Human Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Neural Differentiation of Neural Progenitor Cells
Abstract
:1. Introduction
2. Results and Discussion
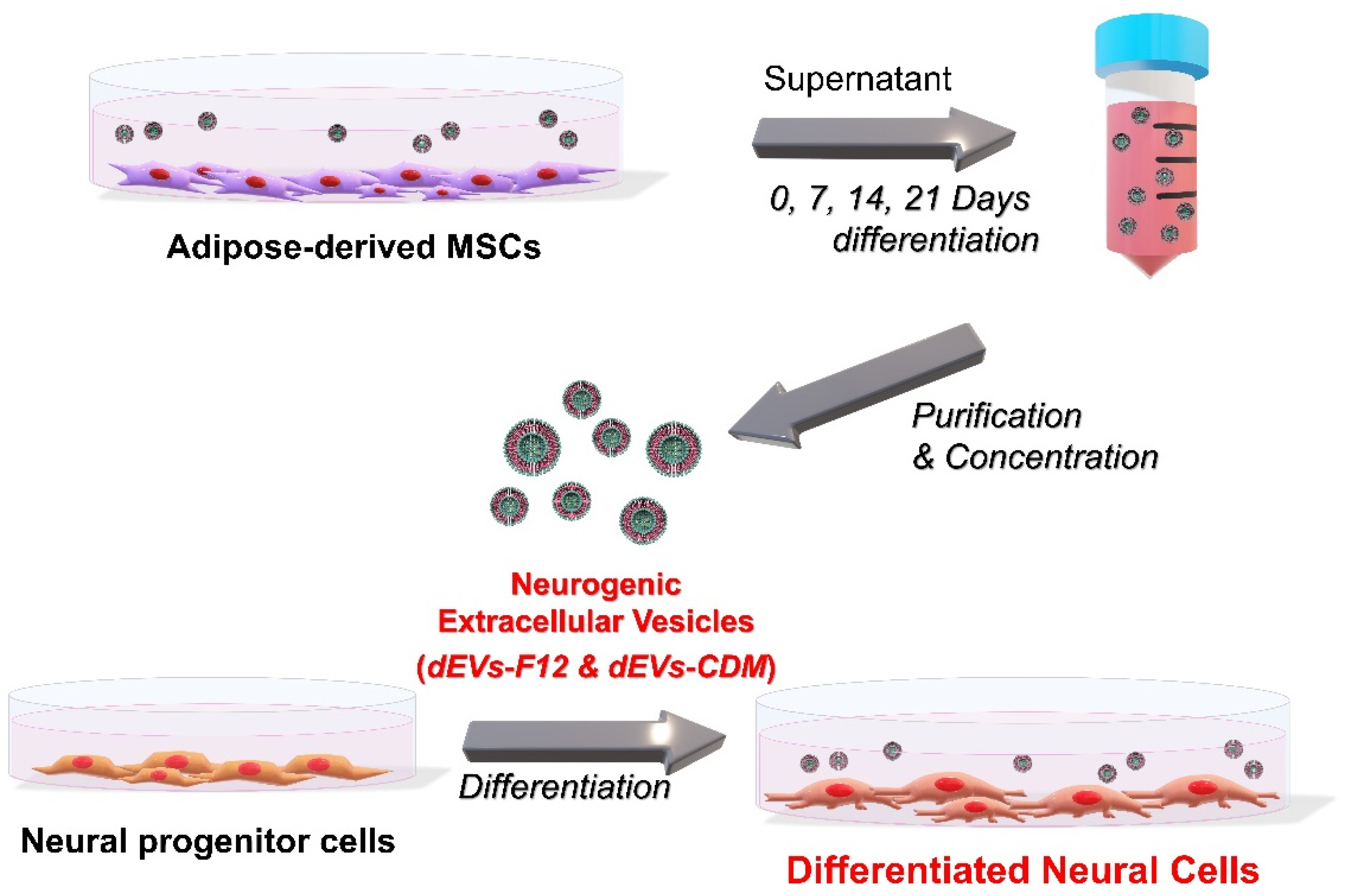
2.1. dEVs Isolation during Neuronal Differentiation of ADMSCs
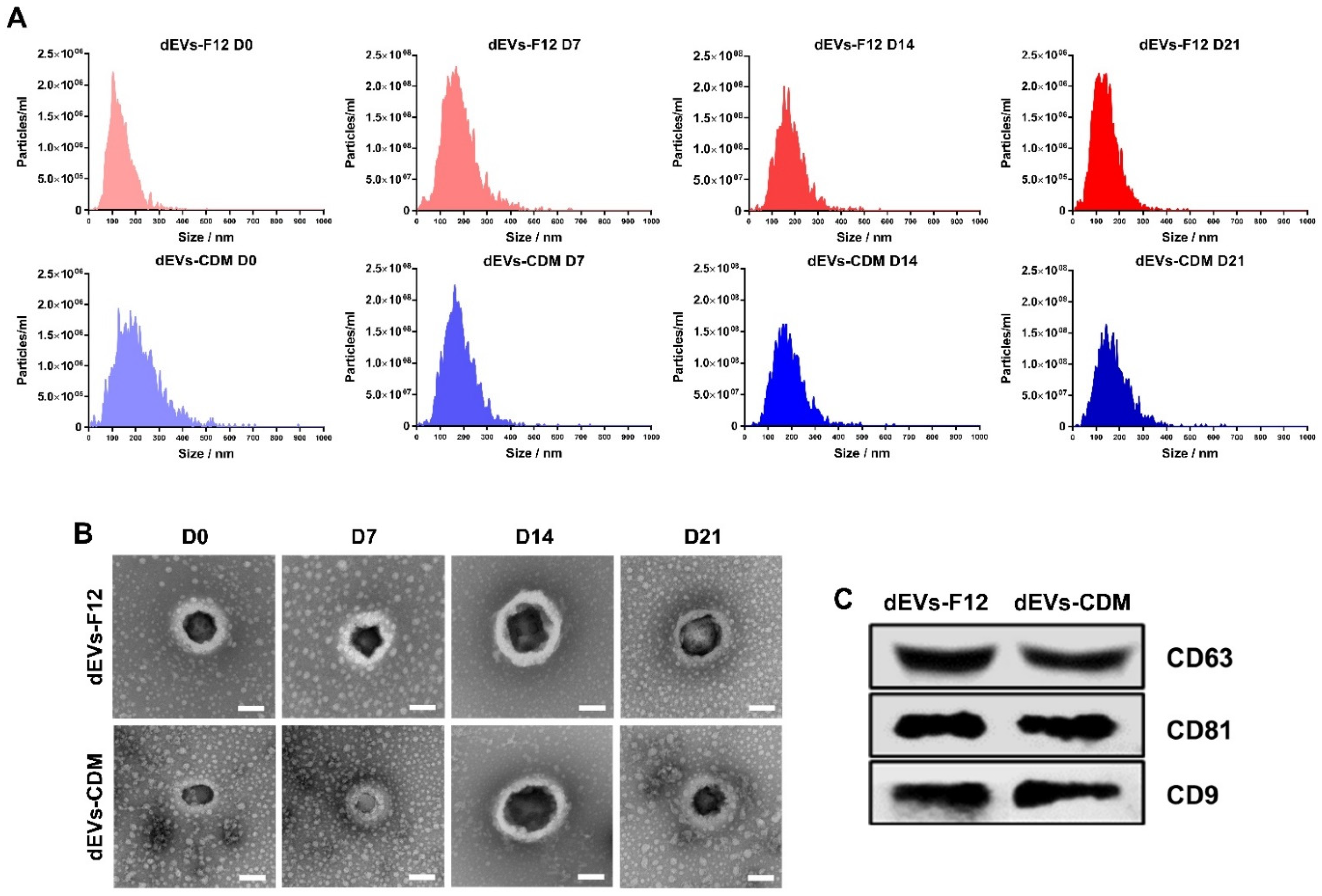
2.2. Characterization of Neuronal Differentiated ADMSC-Derived dEVs
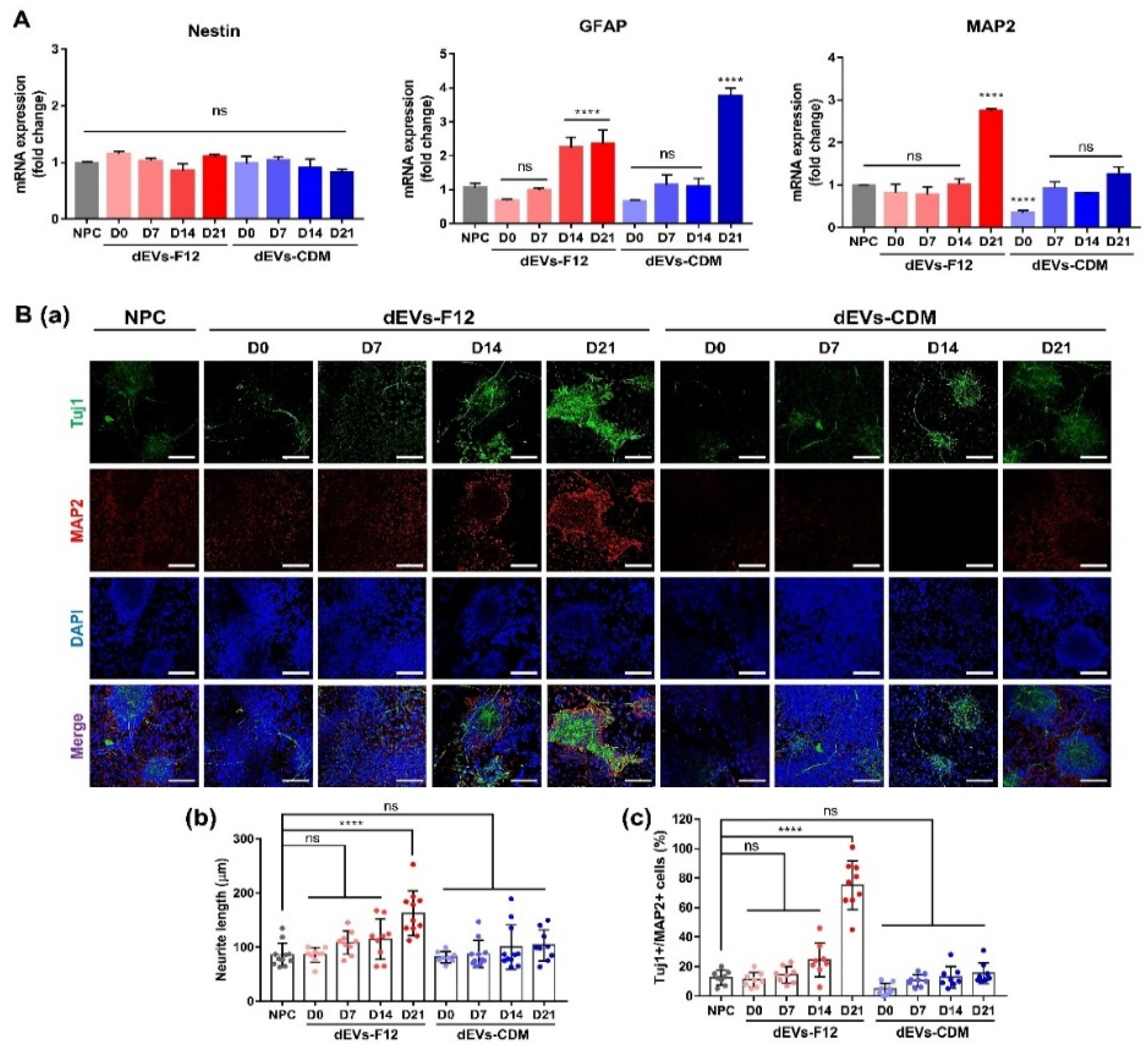
2.3. Enhancing Neurogenesis of NPCs Using dEVs
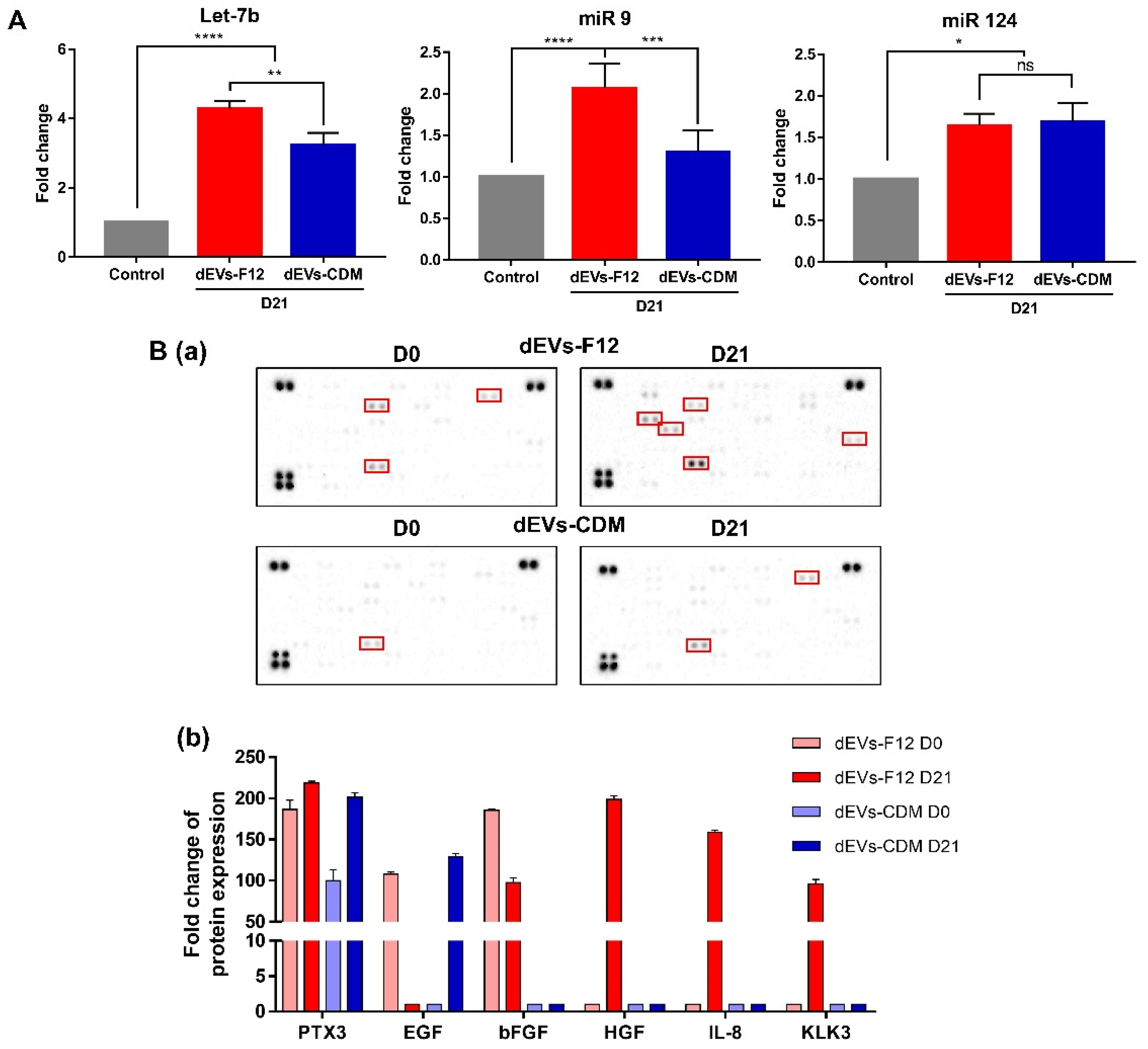
2.4. dEVs-F12 Promotes Neuronal Differentiation of NPCs by Altering Innate miRNA and Protein Composition
3. Materials and Methods
3.1. Neuronal Differentiation of ADMSCs into Neural Lineage Cells
3.2. Neuronal Differentiation of NPCs
3.3. Characterization of Differentiation ADMSC-EVs
3.4. Immunocytochemistry
3.5. Western Blot Analysis
3.6. RT-qPCR Analysis
3.7. Cytokine Array
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NPCs | Neural progenitor cell |
| ADMSC | Adipose-derived mesenchymal stem cells |
| CDM | Chemically defined media |
| TFF | Tangential flow filtration |
| dEV | Neuronal differentiated ADMSC-derived extracellular vesicle (Differentiated EV) |
| NTA | Nanoparticle tracking analysis |
| miRNA | microRNA |
| GFAP | Glial fibrillary acidic protein |
| Tuj1 | Tubulin beta III |
| MAP2 | Microtubule-associated protein 2 |
| PTX3 | Pentraxin 3 |
| bFGF | Fibroblast growth factor-Basic |
| HGF | Hepatocyte growth factor |
| IL-8 | Interleukin 8 |
| KLK3 | Kallikrein 3 |
References
