Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation
Abstract
:1. Introduction
2. Results
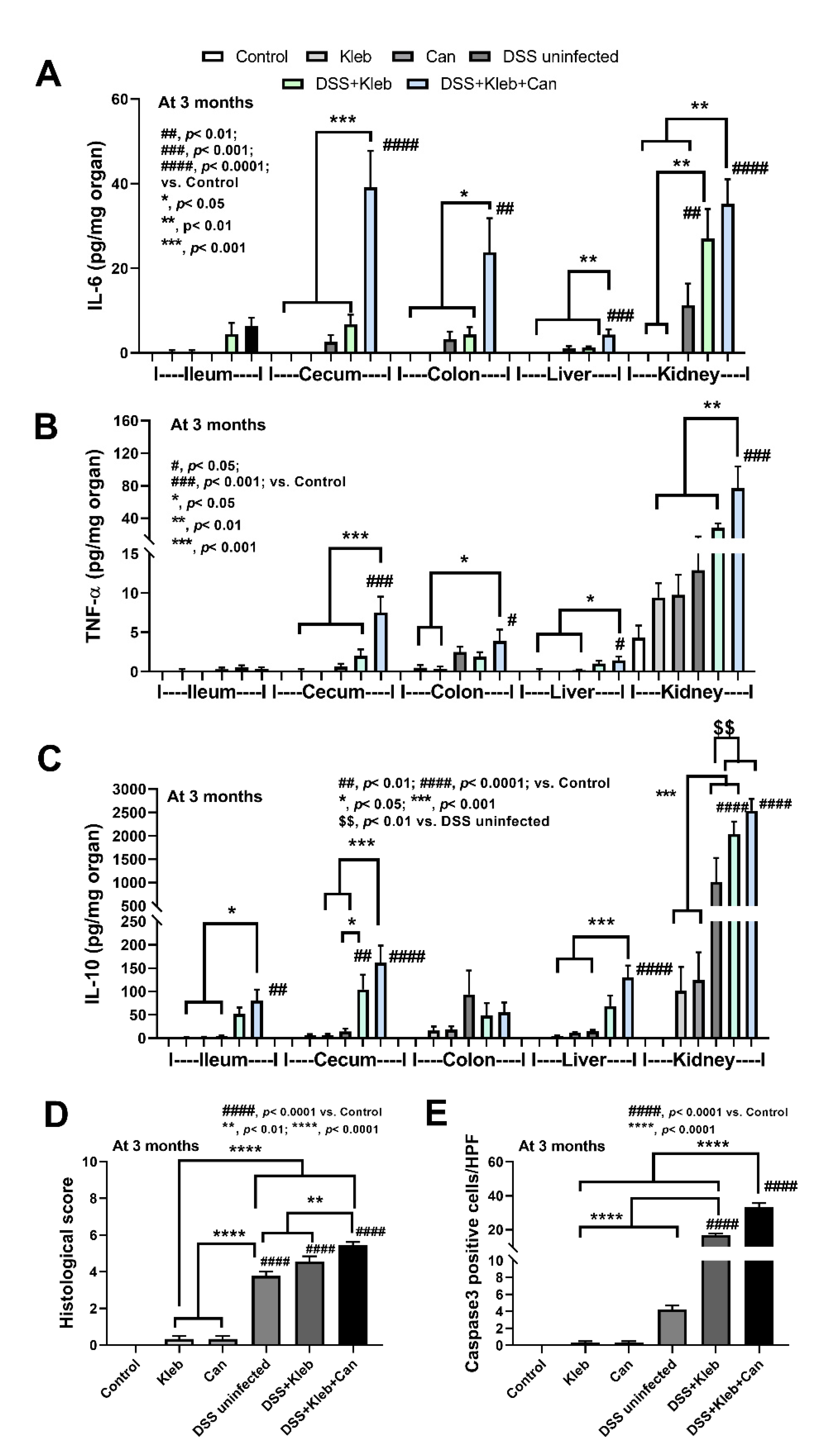
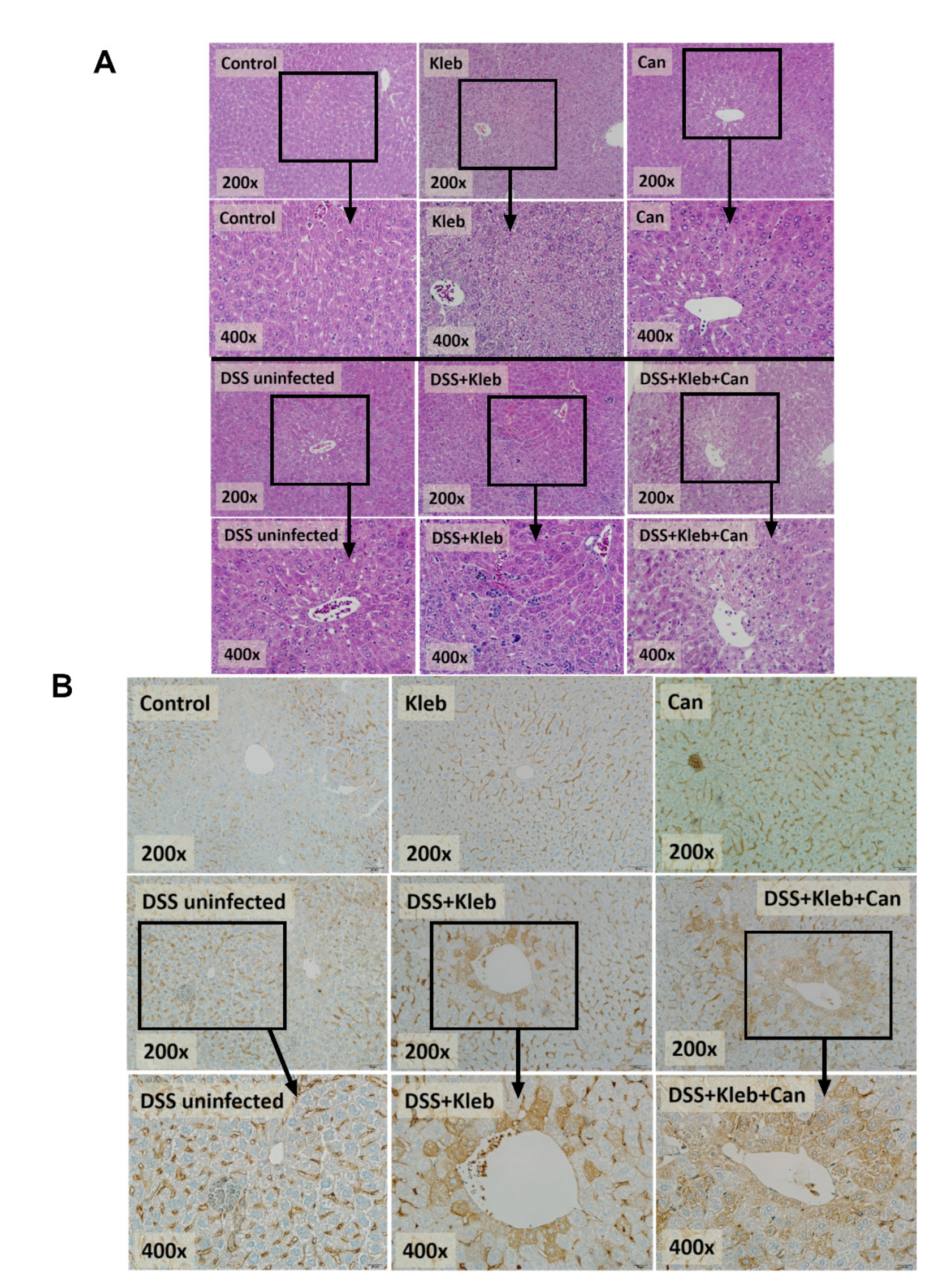
2.1. Candida Administration Worsened Disease Severity in K. pneumoniae-Administered DSS Mice Partly through Gut Barrier Defect and Systemic Inflammation
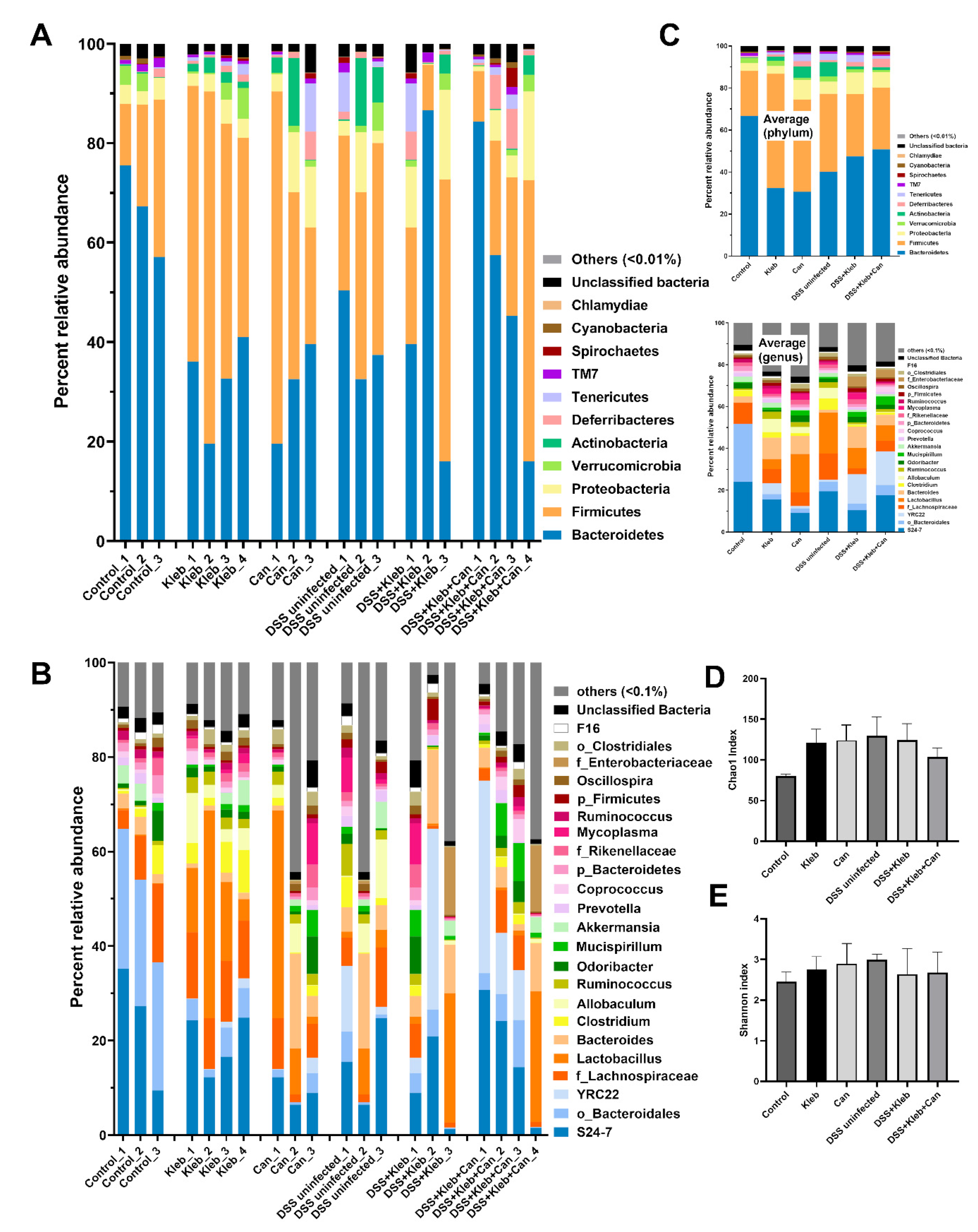
2.2. Candida Slightly Altered Gut Dysbiosis in Klebsiella-Administered DSS Mice
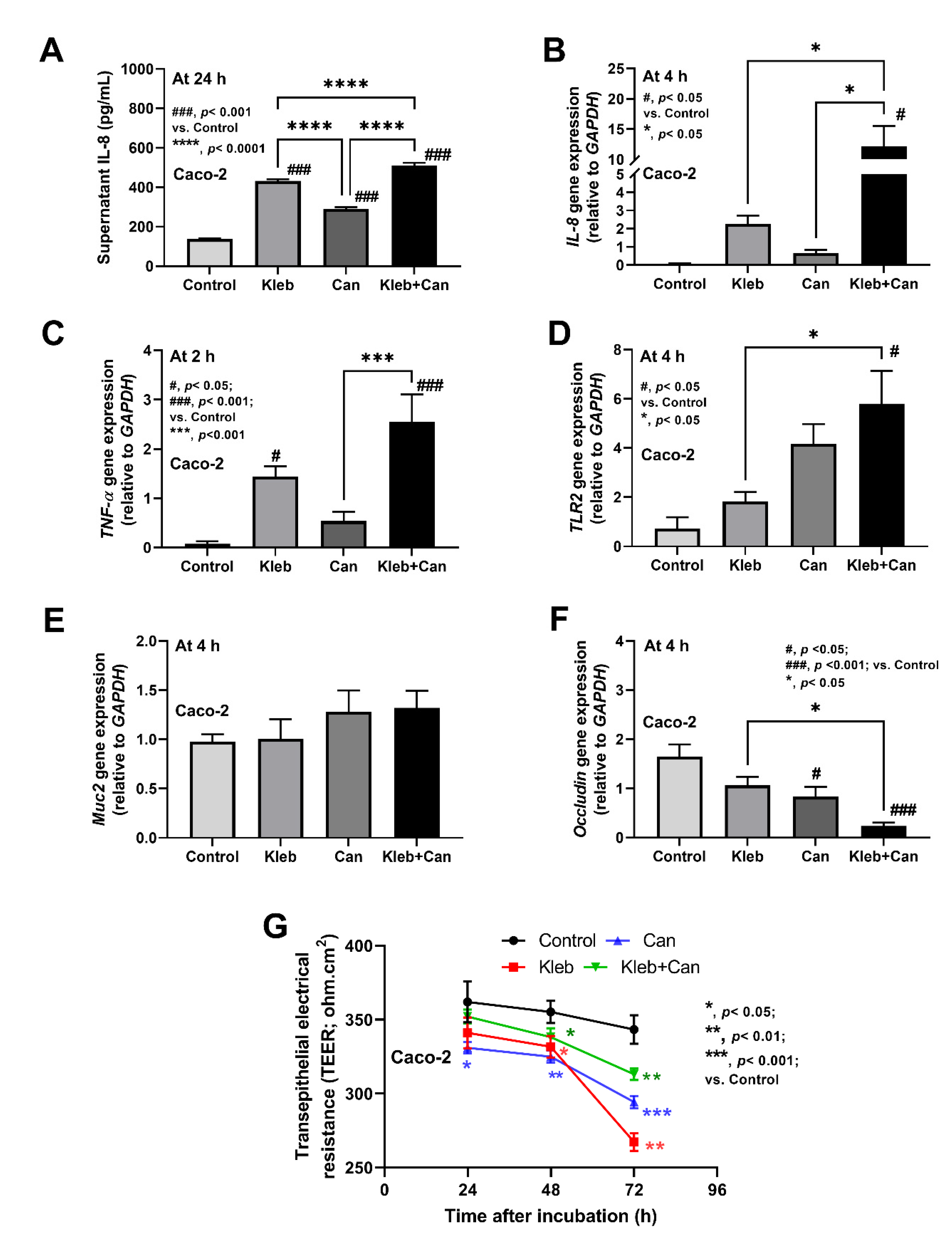
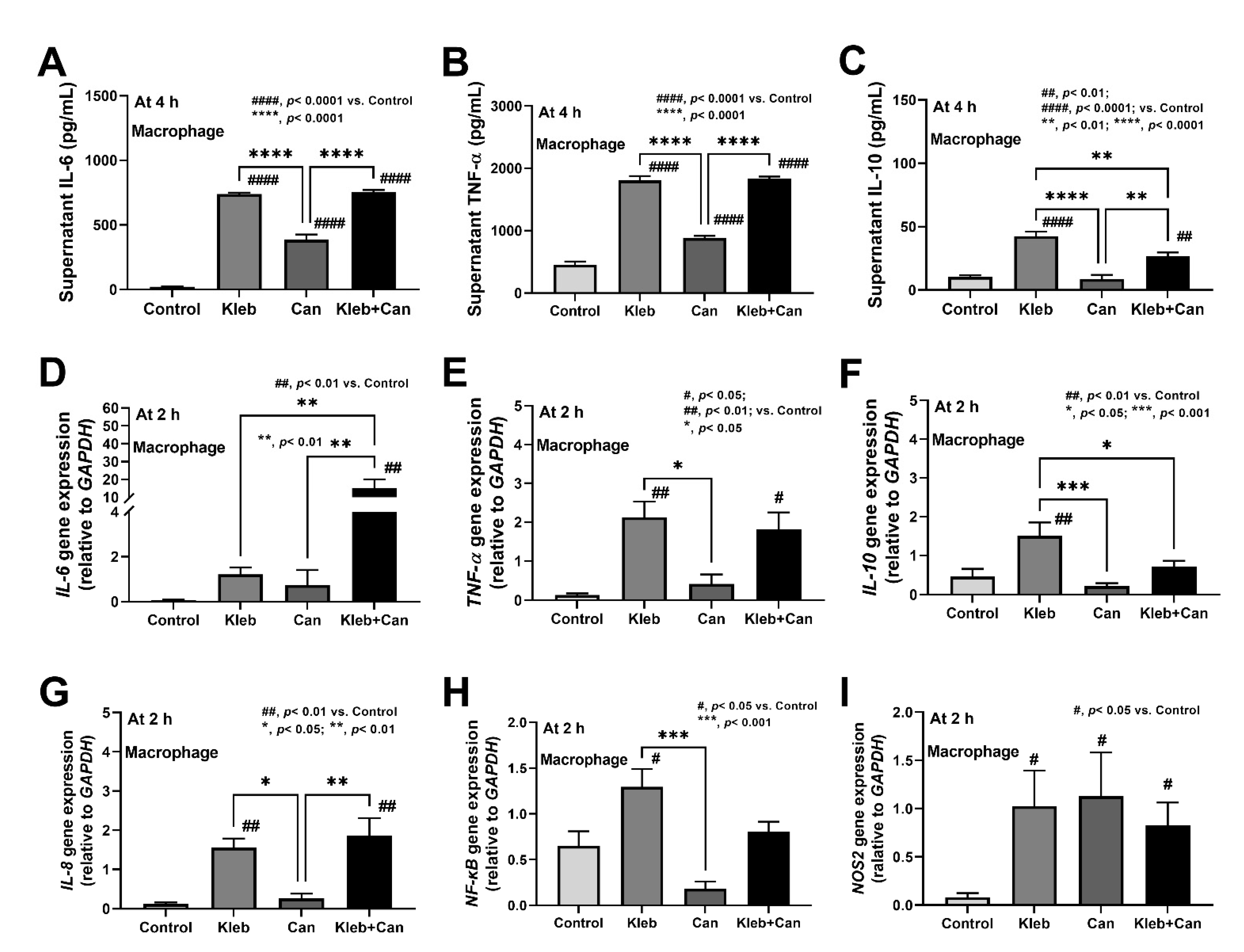
2.3. Molecules of Candida Induced Inflammation against Enterocytes, Hepatocytes, and Macrophages
3. Discussion
3.1. Candida Enhanced Inflammation in Klebsiella-Administered DSS Mice Mainly through the Activations by Microbial Molecules Rather than Gut Dysbiosis
3.2. Candida Enhanced Hyper-Inflammation in the Intestines, Livers, and Macrophages of Klebsiella-Administered DSS Mice
4. Materials and Methods
4.1. Animals and Animal Models
4.1.1. Gut Permeability Determination
4.1.2. Serum and Histological Analyses
4.1.3. Fecal Microbiome Analysis
4.2. The In Vitro Experiments
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, B.; Zhao, Y.; Liu, C.; Chen, Z.; Zhou, D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014, 9, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Ashurst, J.V.; Dawson, A. Klebsiella Pneumonia; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Opoku-Temeng, C.; Kobayashi, S.D.; DeLeo, F.R. Klebsiella pneumoniae capsule polysaccharide as a target for therapeutics and vaccines. Comput. Struct. Biotechnol. J. 2019, 17, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, V.A.K.; Zhao, Y.; Shao, F. Innate immunity to intracellular LPS. Nat. Immunol. 2019, 20, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Mayr, F.B.; Yende, S.; Angus, D.C. Epidemiology of severe sepsis. Virulence 2014, 5, 4–11. [Google Scholar] [CrossRef] [Green Version]
- Leelahavanichkul, A.; Panpetch, W.; Worasilchai, N.; Somparn, P.; Chancharoenthana, W.; Nilgate, S.; Finkelman, M.; Chindamporn, A.; Tumwasorn, S. Evaluation of gastrointestinal leakage using serum (1-->3)-beta-D-glucan in a Clostridium difficile murine model. FEMS Microbiol. Lett. 2016, 363, fnw204. [Google Scholar] [CrossRef] [Green Version]
