Synthesis, In Vitro, and Computational Studies of PTP1B Phosphatase Inhibitors Based on Oxovanadium(IV) and Dioxovanadium(V) Complexes
Abstract
:1. Introduction
2. Results
2.1. Synthesis
2.2. Inhibitory Activity against PTP1B Phosphatase
- IC50–The obtained half maximal inhibitory concentration for each complexes;
- [S]–substrate concentration (1 mM);
- [Km] = The calculated Michaelis constant (0.7 mM).
2.3. In Silico Studies
2.4. Cytotoxicity against Breast Cancer Cell Lines and Human Keratinocyte Cell Line
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Synthesis
4.3. Recombinant PTP1B Assay
4.4. Cell Lines and Culture
4.5. Cell Viability Assay
4.6. Molecular Modeling
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy (Review). Int. J. Mol. Med. 2017, 40, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bononi, A.; Agnoletto, C.; de Marchi, E.; Marchi, S.; Patergnani, S.; Bonora, M.; Giorgi, C.; Missiroli, S.; Poletti, F.; Rimessi, A.; et al. Protein Kinases and Phosphatases in the Control of Cell Fate. Enzym. Res. 2011, 2011, 329098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lessard, L.; Stuible, M.; Tremblay, M.L. The Two Faces of PTP1B in Cancer. Biochim. Biophys. Acta 2010, 1804, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Medgyesi, D.; Hobeika, E.; Biesen, R.; Kollert, F.; Taddeo, A.; Voll, R.E.; Hiepe, F.; Reth, M. The Protein Tyrosine Phosphatase PTP1B Is a Negative Regulator of CD40 and BAFF-R Signaling and Controls B Cell Autoimmunity. J. Exp. Med. 2014, 211, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eleftheriou, P.; Geronikaki, A.; Petrou, A. PTP1b Inhibition, A Promising Approach for the Treatment of Diabetes Type II. Curr. Top. Med. Chem. 2019, 19, 246–263. [Google Scholar] [CrossRef]
