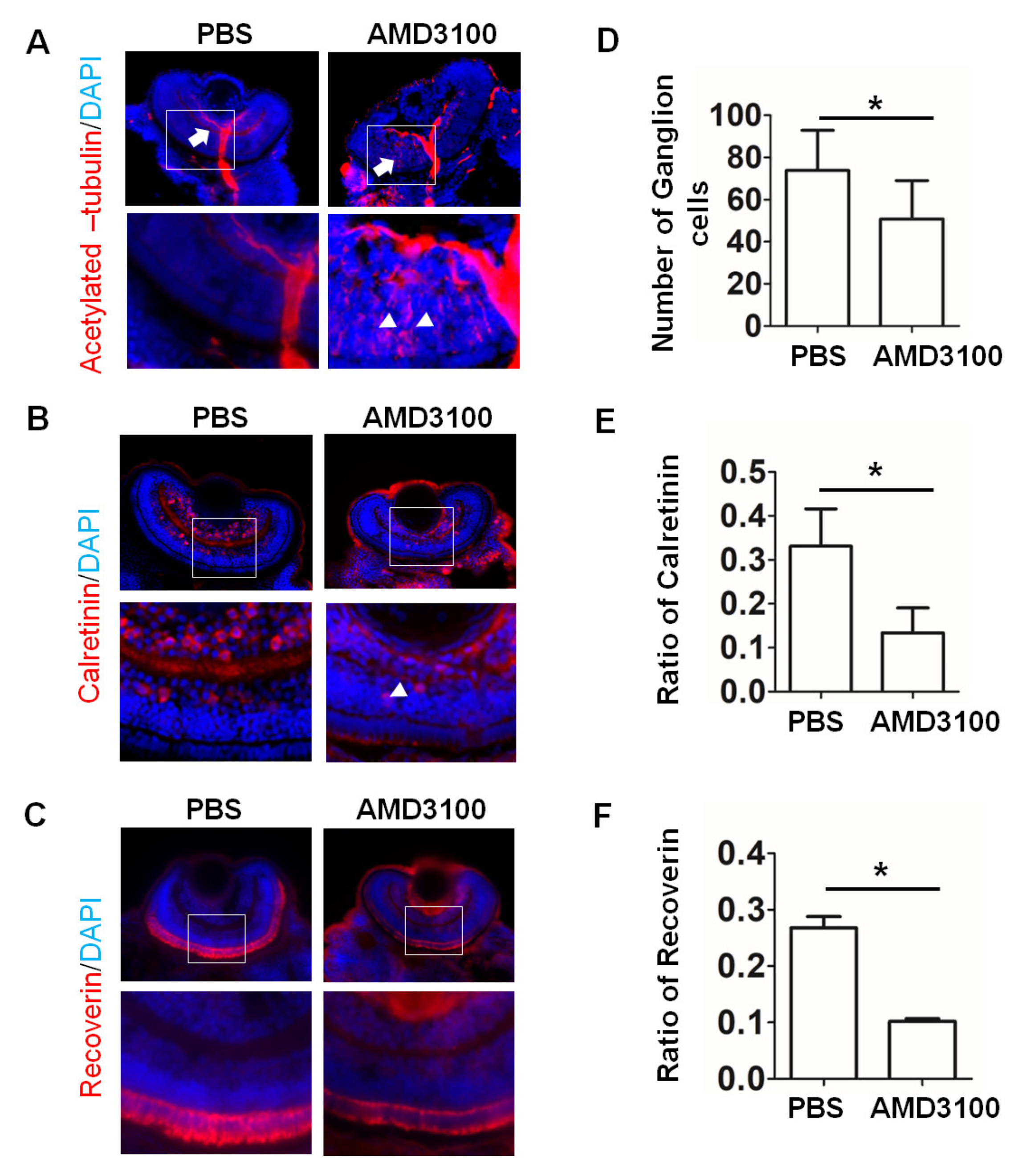
2.9. AMD3100 Treatment of Zebrafish Induced Changes in the Retina Similar to Those Detected in Retinal Organoids
3. Discussion
4. Materials and Methods
4.1. hESC Culture
4.2. Differentiation of hESC into 3D Retinal Organoids
4.3. Immunofluorescence Analysis
4.4. Quantitative Analysis of Immunofluorescence Staining
4.5. Migration Assay
4.6. Zebrafish
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, J.; Bai, L.; Pan, H.; Feng, H.; Clevers, H.; Zeng, Y.A. Long-Term Expansion of Pancreatic Islet Organoids from Resident Procr+ Progenitors. Cell 2020, 180, 1198–1211.e19. [Google Scholar] [CrossRef]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.-H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef]
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Baye, L.M.; Link, B.A. Nuclear migration during retinal development. Brain Res. 2008, 1192, 29–36. [Google Scholar] [CrossRef]
- Assawachananont, J.; Mandai, M.; Okamoto, S.; Yamada, C.; Eiraku, M.; Yonemura, S.; Sasai, Y.; Takahashi, M. Transplantation of Embryonic and Induced Pluripotent Stem Cell-Derived 3D Retinal Sheets into Retinal Degenerative Mice. Stem Cell Rep. 2014, 2, 662–674. [Google Scholar] [CrossRef]
- Fligor, C.M.; Langer, K.B.; Sridhar, A.; Ren, Y.; Shields, P.K.; Edler, M.C.; Ohlemacher, S.K.; Sluch, V.M.; Zack, D.J.; Zhang, C.; et al. Three-Dimensional Retinal Organoids Facilitate the Investigation of Retinal Ganglion Cell Development, Organization and Neurite Outgrowth from Human Pluripotent Stem Cells. Sci. Rep. 2018, 8, 14520. [Google Scholar] [CrossRef]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krok, J.; Szikra, T.; et al. Cell types of the human retina and its organoids at single-cell resolution. Cell 2020, 182, 1623–1640. [Google Scholar] [CrossRef]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659.e4. [Google Scholar] [CrossRef]
- Lane, A.; Jovanovic, K.; Shortall, C.; Ottaviani, D.; Panes, A.B.; Schwarz, N.; Guarascio, R.; Hayes, M.J.; Palfi, A.; Chadderton, N.; et al. Modeling and rescue of RP2 retinitis pigmentosa using iPSC-derived retinal organoids. Stem Cell Rep. 2020, 15, 67–79. [Google Scholar] [CrossRef]
- Huang, K.-C.; Wang, M.-L.; Chen, S.-J.; Kuo, J.-C.; Wang, W.-J.; Nguyen, P.N.N.; Wahlin, K.J.; Lu, J.; Tran, A.A.; Shi, M.; et al. Morphological and Molecular Defects in Human Three-Dimensional Retinal Organoid Model of X-Linked Juvenile Retinoschisis. Stem Cell Rep. 2019, 13, 906–923. [Google Scholar] [CrossRef]