- Panpetch, W.; Chancharoenthana, W.; Bootdee, K.; Nilgate, S.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Lactobacillus rhamnosus L34 Attenuates Gut Translocation-Induced Bacterial Sepsis in Murine Models of Leaky Gut. Infect. Immun. 2018, 86, e00700-17–e00717. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou-Olivgeris, M.; Spiliopoulou, A.; Fligou, F.; Manolopoulou, P.; Spiliopoulou, I.; Vrettos, T.; Dodou, V.; Filos, K.S.; Anastassiou, E.D.; Marangos, M.; et al. Association of KPC-producing Klebsiella pneumoniae colonization or infection with Candida isolation and selection of non-albicans species. Diagn. Microbiol. Infect. Dis. 2014, 80, 227–232. [Google Scholar] [CrossRef]
- Dimopoulos, G.; Ntziora, F.; Rachiotis, G.; Armaganidis, A.; Falagas, M.E. Candida albicans versus non-albicans intensive care unit-acquired bloodstream infections: Differences in risk factors and outcome. Anesth. Analg. 2008, 106, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Eggimann, P.; Pittet, D. Candida colonization index and subsequent infection in critically ill surgical patients: 20 years later. Intensive Care Med. 2014, 40, 1429–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amornphimoltham, P.; Yuen, P.S.T.; Star, R.A.; Leelahavanichkul, A. Gut leakage of fungal-derived inflammatory mediators: Part of a gut-liver-kidney axis in bacterial sepsis. Dig. Dis. Sci. 2019, 64, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Worasilchai, N.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Gastrointestinal colonization of Candida albicans increases serum (1-->3)-beta-D-glucan, without candidemia, and worsens cecal ligation and puncture sepsis in murine model. Shock 2018, 49, 62–70. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Palasuk, M.; Hiengrach, P.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019, 14, e0210798. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Finkelman, M.; Worasilchai, N.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1-->3)-beta-D-glucan. PLoS ONE 2017, 12, e0181439. [Google Scholar] [CrossRef] [Green Version]