- Choi, S.J.; Park, S.Y.; Shin, Y.H.; Heo, S.-H.; Kim, K.-H.; Lee, H.I.; Kim, J.K. Mesenchymal stem cells derived from wharton’s jelly can differentiate into schwann cell-like cells and promote peripheral nerve regeneration in acellular nerve grafts. Tissue Eng. Regen. Med. 2021, 18, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.-S.; Kim, D.-S.; Han, Y.; Son, H.; Chung, Y.-H.; Min, J.; Kim, T.-H. Three-dimensional graphene–rgd peptide nanoisland composites that enhance the osteogenesis of human adipose-derived mesenchymal stem cells. Int. J. Mol. Sci. 2018, 19, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-S.; Lee, J.-K.; Kim, J.H.; Lee, J.; Kim, D.S.; An, S.; Park, S.-B.; Kim, T.-H.; Rim, J.S.; Lee, S.; et al. Advanced PLGA hybrid scaffold with a bioactive PDRN/BMP2 nanocomplex for angiogenesis and bone regeneration using human fetal MSCs. Sci. Adv. 2021, 7, eabj1083. [Google Scholar] [CrossRef]
- Kim, I.G.; Cho, H.; Shin, J.; Cho, J.H.; Cho, S.-W.; Chung, E.-J. Regeneration of irradiation-damaged esophagus by local delivery of mesenchymal stem-cell spheroids encapsulated in a hyaluronic-acid-based hydrogel. Biomater. Sci. 2021, 9, 2197–2208. [Google Scholar] [CrossRef] [PubMed]
- Schwieger, J.; Hamm, A.; Gepp, M.M.; Schulz, A.; Hoffmann, A.; Lenarz, T.; Scheper, V. Alginate-encapsulated brain-derived neurotrophic factor–overexpressing mesenchymal stem cells are a promising drug delivery system for protection of auditory neurons. J. Tissue Eng. 2020, 11, 2041731420911313. [Google Scholar] [CrossRef]
- Urrutia, D.N.; Caviedes, P.; Mardones, R.; Minguell, J.J.; Vega-Letter, A.M.; Jofre, C.M. Comparative study of the neural differentiation capacity of mesenchymal stromal cells from different tissue sources: An approach for their use in neural regeneration therapies. PLoS ONE 2019, 14, e0213032. [Google Scholar] [CrossRef]