- Singh, R.K.; Nasonkin, I.O. Limitations and Promise of Retinal Tissue From Human Pluripotent Stem Cells for Developing Therapies of Blindness. Front. Cell. Neurosci. 2020, 14, 179. [Google Scholar] [CrossRef]
- Chen, Z.; Pan, X.; Georgakilas, A.G.; Chen, P.; Hu, H.; Yang, Y.; Tian, S.; Xia, L.; Zhang, J.; Cai, X.; et al. Tetramethylpyrazine (TMP) protects cerebral neurocytes and inhibits glioma by down regulating chemokine receptor CXCR4 expression. Cancer Lett. 2013, 336, 281–289. [Google Scholar] [CrossRef]
- Pujic, Z.; Omori, Y.; Tsujikawa, M.; Thisse, B.; Thisse, C.; Malicki, J. Reverse genetic analysis of neurogenesis in the zebrafish retina. Dev. Biol. 2006, 293, 330–347. [Google Scholar] [CrossRef]
- Das, A.V.; Edakkot, S.; Thoreson, W.B.; James, J.; Bhattacharya, S.; Ahmad, I. Membrane properties of retinal stem cells/progenitors. Prog. Retin. Eye Res. 2005, 24, 663–681. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, D.; Deng, X.; Chen, Q.; Huang, Y.; Peng, H.; Li, Y.; Jia, B.; Thoreson, W.B.; Ding, W.; et al. CXCL12 Enhances Human Neural Progenitor Cell Survival Through a CXCR7- and CXCR4-Mediated Endocytotic Signaling Pathway. Stem Cells 2012, 30, 2571–2583. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, S.H.; Baribaud, F.; Coughlan, C.M.; Sunshine, M.J.; Lee, V.M.; Doms, R.W.; Littman, D.R.; Raper, J.A. The chemokine stromal cell-derived factor-1 promotes the survival of embryonic retinal ganglion cells. J. Neurosci. 2003, 23, 4601–4612. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shirabe, K.; Thisse, C.; Thisse, B.; Okamoto, H.; Masai, I.; Kuwada, J.Y. Chemokine signaling guides axons within the retina in zebrafish. J. Neurosci. 2005, 25, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, R.; Kholodenko, I.; Sorokin, V.; Tolmazova, A.; Sazonova, O.; Buzdin, A. Anti-apoptotic effect of retinoic acid on retinal progenitor cells mediated by a protein kinase A-dependent mechanism. Cell Res. 2007, 17, 151–162. [Google Scholar] [CrossRef]
- Barton, K.M.; Levine, E.M. Expression patterns and cell cycle profiles of PCNA, MCM6, cyclin D1, cyclin A2, cyclin B1, and phosphorylated histone H3 in the developing mouse retina. Dev. Dyn. 2008, 237, 672–682. [Google Scholar] [CrossRef]