- Panpetch, W.; Sawaswong, V.; Chanchaem, P.; Ondee, T.; Dang, C.P.; Payungporn, S.; Tumwasorn, S.; Leelahavanichkul, A. Corrigendum: Candida administration worsens cecal ligation and puncture-induced sepsis in obese mice through gut dysbiosis enhanced systemic inflammation, impact of pathogen-associated molecules from gut translocation and saturated fatty acid. Front. Immunol. 2020, 11, 613095. [Google Scholar] [CrossRef]
- Costa, R.J.S.; Snipe, R.M.J.; Kitic, C.M.; Gibson, P.R. Systematic review: Exercise-induced gastrointestinal syndrome-implications for health and intestinal disease. Aliment. Pharmacol. Ther. 2017, 46, 246–265. [Google Scholar] [CrossRef] [Green Version]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut. Microbes. 2020, 11, 465–480. [Google Scholar] [CrossRef]
- Yamada, M.; Ohkusa, T.; Okayasu, I. Occurrence of dysplasia and adenocarcinoma after experimental chronic ulcerative colitis in hamsters induced by dextran sulphate sodium. Gut 1992, 33, 1521–1527. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Zhang, S.; Tang, K.; Zhou, F.; Zheng, C.; Zhang, K.; Cai, J.; Zhou, H.; Wang, Y.; Tian, B.; et al. Clinical characteristics, risk factors and outcomes of mixed Candida albicans/bacterial bloodstream infections. BMC Infect. Dis. 2020, 20, 810. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Kullapanich, C.; Dang, C.P.; Visitchanakun, P.; Saisorn, W.; Wongphoom, J.; Wannigama, D.L.; Thim-Uam, A.; Patarakul, K.; Somboonna, N.; et al. Candida administration worsens uremia-induced gut leakage in bilateral nephrectomy mice, an impact of gut fungi and organismal molecules in uremia. mSystems 2021, 6, e01187-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Dou, X.; Wang, Q.; Guan, Z.; Cai, Y.; Liao, X. Isolation of beta-1,3-glucanase-producing microorganisms from Poria cocos cultivation soil via molecular biology. Molecules 2018, 23, 1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueki, A.; Takehara, T.; Ishioka, G.; Kaku, N.; Ueki, K. beta-1,3-Glucanase production as an anti-fungal enzyme by phylogenetically different strains of the genus Clostridium isolated from anoxic soil that underwent biological disinfestation. Appl. Microbiol. Biotechnol. 2020, 104, 5563–5578. [Google Scholar] [CrossRef]