- Cho, Y.-W.; Kim, D.-S.; Suhito, I.R.; Han, D.K.; Lee, T.; Kim, T.-H. Enhancing neurogenesis of neural stem cells using homogeneous nanohole pattern-modified conductive platform. Int. J. Mol. Sci. 2020, 21, 191. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Rhim, W.-K.; Yoo, Y.-I.; Kim, D.-S.; Ko, K.-W.; Heo, Y.; Park, C.G.; Han, D.K. Defined msc exosome with high yield and purity to improve regenerative activity. J. Tissue Eng. 2021, 12, 20417314211008626. [Google Scholar] [CrossRef]
- Upadhya, R.; Madhu, L.N.; Attaluri, S.; Gitaí, D.L.G.; Pinson, M.R.; Kodali, M.; Shetty, G.; Zanirati, G.; Kumar, S.; Shuai, B.; et al. Extracellular vesicles from human ipsc-derived neural stem cells: Mirna and protein signatures, and anti-inflammatory and neurogenic properties. J. Extracell. Vesicles 2020, 9, 1809064. [Google Scholar] [CrossRef]
- Kim, J.Y.; Rhim, W.-K.; Seo, H.J.; Lee, J.Y.; Park, C.G.; Han, D.K. Comparative analysis of msc-derived exosomes depending on cell culture media for regenerative bioactivity. Tissue Eng. Regen. Med. 2021, 18, 355–367. [Google Scholar] [CrossRef]
- Kuang, Y.; Zheng, X.; Zhang, L.; Ai, X.; Venkataramani, V.; Kilic, E.; Hermann, D.M.; Majid, A.; Bähr, M.; Doeppner, T.R. Adipose-derived mesenchymal stem cells reduce autophagy in stroke mice by extracellular vesicle transfer of mir-25. J. Extracell. Vesicles 2020, 10, e12024. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhao, F.; Zhou, L.; Liu, J.; Xu, L.; Dou, Q.; Xu, Z.; Jia, R. Therapeutic potential of adipose-derived mesenchymal stem cell exosomes in tissue-engineered bladders. J. Tissue Eng. 2021, 12, 20417314211001545. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.T.; Toh, W.S.; Lai, R.C.; Lim, S.K. Practical considerations in transforming msc therapy for neurological diseases from cell to ev. Exp. Neurol. 2022, 349, 113953. [Google Scholar] [CrossRef] [PubMed]