- Singhal, S.; Bhatia, B.; Jayaram, H.; Becker, S.; Jones, M.F.; Cottrill, P.B.; Khaw, P.T.; Salt, T.E.; Limb, G.A. Human Müller glia with stem cell characteristics differentiate into retinal ganglion cell (RGC) precursors in vitro and partially restore RGC function in vivo following transplantation. Stem Cells Transl. Med. 2012, 1, 188–199. [Google Scholar] [CrossRef]
- Luo, Z.; Xu, C.; Li, K.; Xian, B.; Liu, Y.; Li, K.; Liu, Y.; Rong, H.; Tang, M.; Hu, D.; et al. Islet1 and Brn3 Expression Pattern Study in Human Retina and hiPSC-Derived Retinal Organoid. Stem Cells Int. 2019, 2019, 8786396. [Google Scholar] [CrossRef]
- Lieberam, I.; Agalliu, D.; Nagasawa, T.; Ericson, J.; Jessell, T.M. A Cxcl12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron 2005, 47, 667–679. [Google Scholar] [CrossRef]
- Yang, S.; Edman, L.C.; Sánchez-Alcañiz, J.A.; Fritz, N.; Bonilla, S.; Hecht, J.; Uhlén, P.; Pleasure, S.J.; Villaescusa, J.C.; Marín, O.; et al. Cxcl12/Cxcr4 signaling controls the migration and process orientation of A9-A10 dopaminergic neurons. Development 2013, 140, 4554–4564. [Google Scholar] [CrossRef]
- Huang, G.J.; Edwards, A.; Tsai, C.Y.; Lee, Y.S.; Peng, L.; Era, T.; Hirabayashi, Y.; Tsai, C.; Nishikawa, S.; Iwakura, Y.; et al. Ectopic cerebellar cell migration causes maldevelopment of Purkinje cells and abnormal motor behaviour in Cxcr4 null mice. PLoS ONE 2014, 9, e86471. [Google Scholar] [CrossRef]
- Chen, P.; Yang, Y.; Chen, Z.; Qiu, J.; Yu, N.; Tang, M.; Wang, Q.; Ge, J.; Yu, K.; Zhuang, J. Nuclear respiratory factor-1 (NRF-1) regulates transcription of the CXC receptor 4 (CXCR4) in the rat retina. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4662–4669. [Google Scholar] [CrossRef]
- Nishida, A.; Furukawa, A.; Koike, C.; Tano, Y.; Aizawa, S.; Matsuo, I.; Furukawa, T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003, 6, 1255–1263. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Aparicio, J.G.; Hopp, H.; Choi, A.; Comar, J.M.; Liao, V.C.; Harutyunyan, N.; Lee, T.C. Temporal expression of CD184 (CXCR4) and CD171 (L1CAM) identifies distinct early developmental stages of human retinal ganglion cells in embryonic stem cell derived retina. Exp. Eye Res. 2017, 154, 177–189. [Google Scholar] [CrossRef]
- López-Bendito, G.; Sánchez-Alcaniz, J.A.; Pla, R.; Borrell, V.; Picó, E.; Valdeolmillos, M.; Marín, O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 2008, 28, 1613–1624. [Google Scholar] [CrossRef]