- Hiengrach, P.; Panpetch, W.; Worasilchai, N.; Chindamporn, A.; Tumwasorn, S.; Jaroonwitchawan, T.; Wilantho, A.; Chatthanathon, P.; Somboonna, N.; Leelahavanichkul, A. Administration of Candida albicans to dextran sulfate solution treated mice causes intestinal dysbiosis, emergence and dissemination of intestinal Pseudomonas aeruginosa and lethal sepsis. Shock 2020, 53, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Azman, A.S.; Mawang, C.I.; Khairat, J.E.; AbuBakar, S. Actinobacteria—A promising natural source of anti-biofilm agents. Int. Microbiol. 2019, 22, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Visitchanakun, P.; Panpetch, W.; Saisorn, W.; Chatthanathon, P.; Wannigama, D.L.; Thim-Uam, A.; Svasti, S.; Fucharoen, S.; Somboonna, N.; Leelahavanichkul, A. Increased susceptibility to dextran sulfate-induced mucositis of iron-overload beta-thalassemia mice, another endogenous cause of septicemia in thalassemia. Clin. Sci 2021, 135, 1467–1486. [Google Scholar] [CrossRef]
- Berry, D.; Schwab, C.; Milinovich, G.; Reichert, J.; Ben Mahfoudh, K.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091–2106. [Google Scholar] [CrossRef] [Green Version]
- Heisel, T.; Montassier, E.; Johnson, A.; Al-Ghalith, G.; Lin, Y.W.; Wei, L.N.; Knights, D.; Gale, C.A. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2017, 2, e00351-17. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.Y. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell. 2013, 12, 1416–1422. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.M.; de Paula, T.O.; Sarmiento, M.R.A.; de Oliveira, M.G.; Pereira, M.L.M.; Toledo, I.V.; Nascimento, T.C.; Ferreira-Machado, A.B.; Silva, V.L.; Diniz, C.G. Fungal diversity of human gut microbiota among eutrophic, overweight, and obese individuals based on aerobic culture-dependent approach. Curr. Microbiol. 