- Ahmadvand Koohsari, S.; Absalan, A.; Azadi, D. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles attenuate experimental autoimmune encephalomyelitis via regulating pro and anti-inflammatory cytokines. Sci. Rep. 2021, 11, 11658. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Zhang, L. Microrna-129-5p shuttled by mesenchymal stem cell-derived extracellular vesicles alleviates intervertebral disc degeneration via blockade of lrg1-mediated p38 mapk activation. J. Tissue Eng. 2021, 12, 20417314211021679. [Google Scholar] [CrossRef]
- Phinney, D.G.; di Giuseppe, M.; Njah, J.; Sala, E.; Shiva, S.; Croix, C.M.S.; Stolz, D.B.; Watkins, S.C.; Di, Y.P.; Leikauf, G.D.; et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle micrornas. Nat. Commun. 2015, 6, 8472. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhao, X. Micrornas in adult and embryonic neurogenesis. Neuromol. Med. 2009, 11, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Nishino, J.; Kim, I.; Chada, K.; Morrison, S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16ink4a and p19arf expression. Cell 2008, 135, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Stappert, L.; Klaus, F.; Brüstle, O. Micrornas engage in complex circuits regulating adult neurogenesis. Front. Neurosci. 2018, 12, 707. [Google Scholar] [CrossRef]