- Byeon, H.J.; Kim, J.Y.; Ko, J.; Lee, E.J.; Don, K.; Yoon, J.S. Protein Tyrosine Phosphatase 1B as a Therapeutic Target for Graves’ Orbitopathy in an in Vitro Model. PLoS ONE 2020, 15, e0237015. [Google Scholar] [CrossRef]
- Villamar-Cruz, O.; Loza-Mejia, M.A.; Arias-Romero, L.E.; Camacho-Arroyo, I. Recent Advances in PTP1B Signaling in Metabolism and Cancer. Biosci. Rep. 2021, 41, BSR20211994. [Google Scholar] [CrossRef]
- Hilmarsdottir, B.; Briem, E.; Halldorsson, S.; Kricker, J.; Ingthorsson, S.; Gustafsdottir, S.; Mælandsmo, G.M.; Magnusson, M.K.; Gudjonsson, T. Inhibition of PTP1B Disrupts Cell–Cell Adhesion and Induces Anoikis in Breast Epithelial Cells. Cell Death Dis. 2017, 8, e2769. [Google Scholar] [CrossRef] [Green Version]
- Garner, W.B.; Smith, B.D.; Shabason, J.E.; Williams, G.R.; Martin, M.Y.; Ballo, M.T.; VanderWalde, N.A. Predicting Future Cancer Incidence by Age and Gender. J. Clin. Oncol. 2019, 37, 1559. [Google Scholar] [CrossRef]
- Sivaganesh, V.; Sivaganesh, V.; Scanlon, C.; Iskander, A.; Maher, S.; Lê, T.; Peethambaran, B. Protein Tyrosine Phosphatases: Mechanisms in Cancer. Int. J. Mol. Sci. 2021, 22, 12865. [Google Scholar] [CrossRef]
- Pei, Z.; Liu, G.; Lubben, T.; Szczepankiewicz, B. Inhibition of Protein Tyrosine Phosphatase 1B as a Potential Treatment of Diabetes and Obesity. Curr. Pharm. Des. 2005, 10, 3481–3504. [Google Scholar] [CrossRef] [PubMed]