2018, 75, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Heinsbroek, S.E.; Williams, D.L.; Welting, O.; Meijer, S.L.; Gordon, S.; de Jonge, W.J. Orally delivered beta-glucans aggravate dextran sulfate sodium (DSS)-induced intestinal inflammation. Nutr. Res. 2015, 35, 1106–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinan, J.; Wang, S.; Hazbun, T.R.; Yadav, H.; Thangamani, S. Antibiotic-induced decreases in the levels of microbial-derived short-chain fatty acids correlate with increased gastrointestinal colonization of Candida albicans. Sci. Rep. 2019, 9, 8872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, D.; Weinstock, A.; Antharam, V.C.; Gu, H.; Jasbi, P.; Shi, X.; Dirks, B.; Krajmalnik-Brown, R.; Maldonado, J.; Guinan, J.; et al. Antibiotic-induced gut metabolome and microbiome alterations increase the susceptibility to Candida albicans colonization in the gastrointestinal tract. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef] [Green Version]
- Jawhara, S.; Thuru, X.; Standaert-Vitse, A.; Jouault, T.; Mordon, S.; Sendid, B.; Desreumaux, P.; Poulain, D. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J. Infect. Dis. 2008, 197, 972–980. [Google Scholar] [CrossRef] [Green Version]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Dharmani, P.; Srivastava, V.; Kissoon-Singh, V.; Chadee, K. Role of intestinal mucins in innate host defense mechanisms against pathogens. J. Innate. Immun. 2009, 1, 123–135. [Google Scholar] [CrossRef]
- Panpetch, W.; Visitchanakun, P.; Saisorn, W.; Sawatpanich, A.; Chatthanathon, P.; Somboonna, N.; Tumwasorn, S.; Leelahavanichkul, A. Lactobacillus rhamnosus attenuates Thai chili extracts induced gut inflammation and dysbiosis despite capsaicin bactericidal effect against the probiotics, a possible toxicity of high dose capsaicin. PLoS ONE 2021, 16, e0261189. [Google Scholar] [CrossRef]
- Correa, R.; de Oliveira Santos, I.; Braz-de-Melo, H.A.; de Sant’Ana, L.P.; das Neves Almeida, R.; Pasquarelli-do-Nascimento, G.; Prado, P.S.; Kobinger, G.P.; Maurice, C.F.; Magalhaes, K.G. Gut microbiota modulation induced by Zika virus infection in immunocompetent mice. Sci. Rep. 2021, 11, 1421. [Google Scholar] [CrossRef]