- Conaco, C.; Otto, S.; Han, J.-J.; Mandel, G. Reciprocal actions of rest and a microrna promote neuronal identity. Proc. Natl. Acad. Sci. USA 2006, 103, 2422–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Tang, C.; Zhang, J.; Li, Z.; Zhang, X.; Miron, R.J.; Zhang, Y. Extracellular vesicles derived from the mid-to-late stage of osteoblast differentiation markedly enhance osteogenesis in vitro and in vivo. Biochem. Biophys. Res. Commun. 2019, 514, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Zhai, M.; Zhu, Y.; Yang, M.; Mao, C. Human mesenchymal stem cell derived exosomes enhance cell-free bone regeneration by altering their mirnas profiles. Adv. Sci. 2020, 7, 2001334. [Google Scholar] [CrossRef]
- Liu, D.; Shu, M.; Liu, W.; Shen, Y.; Long, G.; Zhao, Y.; Hou, X.; Xiao, Z.; Dai, J.; Li, X. Binary scaffold facilitates in situ regeneration of axons and neurons for complete spinal cord injury repair. Biomater. Sci. 2021, 9, 2955–2971. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, A.; Mazzon, E. Dental mesenchymal stem cell secretome: An intriguing approach for neuroprotection and neuroregeneration. Int. J. Mol. Sci. 2022, 23, 456. [Google Scholar] [CrossRef] [PubMed]
- Eng, L.F. Glial fibrillary acidic protein (gfap): The major protein of glial intermediate filaments in differentiated astrocytes. J. Neuroimmunol. 1985, 8, 203–214. [Google Scholar] [CrossRef]
- Biancotti, J.C.; Walker, K.A.; Jiang, G.; di Bernardo, J.; Shea, L.D.; Kunisaki, S.M. Hydrogel and neural progenitor cell delivery supports organotypic fetal spinal cord development in an ex vivo model of prenatal spina bifida repair. J. Tissue Eng. 2020, 11, 2041731420943833. [Google Scholar] [CrossRef]
- Ferreiro, I.; Genevois, C.; Konate, K.; Vivès, E.; Boisguérin, P.; Deshayes, S.; Couillaud, F. In vivo follow-up of gene inhibition in solid tumors using peptide-based nanoparticles for sirna delivery. Pharmaceutics 2021, 13, 749. [Google Scholar] [CrossRef]