- Irving, E.; Stoker, A.W. Vanadium Compounds as PTP Inhibitors. Molecules 2017, 22, 2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Ivanov, S.; Lagunin, A.; Goel, R.K. Attenuation of Hyperhomocysteinemia Induced Vascular Dementia by Sodium Orthovanadate Perhaps via PTP1B: Pertinent Downstream Outcomes. Behav. Brain Res. 2019, 364, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Takakura, K.; Beckman, J.S.; MacMillan-Crow, L.A.; Crow, J.P. Rapid and Irreversible Inactivation of Protein Tyrosine Phosphatases PTP1B, CD45, and LAR by Peroxynitrite. Arch. Biochem. Biophys. 1999, 369, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Weiser, D.C.; Shenolikar, S. Use of Protein Phosphatase Inhibitors. Curr. Protoc. Mol. Biol. 2003, 62, 18.10.1–18.10.13. [Google Scholar] [CrossRef] [PubMed]
- Bialy, L.; Waldmann, H. Inhibitors of Protein Tyrosine Phosphatases: Next-Generation Drugs? Angew. Chem. Int. 2005, 44, 3814–3839. [Google Scholar] [CrossRef]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.F.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.A.; Müller, S.; Knapp, S. Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Fantus, I.G. Inhibition of the Protein Tyrosine Phosphatase PTP1B: Potential Therapy for Obesity, Insulin Resistance and Type-2 Diabetes Mellitus. Best Prac. Res. Clin. Endocrinol. Metab. 2007, 21, 621–640. [Google Scholar] [CrossRef]
- Panzhinskiy, E.; Ren, J.; Nair, S. Protein Tyrosine Phosphatase 1B and Insulin Resistance: Role of Endoplasmic Reticulum Stress/Reactive Oxygen Species/Nuclear Factor Kappa B Axis. PLoS ONE 2013, 8, e77228. [Google Scholar] [CrossRef]
