3D Chromatin Organization Involving MEIS1 Factor in the cis-Regulatory Landscape of GJB2
Abstract
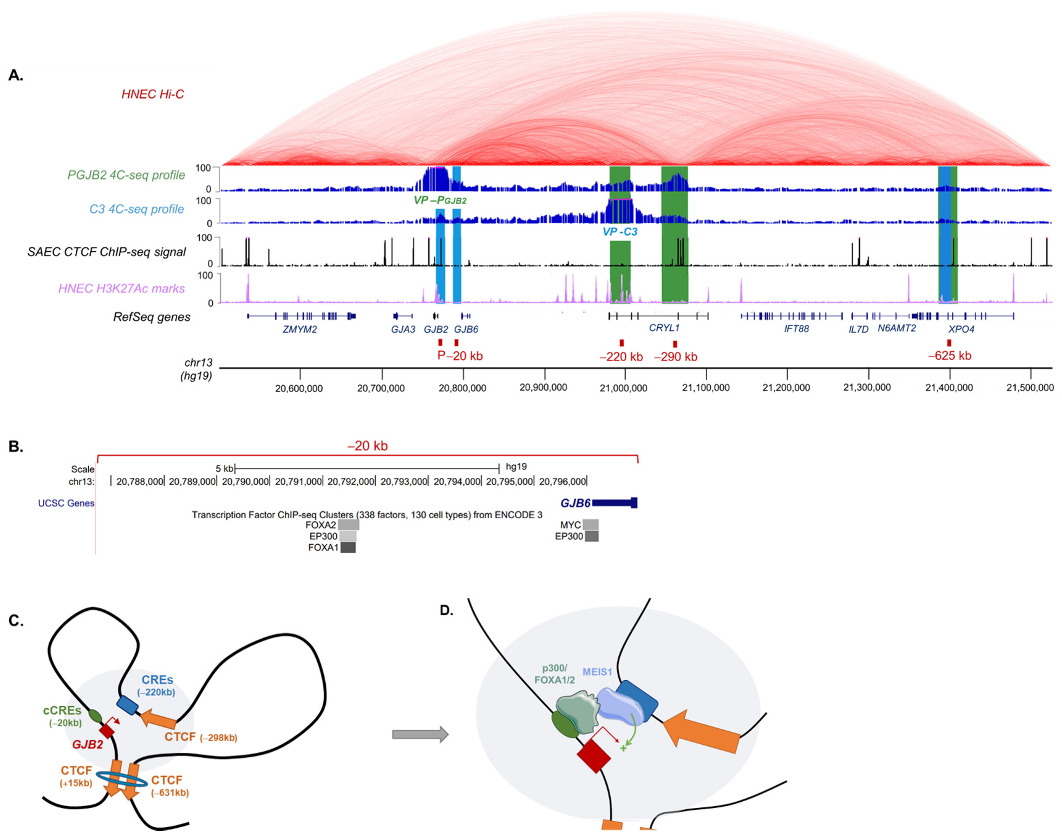
:1. Introduction
2. Results
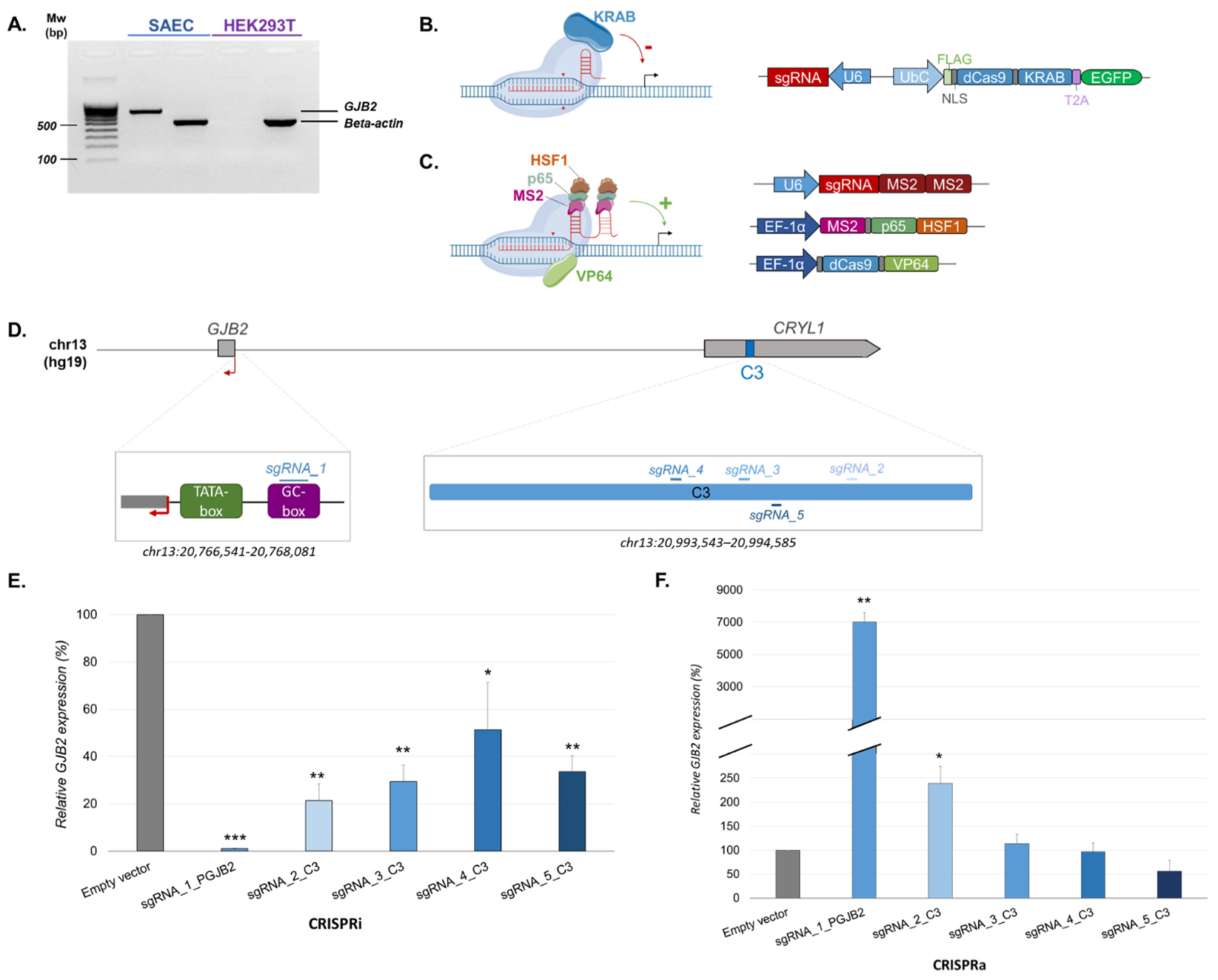
2.1. CRISPR Analyses Confirm GJB2 Biological Enhancers
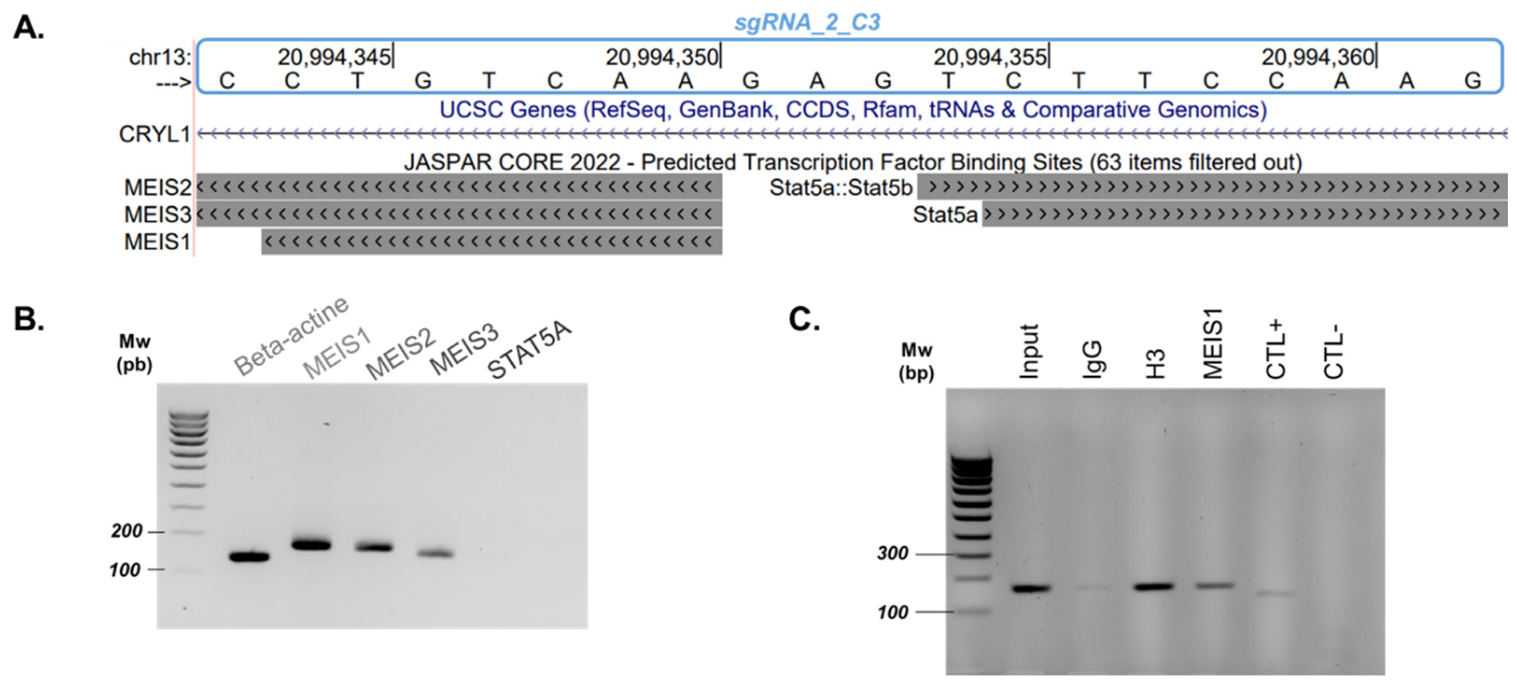
2.2. MEIS1 Transcription Factor Contributes to GJB2 Expression
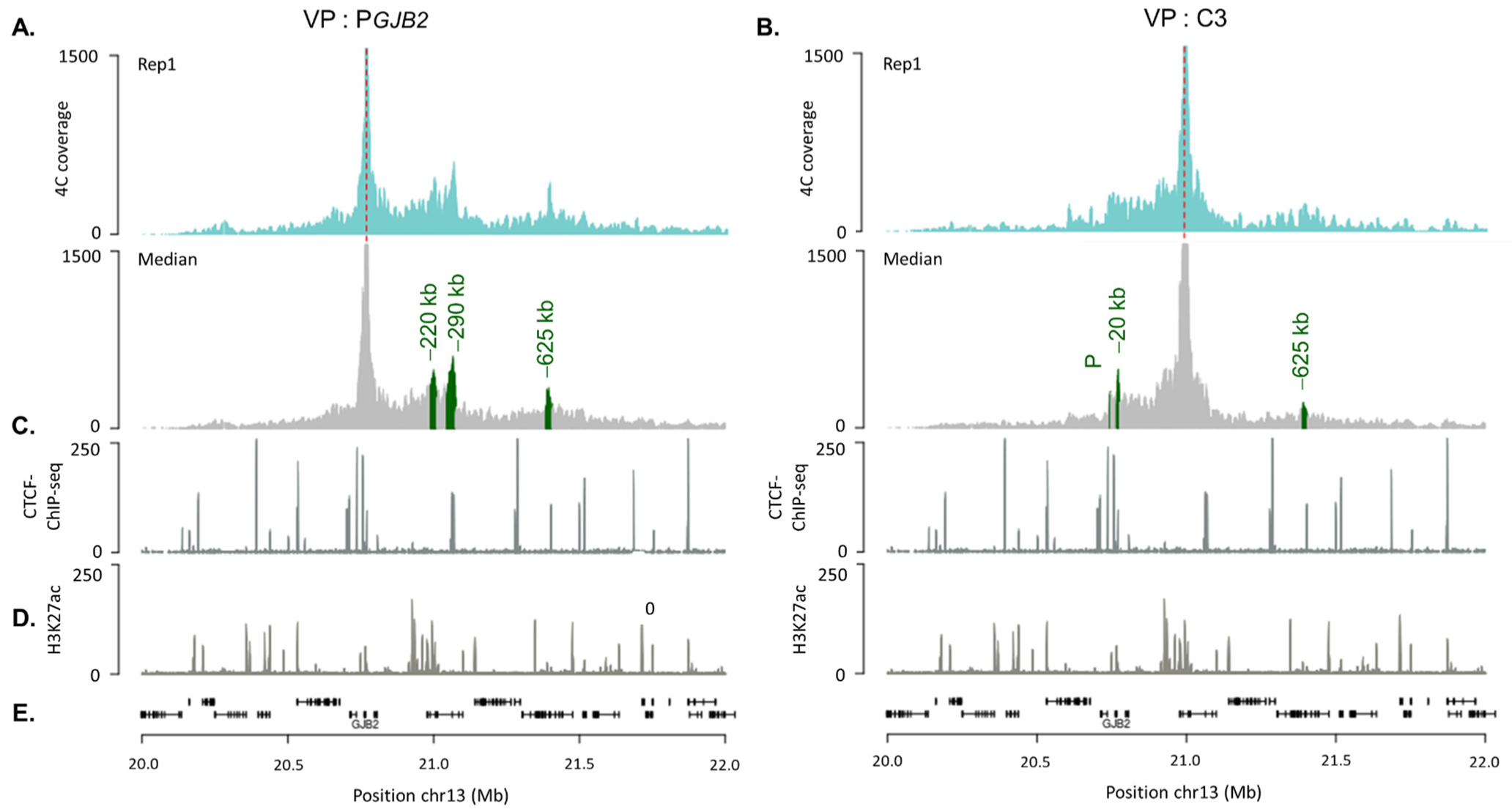
2.3. DFNB1 Chromatin Organization
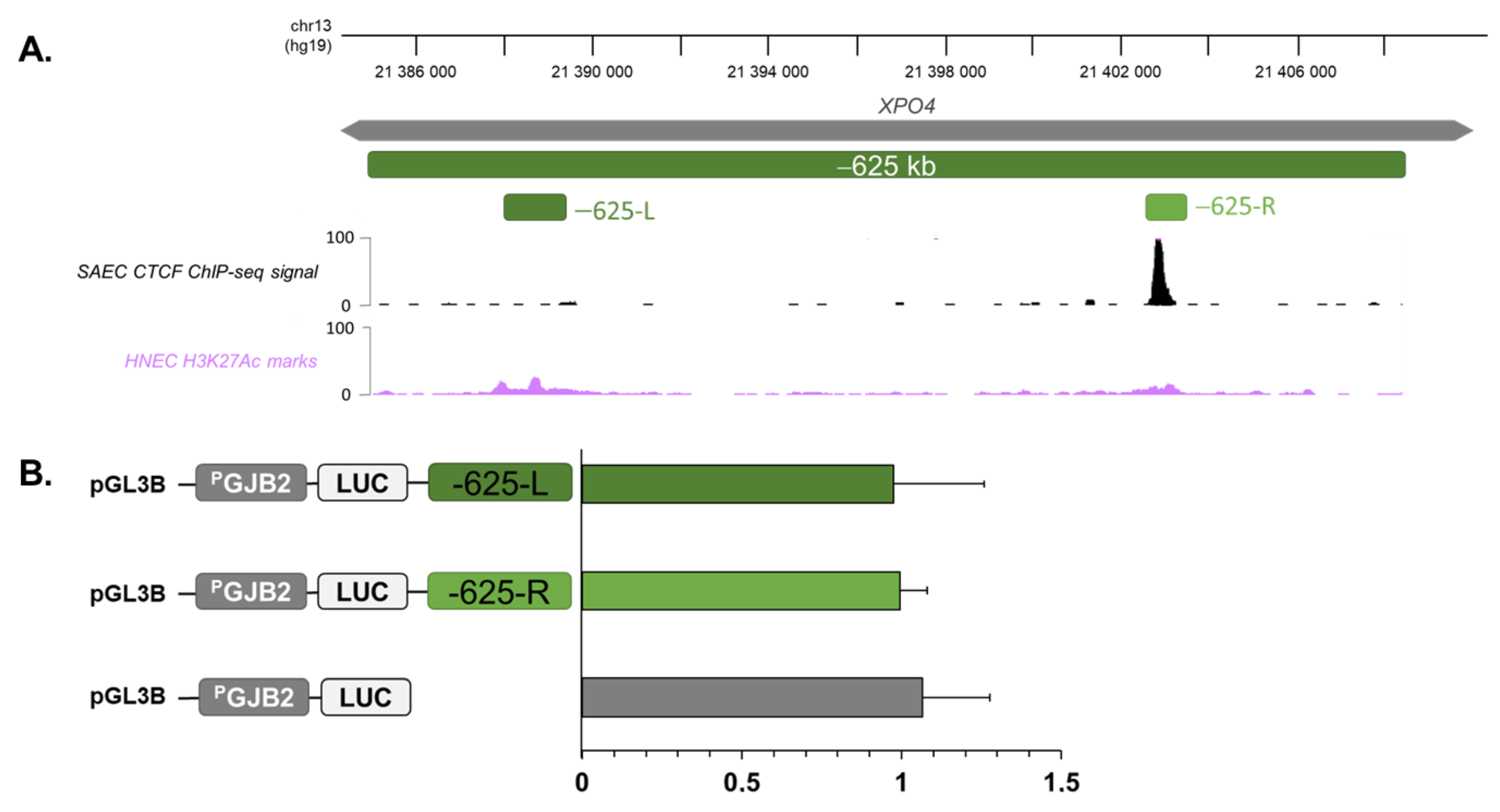