- Wislet-Gendebien, S.; Hans, G.; Leprince, P.; Rigo, J.M.; Moonen, G.; Rogister, B. Plasticity of cultured mesenchymal stem cells: Switch from nestin-positive to excitable neuron-like phenotype. Stem Cells 2005, 23, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (misev2018): A position statement of the international society for extracellular vesicles and update of the misev2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-S.; Lee, D.R.; Kim, H.-S.; Yoo, J.-E.; Jung, S.J.; Lim, B.Y.; Jang, J.; Kang, H.-C.; You, S.; Hwang, D.-Y.; et al. Highly pure and expandable psa-ncam-positive neural precursors from human ESC and IPSC-derived neural rosettes. PLoS ONE 2012, 7, e39715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.-F.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor tlx signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Luo, M.; Ni, N.; Den, Y.; Xia, J.; Chen, J.; Ji, J.; Zhou, X.; Fan, X.; Gu, P. Reciprocal actions of microrna-9 and tlx in the proliferation and differentiation of retinal progenitor cells. Stem Cells Dev. 2014, 23, 2771–2781. [Google Scholar] [CrossRef]
- Kapsimali, M.; Kloosterman, W.P.; de Bruijn, E.; Rosa, F.; Plasterk, R.H.A.; Wilson, S.W. Micrornas show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007, 8, R173. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Hu, D.-H.; Yin, J.Q.; Xu, R.-X. Molecular mechanisms of transdifferentiation of adipose-derived stem cells into neural cells: Current status and perspectives. Stem Cells Int. 2018, 2018, 5630802. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, H.; Zheng, J.-F.; Guo, Z.-D.; Huang, Z.-J.; Wu, Y.; Zhong, J.-J.; Sun, X.-C.; Cheng, C.-J. Pentraxin 3 contributes to neurogenesis after traumatic brain injury in mice. Neural Regen. Res. 2020, 15, 2318–2326. [Google Scholar]