- Yuan, C.; Lu, L.; Gao, X.; Wu, Y.; Guo, M.; Li, Y.; Fu, X.; Zhu, M. Ternary Oxovanadium(IV) Complexes of ONO-Donor Schiff Base and Polypyridyl Derivatives as Protein Tyrosine Phosphatase Inhibitors: Synthesis, Characterization, and Biological Activities. J. Biol. Inorg. Chem. 2009, 14, 841–851. [Google Scholar] [CrossRef]
- Yuan, C.; Lu, L.; Wu, Y.; Liu, Z.; Guo, M.; Xing, S.; Fu, X.; Zhu, M. Synthesis, Characterization, and Protein Tyrosine Phosphatases Inhibition Activities of Oxovanadium(IV) Complexes with Schiff Base and Polypyridyl Derivatives. J. Inorg. Biochem. 2010, 104, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Drzeżdżon, J.; Pawlak, M.; Matyka, N.; Sikorski, A.; Gawdzik, B.; Jacewicz, D. Relationship between Antioxidant Activity and Ligand Basicity in the Dipicolinate Series of Oxovanadium(Iv) and Dioxovanadium(v) Complexes. Int. J. Mol. Sci. 2021, 22, 9886. [Google Scholar] [CrossRef]
- Chatterjee, M.; Ghosh, S.; Wu, B.-M.; Mak, T.C.W. A structural and electrochemical study of some oxovanadium(IV) heterochelate complexes. Polyhedron 1998, 17, 1369–1374. [Google Scholar] [CrossRef]
- Pranczk, J.; Wyrzykowski, D.; Jacewicz, D.; Sikorski, A.; Tesmar, A.; Chmurzyński, L. Structural, Physico-Chemical and Antioxidant Characteristics of 2,2′-Bipyridyl(Iminodiacetato)Oxidovanadium(IV) Dihydrate. Polyhedron 2015, 100, 74–81. [Google Scholar] [CrossRef]
- Drzeżdżon, J.; Pawlak, M.; Gawdzik, B.; Wypych, A.; Kramkowski, K.; Kowalczyk, P.; Jacewicz, D. Dipicolinate Complexes of Oxovanadium(IV) and Dioxovanadium(V) with 2-Phenylpyridine and 4,4′-Dimethoxy-2,2′-Bipyridyl as New Precatalysts for Olefin Oligomerization. Materials 2022, 15, 1379. [Google Scholar] [CrossRef]