- Levison, M.E.; Pitsakis, P.G. Susceptibility to experimental Candida albicans urinary tract infection in the rat. J. Infect. Dis. 1987, 155, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.R.; Pan, Y.J.; Liu, J.Y.; Chen, C.T.; Lin, T.L.; Wang, J.T. Klebsiella pneumoniae translocates across the intestinal epithelium via Rho GTPase- and phosphatidylinositol 3-kinase/Akt-dependent cell invasion. Infect. Immun. 2015, 83, 769–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sears, C.L. Molecular physiology and pathophysiology of tight junctions V. assault of the tight junction by enteric pathogens. Am. J. Physiol. Gastrointest Liver Physiol. 2000, 279, G1129–G1134. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, B.I.; Mostov, K.; Engel, J.N. Interaction of bacterial pathogens with polarized epithelium. Annu. Rev. Microbiol. 2001, 55, 407–435. [Google Scholar] [CrossRef]
- Balzan, S.; de Almeida Quadros, C.; de Cleva, R.; Zilberstein, B.; Cecconello, I. Bacterial translocation: Overview of mechanisms and clinical impact. J. Gastroenterol. Hepatol. 2007, 22, 464–471. [Google Scholar] [CrossRef]
- Wieland, C.W.; van Lieshout, M.H.; Hoogendijk, A.J.; van der Poll, T. Host defence during Klebsiella pneumonia relies on haematopoietic-expressed Toll-like receptors 4 and 2. Eur. Respir. J. 2011, 37, 848–857. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.M.; Inamine, T.; Hochrath, K.; Chen, P.; Wang, L.; Llorente, C.; Bluemel, S.; Hartmann, P.; Xu, J.; Koyama, Y.; et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Investig. 2017, 127, 2829–2841. [Google Scholar] [CrossRef] [Green Version]
- Villar, C.C.; Zhao, X.R. Candida albicans induces early apoptosis followed by secondary necrosis in oral epithelial cells. Mol. Oral. Microbiol. 2010, 25, 215–225. [Google Scholar] [CrossRef]
- Villar, C.C.; Chukwuedum Aniemeke, J.; Zhao, X.R.; Huynh-Ba, G. Induction of apoptosis in oral epithelial cells by Candida albicans. Mol. Oral. Microbiol. 2012, 27, 436–448. [Google Scholar] [CrossRef]
- Wu, H.; Downs, D.; Ghosh, K.; Ghosh, A.K.; Staib, P.; Monod, M.; Tang, J. Candida albicans secreted aspartic proteases 4-6 induce apoptosis of epithelial cells by a novel Trojan horse mechanism. FASEB J. 2013, 27, 2132–2144. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, O.; Elias, M.; Peretz, A.; Saliba, W. Role of antibiotics for treatment of inflammatory bowel disease. World J. Gastroenterol. 