- Rodriguez-Grande, B.; Varghese, L.; Molina-Holgado, F.; Rajkovic, O.; Garlanda, C.; Denes, A.; Pinteaux, E. Pentraxin 3 mediates neurogenesis and angiogenesis after cerebral ischaemia. J. Neuroinflamm. 2015, 12, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-Y.; Wang, Z.-G.; Wu, F.-Z.; Kong, X.-X.; Yang, J.; Lin, B.-B.; Zhu, S.-P.; Lin, L.; Gan, C.-S.; Fu, X.-B.; et al. Regulation of autophagy and ubiquitinated protein accumulation by bfgf promotes functional recovery and neural protection in a rat model of spinal cord injury. Mol. Neurobiol. 2013, 48, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Desole, C.; Gallo, S.; Vitacolonna, A.; Montarolo, F.; Bertolotto, A.; Vivien, D.; Comoglio, P.; Crepaldi, T. HGF and MET: From brain development to neurological disorders. Front. Cell Dev. Biol. 2021, 9, 683609. [Google Scholar] [CrossRef] [PubMed]
- Radulovic, M.; Yoon, H.; Larson, N.; Wu, J.; Linbo, R.; Burda, J.E.; Diamandis, E.P.; Blaber, S.I.; Blaber, M.; Fehlings, M.G.; et al. Kallikrein cascades in traumatic spinal cord injury: In vitro evidence for roles in axonopathy and neuron degeneration. J. Neuropathol. Exp. Neurol. 2013, 72, 1072–1089. [Google Scholar] [CrossRef] [Green Version]
- Araujo, D.M.; Cotman, C.W. Trophic effects of interleukin-4, -7 and -8 on hippocampal neuronal cultures, Potential involvement of glial-derived factors. Brain Res. 1993, 600, 49–55. [Google Scholar] [CrossRef]