- Kamerlin, S.C.; Rucker, R.; Boresch, S. A targeted molecular dynamics study of WPD loop movement in PTP1B. Biochem. Biophys. Res. Commun. 2006, 345, 1161–1166. [Google Scholar] [CrossRef]
- Brandão, T.A.; Hengge, A.C.; Johnson, S.J. Insights into the reaction of protein-tyrosine phosphatase 1B: Crystal structures for transition state analogs of both catalytic steps. J. Biol. Chem. 2010, 21, 15874-83. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Xie, L.; Yang, F.; Wang, W.; Zhou, Q.; Xiang, M.; Zhou, S.; Lv, W.; Jia, Y.; Pokhrel, L.; et al. Recent advance on PTP1B inhibitors and their biomedical applications. Eur. J. Med. Chem. 2020, 199, 112376. [Google Scholar] [CrossRef]
- Zhao, Y.; Cao, Y.; Chen, H.; Zhuang, F.; Wu, C.; Yoon, G.; Zhu, W.; Su, Y.; Zheng, S.; Liu, Z.; et al. Synthesis, biological evaluation, and molecular docking study of novel allyl-retrochalcones as a new class of protein tyrosine phosphatase 1B inhibitors. Bioorg. Med. Chem. 2019, 27, 963–977. [Google Scholar] [CrossRef]
- Shen, R.; Crean, R.M.; Johnson, S.J.; Kamerlin, S.C.; Hengge, A.C. Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases. JACS Au 2021, 1, 646–659. [Google Scholar] [CrossRef]
- Kowalski, S.; Wyrzykowski, D.; Inkielewicz-Stȩpniak, I. Molecular and Cellular Mechanisms of Cytotoxic Activity of Vanadium Compounds against Cancer Cells. Molecules 2020, 25, 1757. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, E.; Birla Singh, K.; Massarotti, A.; Hogstrand, C.; Maret, W. The Metal Face of Protein Tyrosine Phosphatase 1B. Coord. Chem. Rev. 2016, 70, 327–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, L.; Chang, W.; Zhu, S.; Zhang, Y.; Chen, P.; Zhang, H.; Zhao, H.; Zha, J.; Jiang, S.; Tao, L.; et al. Exploring Anticancer Activities and Structure—Activity Relationships of Binuclear Oxidovanadium(IV) Complexes. ACS Appl. Bio Mater. 2021, 4, 8571–8583. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Zhao, H.; Tao, L.; Li, X.; Zhou, Z.; Sun, Y.; Chen, C.; Wei, D.; Liu, Y.; Diao, G. Synthesis, in vitro cytotoxicity, and structure-activity relationships (SAR) of multidentate oxidovanadium(iv) complexes as anticancer agents. Dalton Trans. 2018, 47, 10035–10045. [Google Scholar] [CrossRef]
- Kuban-Jankowska, A.; Kostrzewa, T.; Musial, C.; Barone, G.; Lo-Bosco, G.; Lo-Celso, F.; Gorska-Ponikowska, M. Green Tea Catechins Induce Inhibition of PTP1B Phosphatase in Breast Cancer Cells with Potent Anti-Cancer Properties: In Vitro Assay, Molecular Docking, and Dynamics Studies. Antioxidants 2020, 9, 1208. [Google Scholar] [CrossRef]
- Diluvio, G.; del Gaudio, F.; Giuli, M.V.; Franciosa, G.; Giuliani, E.; Palermo, R.; Besharat, Z.M.; Pignataro, M.G.; Vacca, A.; D’Amati, G.; et al. NOTCH3 Inactivation Increases Triple Negative Breast Cancer Sensitivity to Gefitinib by Promoting EGFR Tyrosine Dephosphorylation and Its Intracellular Arrest. Oncogenesis 2018, 7, 42. [Google Scholar] [CrossRef]
- Yu, M.; Liu, Z.; Liu, Y.; Zhou, X.; Sun, F.; Liu, Y.; Li, L.; Hua, S.; Zhao, Y.; Gao, H.; et al. PTP1B Markedly Promotes Breast Cancer Progression and Is Regulated by MiR-193a-3p. FEBS J. 2019, 286, 1136–1153. [Google Scholar] [CrossRef]