2016, 22, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Hager, C.L.; Ghannoum, M.A. The mycobiome: Role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig. Liver Dis. 2017, 49, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Li, X.; Shen, J.; Feng, Q. Fecal microbiota transplantation for the treatment of inflammatory bowel disease: An update. Front. Pharmacol. 2020, 11, 574533. [Google Scholar] [CrossRef]
- Welp, A.L.; Bomberger, J.M. Bacterial community interactions during chronic respiratory disease. Front. Cell Infect. Microbiol. 2020, 10, 213. [Google Scholar] [CrossRef]
- Bloomfield, M.; Parackova, Z.; Cabelova, T.; Pospisilova, I.; Kabicek, P.; Houstkova, H.; Sediva, A. Anti-IL6 autoantibodies in an infant with CRP-less septic shock. Front. Immunol. 2019, 10, 2629. [Google Scholar] [CrossRef]
- Cai, B.; Cai, J.P.; Luo, Y.L.; Chen, C.; Zhang, S. The specific roles of JAK/STAT signaling pathway in sepsis. Inflammation 2015, 38, 1599–1608. [Google Scholar] [CrossRef]
- Kim, J.J.; Shajib, M.S.; Manocha, M.M.; Khan, W.I. Investigating intestinal inflammation in DSS-induced model of IBD. J. Vis. Exp. 2012. [Google Scholar] [CrossRef] [Green Version]
- Leelahavanichkul, A.; Somparn, P.; Panich, T.; Chancharoenthana, W.; Wongphom, J.; Pisitkun, T.; Hirankarn, N.; Eiam-Ong, S. Serum miRNA-122 in acute liver injury induced by kidney injury and sepsis in CD-1 mouse models. Hepatol. Res. 2015, 45, 1341–1352. [Google Scholar] [CrossRef]
- Visitchanakun, P.; Saisorn, W.; Wongphoom, J.; Chatthanathon, P.; Somboonna, N.; Svasti, S.; Fucharoen, S.; Leelahavanichkul, A. Gut leakage enhances sepsis susceptibility in iron-overloaded beta-thalassemia mice through macrophage hyperinflammatory responses. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G966–G979. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Phuengmaung, P.; Cheibchalard, T.; Somboonna, N.; Leelahavanichkul, A.; Tumwasorn, S. Lacticaseibacillus casei strain T21 attenuates Clostridioides difficile infection in a murine model through reduction of inflammation and gut dysbiosis with decreased toxin lethality and enhanced mucin production. Front. Microbiol. 2021, 12, 745299. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.; Dockrell, D.H. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]