- Hatse, S.; Princen, K.; Bridger, G.; De Clercq, E.; Schols, D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002, 527, 255–262. [Google Scholar] [CrossRef]
- Hitchinson, B.; Eby, J.M.; Gao, X.; Guite-Vinet, F.; Ziarek, J.J.; Abdelkarim, H.; Lee, Y.; Okamoto, Y.; Shikano, S.; Majetschak, M.; et al. Biased antagonism of CXCR4 avoids antagonist tolerance. Sci. Signal 2018, 11, eaat2214. [Google Scholar] [CrossRef]
- De Clercq, E. AMD3100/CXCR4 inhibitor. Front. Immunol. 2015, 6, 276. [Google Scholar] [CrossRef]
- Zhang, M.; Song, A.; Lai, S.; Qiu, L.; Huang, Y.; Chen, Q.; Zhu, B.; Xu, D.; Zheng, J.C. Applications of stripe assay in the study of CXCL12-mediated neural progenitor cell migration and polarization. Biomaterials 2015, 72, 163–171. [Google Scholar] [CrossRef][Green Version]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. 2000. Available online: http://zfin.org/zf_info/zfbook/zfbk.html (accessed on 12 February 2020).
- Kazimi, N.; Cahill, G.M. Development of a circadian melatonin rhythm in embryonic zebrafish. Dev. Brain Res. 1999, 117, 47–52. [Google Scholar] [CrossRef]
- Novoa, B.; Bowman, T.V.; Zon, L.; Figueras, A. LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish Immunol. 2009, 26, 326–331. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Qiu, J.; Chen, S.; Chen, X.; Zhang, J.; Zhuang, J.; Liu, S.; Yang, M.; Zhou, P.; Chen, H.; et al. Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina. Int. J. Mol. Sci. 2022, 23, 7088. https://doi.org/10.3390/ijms23137088
Wu Y, Qiu J, Chen S, Chen X, Zhang J, Zhuang J, Liu S, Yang M, Zhou P, Chen H, et al. Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina. International Journal of Molecular Sciences. 2022; 23(13):7088. https://doi.org/10.3390/ijms23137088
Chicago/Turabian StyleWu, Yihui, Jin Qiu, Shuilian Chen, Xi Chen, Jing Zhang, Jiejie Zhuang, Sian Liu, Meng Yang, Pan Zhou, Haoting Chen, and et al. 2022. "Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina" International Journal of Molecular Sciences 23, no. 13: 7088. https://doi.org/10.3390/ijms23137088
APA StyleWu, Y., Qiu, J., Chen, S., Chen, X., Zhang, J., Zhuang, J., Liu, S., Yang, M., Zhou, P., Chen, H., Yu, K., Ge, J., & Zhuang, J. (2022). Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina. International Journal of Molecular Sciences, 23(13), 7088. https://doi.org/10.3390/ijms23137088