- Jiménez, J.; Škalič, M.; Martínez-Rosell, G.; De Fabritiis, G. KDEEP: Protein-Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J. Chem. Inf. Model. 2018, 58, 287–296. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
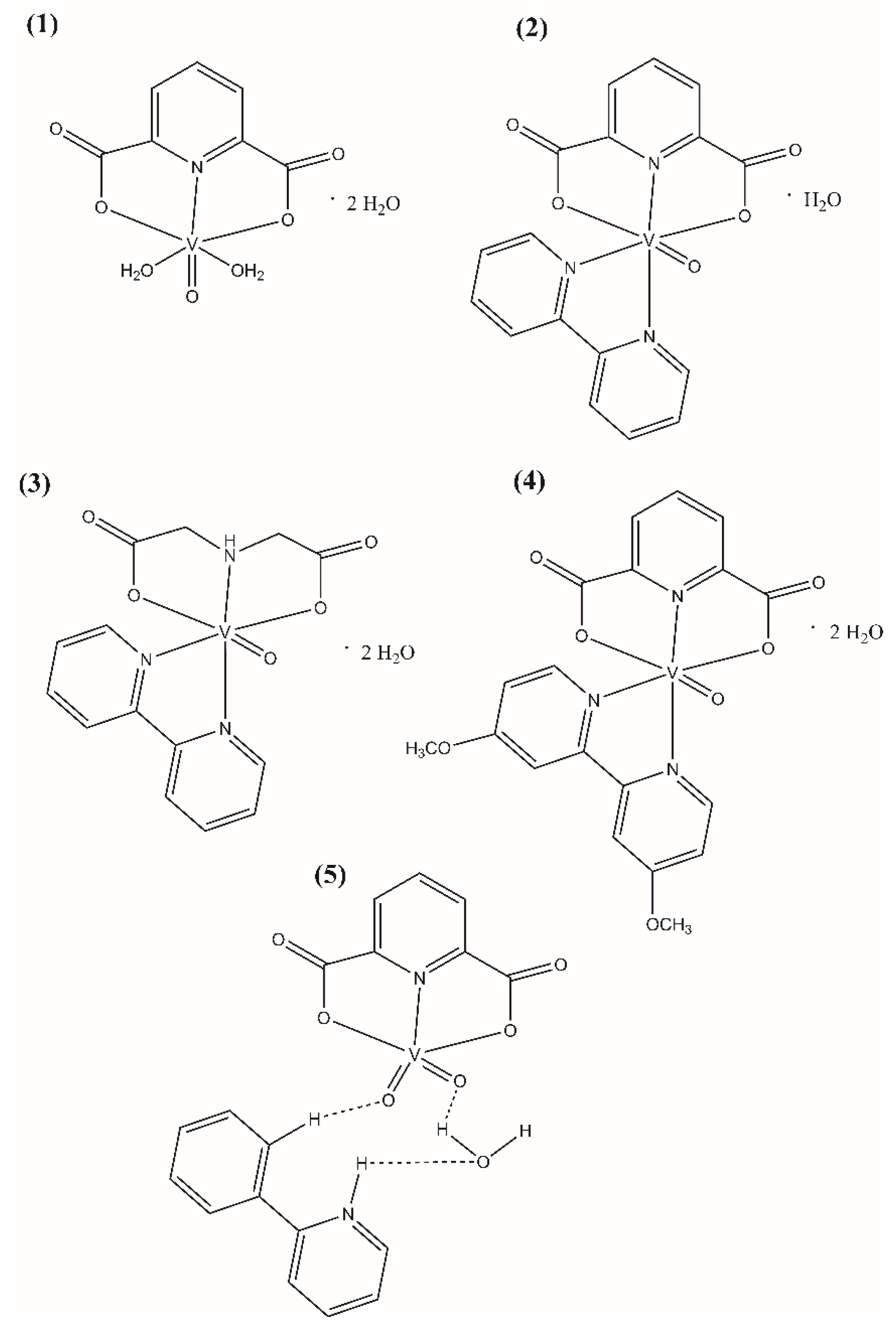

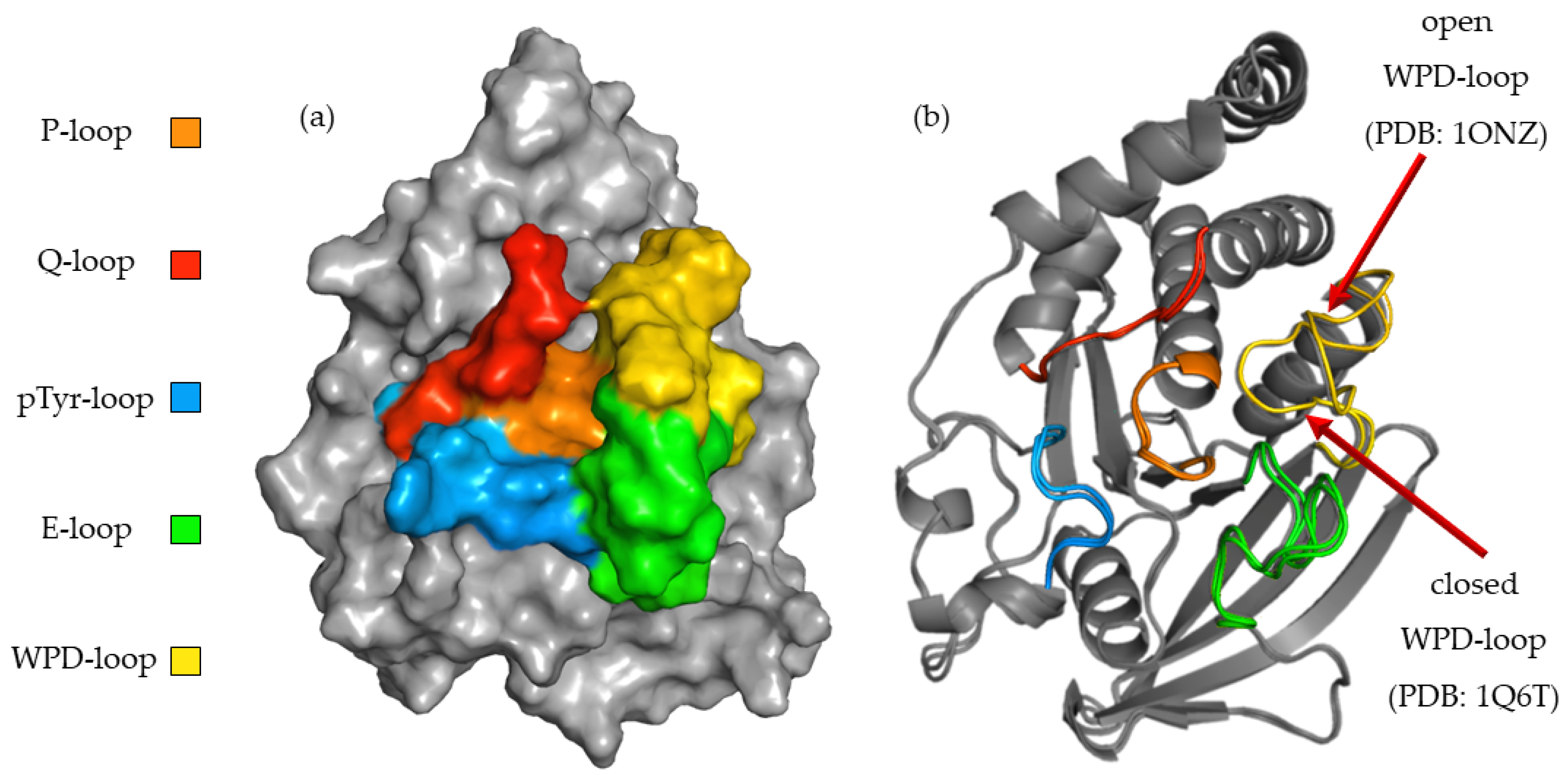
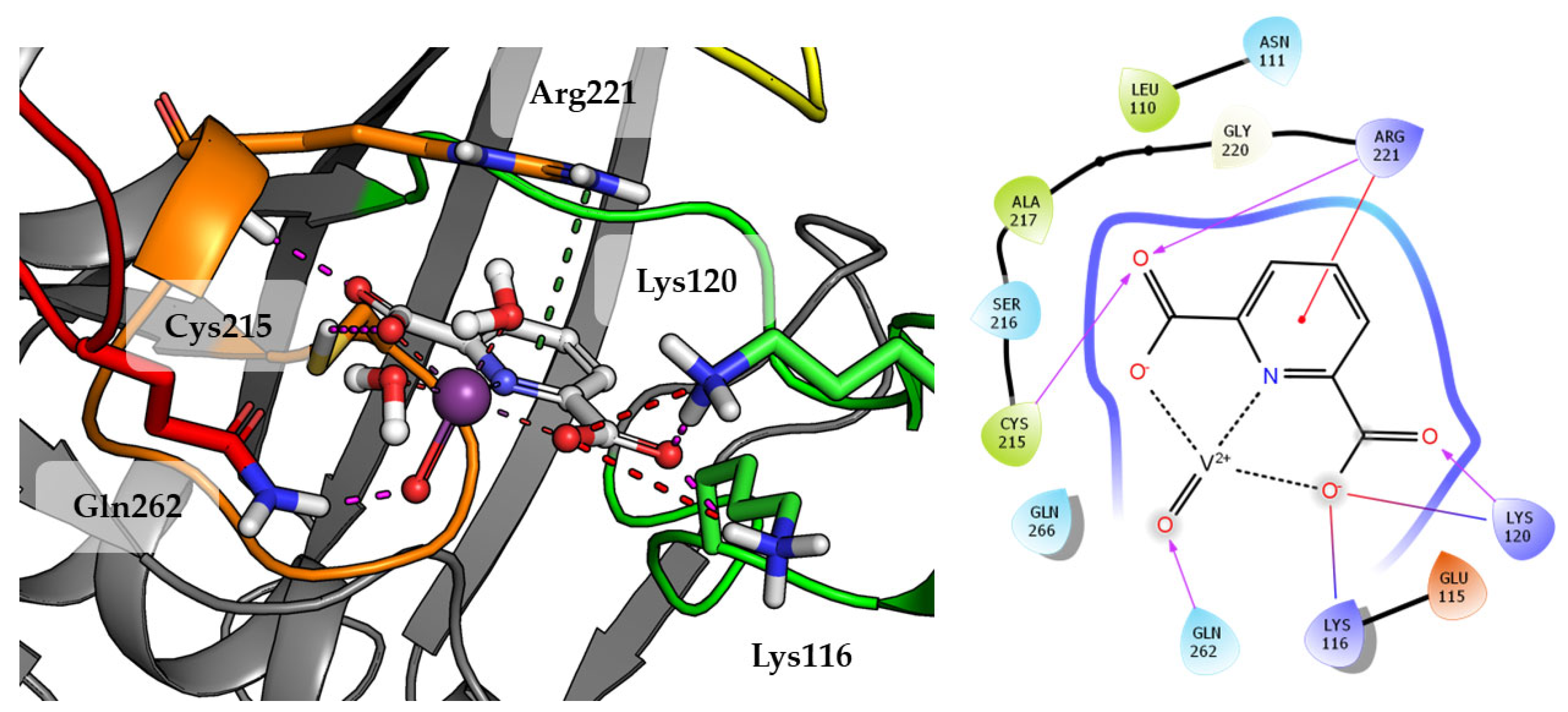
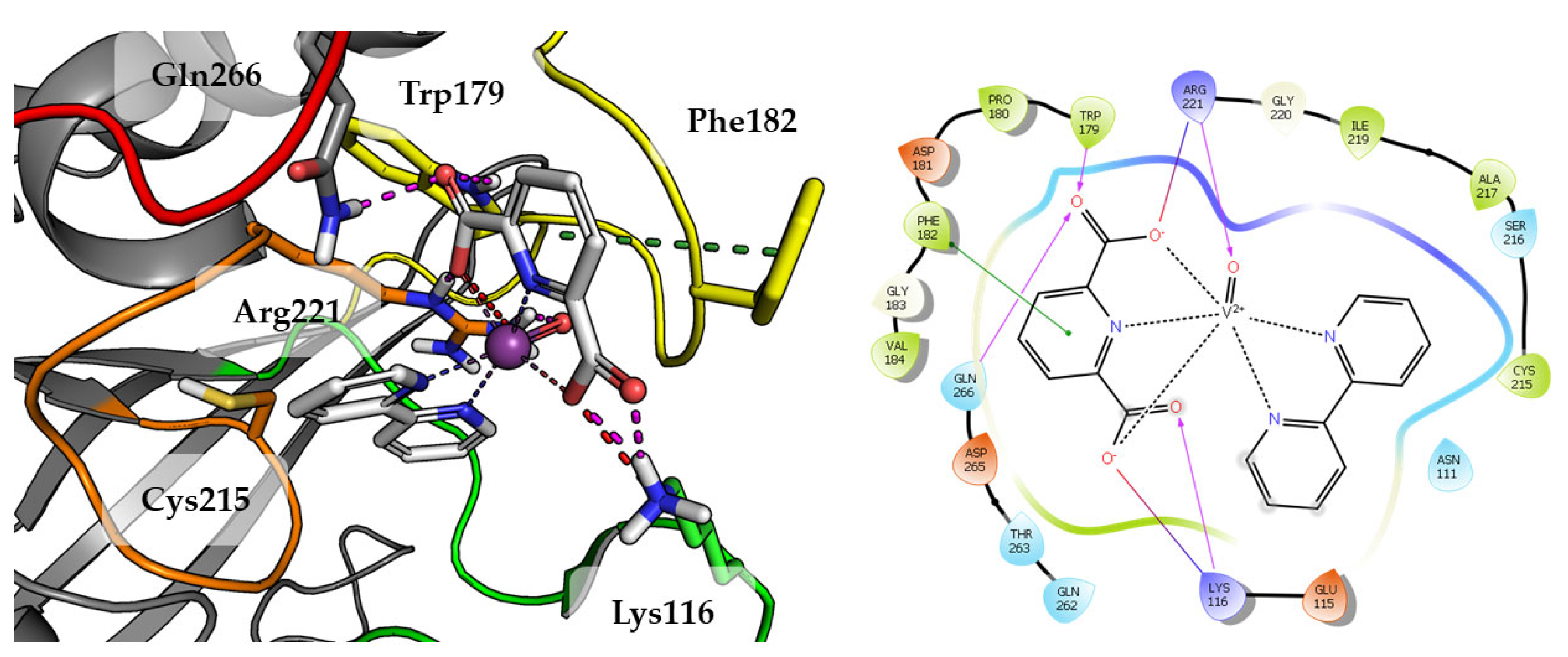



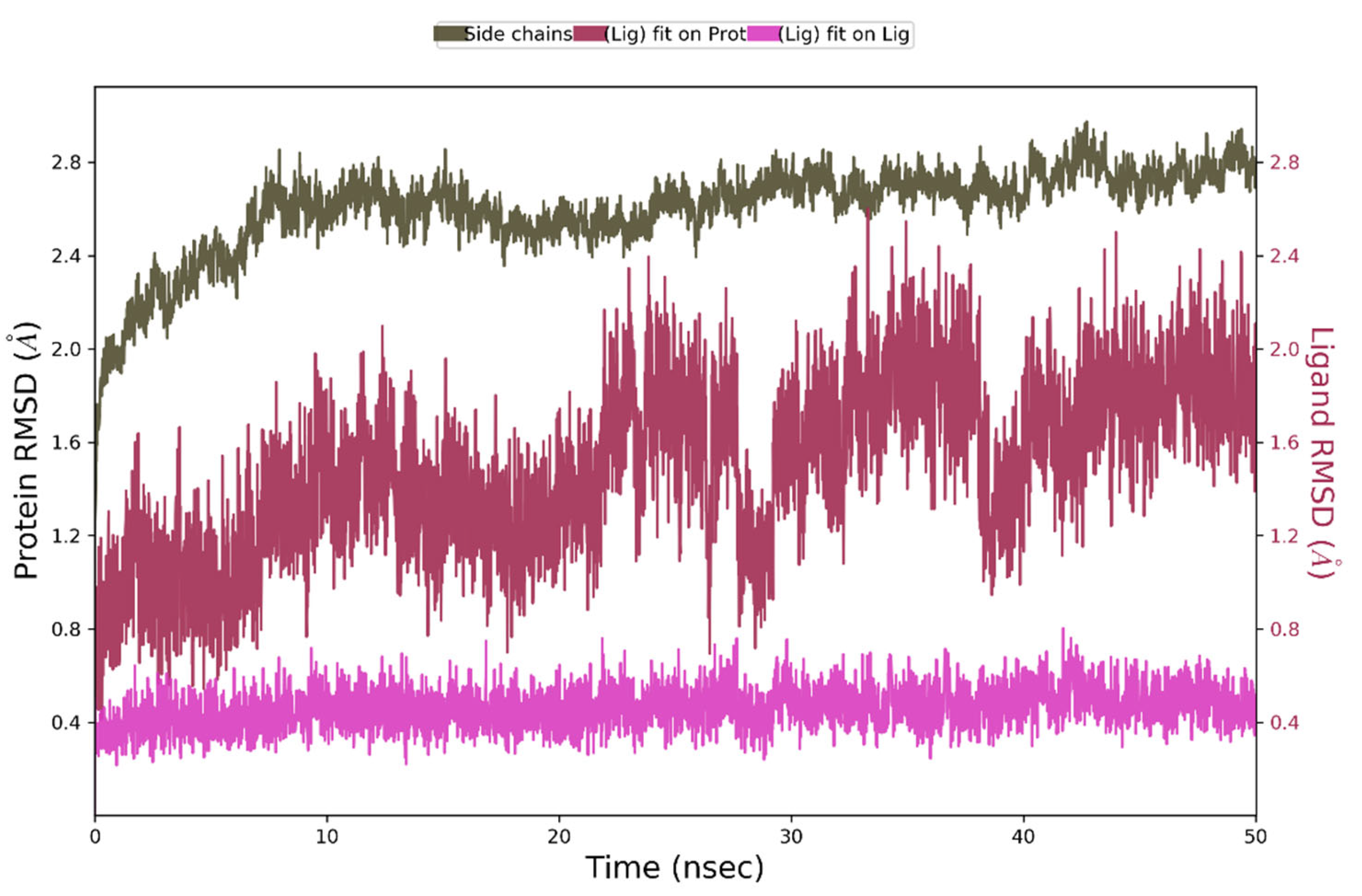
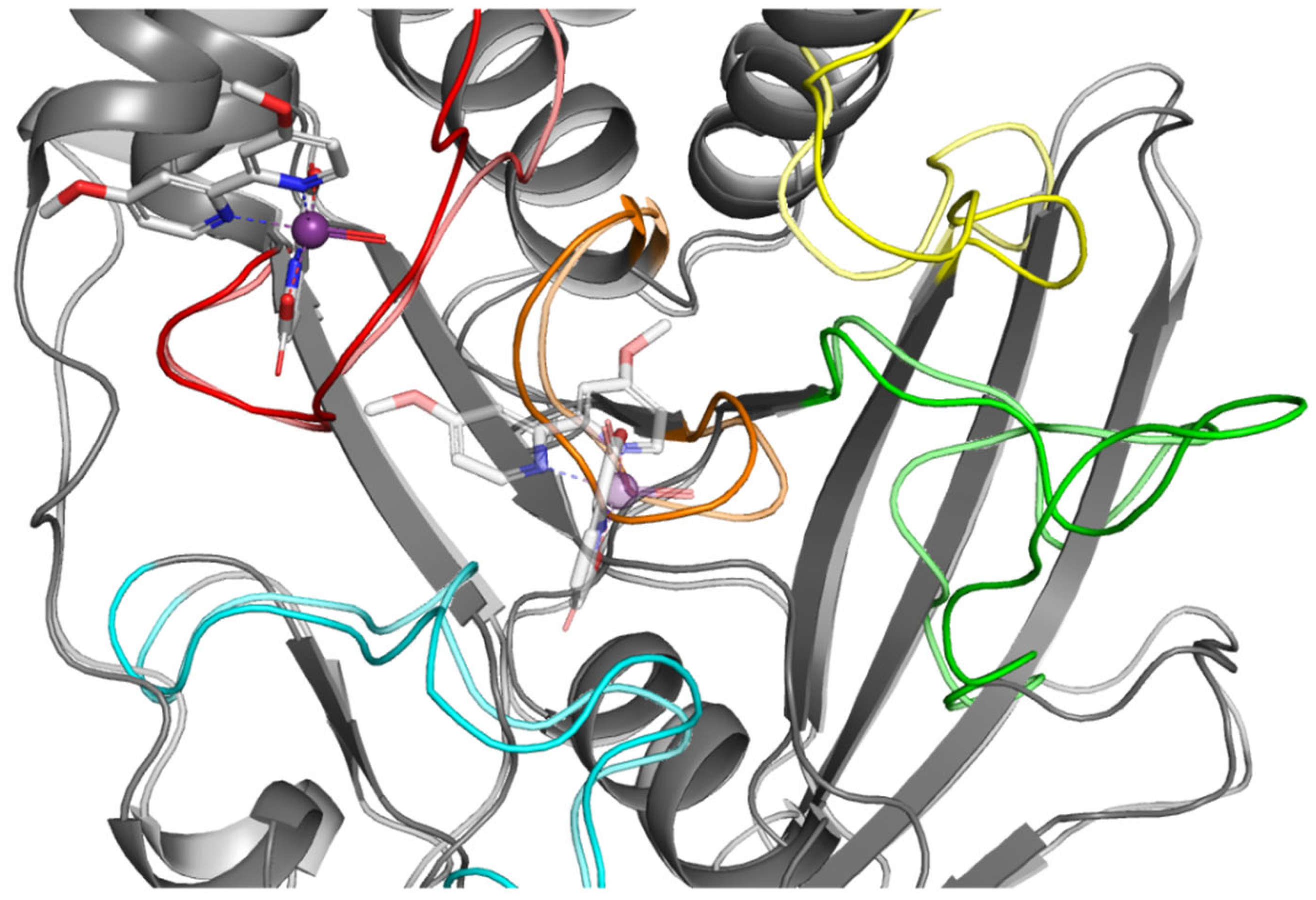
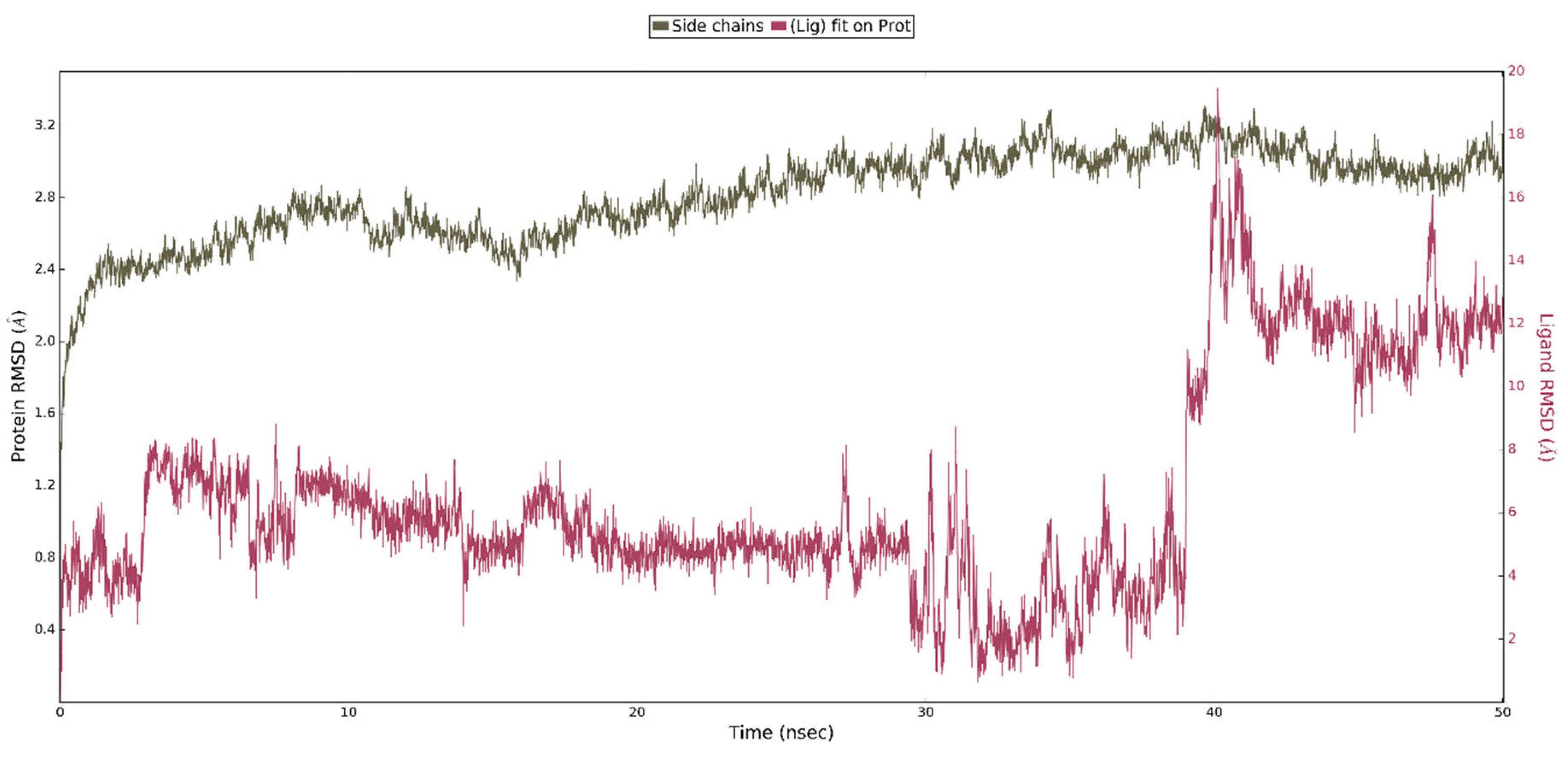


| Compounds | IC50 ± SD [nM] | Ki ± SD [nM] |
|---|---|---|
| 1 | 627.5 ± 39.1 | 256.3 ± 15.3 |
| 2 | 426.3 ± 37.1 | 173.4 ± 17.2 |
| 3 | 542.8 ± 48.4 | 221.4 ± 19.1 |
| 4 | 185.4 ± 9.8 | 74.2 ± 4.1 |
| 5 | 167.2 ± 8.0 | 66.7 ± 3.3 |
| Sodium orthovanadate | 204.1 ± 25.15 | 96.8 ± 15.6 |
| Open | ||||||||||||
| 1ONZ | 3EAX | |||||||||||
| GS | ΔG[kcal/mol] * | Ki[µM] * | RMSD | GS | ΔG[kcal/mol] * | Ki[µM] * | RMSD | |||||
| 1 | 40.80 | −5.03 | 147.91 | 0.1 | 38.86 | −5.81 | 48.98 | 0.16 | ||||
| 2 | 51.52 | −7.65 | 1.23 | 0.12 | 48.42 | −7.08 | 4.57 | 5.19 | ||||
| 3 | 48.04 | −6.68 | 7.94 | 0.62 | 46.65 | −6.57 | 9.55 | 5.48 | ||||
| 4 | 60.94 | −8.36 | 0.28 | 0.09 | 45.56 | −7.74 | 1.17 | 6.93 | ||||
| Closed | ||||||||||||
| 1Q6J | 2FJN | 5KAD | ||||||||||
| GS | ΔG[kcal/mol] * | Ki[µM] * | RMSD | GS | ΔG[kcal/mol] * | Ki[µM] * | RMSD | GS | ΔG[kcal/mol] * | Ki[µM] * | RMSD | |
| 1 | 34.84 | −4.66 | 263.03 | 0.21 | 33.94 | −5.08 | 173.78 | 13.29 | 32.60 | −5.77 | 54.95 | 9.35 |
| 2 | 18.17 | −5.17 | 151.36 | 16.64 | 30.26 | −5.95 | 18.62 | 13.08 | 37.61 | −6.49 | 8.51 | 4.24 |
| 3 | 24.38 | −5.42 | 56.23 | 14.98 | 32.05 | −5.55 | 60.26 | 17.17 | 32.11 | −5.49 | 53.70 | 6.98 |