| Target | Primer Sequence | |
|---|---|---|
| Forward | Reverse | |
| IL-8 | 5′-ACACTGCGCCAACACAGAAATTA-3′ | 5′-TTTGCTTGAAGTTTCACTGGCATC-3′ |
| TNF-α | 5′-CTCTTCTGCCTGCTGCACTTTG-3′ | 5′-ATGGGCTACAGGCTTGTCACTC-3′ |
| IL-6 | 5′-ATGAACTCCTTCTCCACAAGC-3′ | 5′-GTTTTCTGCCAGTGCCTCTTTG-3′ |
| IL-10 | 5′-TCTCCGAGATGCCTTCAGCAGA-3′ | 5′-TCAGACAAGGCTTGGCAACCCA-3′ |
| NF-κB | 5′-ATGGCTTCTATGAGGCTGAG-3′ | 5′-GTTGTTGTTGGTCTGGATGC-3′ |
| Occludin | 5′-CCAATGTCGAGGAGTGGG-3′ | 5′-CGCTGCTGTAACGAGGCT-3′ |
| MUC2 | 5′-CCTGCCGACACCTGCTGCAA-3′ | 5′-ACACCAGTAGAAGGGACAGCACCT-3′ |
| bcl-2 | 5′-GGTGCCACCTGTGGTCCACCT-3′ | 5′-CTTCACTTGTGGCCCAGATAGG-3′ |
| Casp8 | 5′-TTTCTGCCTACAGGGTCATGC-3′ | 5′-TGTCCAACTTTCCTTCTCCCA-3′ |
| Casp9 | 5′-CTCAGACCAGAGATTCGCAAAC-3′ | 5′-GCATTTCCCCTCAAACTCTCAA-3′ |
| NOS2 | 5′-CAGCGGGATGACTTTCCAAG-3′ | 5′-AGGCAAGATTTGGACCTGCA-3′ |
| GAPDH | 5′-GCACCGTCAAGGCTGAGAAC-3′ | 5′-ATGGTGGTGAAGACGCCAGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panpetch, W.; Phuengmaung, P.; Hiengrach, P.; Issara-Amphorn, J.; Cheibchalard, T.; Somboonna, N.; Tumwasorn, S.; Leelahavanichkul, A. Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation. Int. J. Mol. Sci. 2022, 23, 7050. https://doi.org/10.3390/ijms23137050
Panpetch W, Phuengmaung P, Hiengrach P, Issara-Amphorn J, Cheibchalard T, Somboonna N, Tumwasorn S, Leelahavanichkul A. Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation. International Journal of Molecular Sciences. 2022; 23(13):7050. https://doi.org/10.3390/ijms23137050
Chicago/Turabian StylePanpetch, Wimonrat, Pornpimol Phuengmaung, Pratsanee Hiengrach, Jiraphorn Issara-Amphorn, Thanya Cheibchalard, Naraporn Somboonna, Somying Tumwasorn, and Asada Leelahavanichkul. 2022. "Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation" International Journal of Molecular Sciences 23, no. 13: 7050. https://doi.org/10.3390/ijms23137050
APA StylePanpetch, W., Phuengmaung, P., Hiengrach, P., Issara-Amphorn, J., Cheibchalard, T., Somboonna, N., Tumwasorn, S., & Leelahavanichkul, A. (2022). Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation. International Journal of Molecular Sciences, 23(13), 7050. https://doi.org/10.3390/ijms23137050