- Ko, K.-W.; Park, S.-Y.; Lee, E.H.; Yoo, Y.-I.; Kim, D.-S.; Kim, J.Y.; Kwon, T.G.; Han, D.K. Integrated bioactive scaffold with polydeoxyribonucleotide and stem-cell-derived extracellular vesicles for kidney regeneration. ACS Nano 2021, 15, 7575–7585. [Google Scholar] [CrossRef]
- d’Angelo, M.; Cimini, A.; Castelli, V. Insights into the effects of mesenchymal stem cell-derived secretome in parkinson’s disease. Int. J. Mol. Sci. 2020, 21, 5241. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-Y.; Kim, D.-S.; Kim, H.-M.; Lee, J.-K.; Hwang, D.-Y.; Kim, T.-H.; You, S.; Han, D.K. Human Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Neural Differentiation of Neural Progenitor Cells. Int. J. Mol. Sci. 2022, 23, 7047. https://doi.org/10.3390/ijms23137047
Park S-Y, Kim D-S, Kim H-M, Lee J-K, Hwang D-Y, Kim T-H, You S, Han DK. Human Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Neural Differentiation of Neural Progenitor Cells. International Journal of Molecular Sciences. 2022; 23(13):7047. https://doi.org/10.3390/ijms23137047
Chicago/Turabian StylePark, So-Yeon, Da-Seul Kim, Hyun-Mun Kim, Jun-Kyu Lee, Dong-Youn Hwang, Tae-Hyung Kim, Seungkwon You, and Dong Keun Han. 2022. "Human Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Neural Differentiation of Neural Progenitor Cells" International Journal of Molecular Sciences 23, no. 13: 7047. https://doi.org/10.3390/ijms23137047
APA StylePark, S.-Y., Kim, D.-S., Kim, H.-M., Lee, J.-K., Hwang, D.-Y., Kim, T.-H., You, S., & Han, D. K. (2022). Human Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Neural Differentiation of Neural Progenitor Cells. International Journal of Molecular Sciences, 23(13), 7047. https://doi.org/10.3390/ijms23137047