| 4 | 37.55 | −7.28 | 1.95 | 5.01 | 41.39 | −6.95 | 4.37 | 7.66 | 43.66 | −7.08 | 2.57 | 6.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostrzewa, T.; Jończyk, J.; Drzeżdżon, J.; Jacewicz, D.; Górska-Ponikowska, M.; Kołaczkowski, M.; Kuban-Jankowska, A. Synthesis, In Vitro, and Computational Studies of PTP1B Phosphatase Inhibitors Based on Oxovanadium(IV) and Dioxovanadium(V) Complexes. Int. J. Mol. Sci. 2022, 23, 7034. https://doi.org/10.3390/ijms23137034
Kostrzewa T, Jończyk J, Drzeżdżon J, Jacewicz D, Górska-Ponikowska M, Kołaczkowski M, Kuban-Jankowska A. Synthesis, In Vitro, and Computational Studies of PTP1B Phosphatase Inhibitors Based on Oxovanadium(IV) and Dioxovanadium(V) Complexes. International Journal of Molecular Sciences. 2022; 23(13):7034. https://doi.org/10.3390/ijms23137034
Chicago/Turabian StyleKostrzewa, Tomasz, Jakub Jończyk, Joanna Drzeżdżon, Dagmara Jacewicz, Magdalena Górska-Ponikowska, Marcin Kołaczkowski, and Alicja Kuban-Jankowska. 2022. "Synthesis, In Vitro, and Computational Studies of PTP1B Phosphatase Inhibitors Based on Oxovanadium(IV) and Dioxovanadium(V) Complexes" International Journal of Molecular Sciences 23, no. 13: 7034. https://doi.org/10.3390/ijms23137034
APA StyleKostrzewa, T., Jończyk, J., Drzeżdżon, J., Jacewicz, D., Górska-Ponikowska, M., Kołaczkowski, M., & Kuban-Jankowska, A. (2022). Synthesis, In Vitro, and Computational Studies of PTP1B Phosphatase Inhibitors Based on Oxovanadium(IV) and Dioxovanadium(V) Complexes. International Journal of Molecular Sciences, 23(13), 7034. https://doi.org/10.3390/ijms23137034