2.4. Region at −625 kb of GJB2 Promoter Corresponds to an Insulator
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Circular Chromosome Conformation Capture (4C)
4.3. C-Seq Analysis
4.4. CRISPR Interference/Activation
4.4.1. Design and Cloning of gRNA
4.4.2. Lentivirale Production for CRISPRi
4.4.3. Plasmid Transfection for CRISPRa
4.4.4. RT-qPCR
4.5. Chromatin Immunoprecipitation
4.6. Plasmid Constructs
4.7. Luciferase Assays
4.8. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The ENCODE Project Consortium; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Shilatifard, A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014, 21, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Elkon, R.; Agami, R. Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 2017, 35, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Rickels, R.; Shilatifard, A. Enhancer Logic and Mechanics in Development and Disease. Trends Cell Biol. 2018, 28, 608–630. [Google Scholar] [CrossRef]
- Claringbould, A.; Zaugg, J.B. Enhancers in disease: Molecular basis and emerging treatment strategies. Trends Mol. Med. 2021, 27, 1060–1073. [Google Scholar] [CrossRef]
- Gasperini, M.; Tome, J.M.; Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020, 21, 292–310. [Google Scholar] [CrossRef]
- Robson, M.; Ringel, A.R.; Mundlos, S. Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol. Cell 2019, 74, 1110–1122. [Google Scholar] [CrossRef]
- Tanaka-Ouyang, L.; Marlin, S.; Nevoux, J. Les surdités d’origine génétique. Presse Med. 2017, 46, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, D.; Denoyelle, F.; Chauvin, P.; Garabédian, E.-N.; Couderc, R.; Odent, S.; Joannard, A.; Schmerber, S.; Delobel, B.; Leman, J.; et al. Large deletion of the GJB6 gene in deaf patients heterozygous for the GJB2 gene mutation: Genotypic and phenotypic analysis. Am. J. Med. Genet. Part A 2004, 127, 263–267. [Google Scholar] [CrossRef]
- Kiang, D.T.; Jin, N.; Tu, Z.-J.; Lin, H.H. Upstream genomic sequence of the human connexin26 gene. Gene 1997, 199, 165–171. [Google Scholar] [CrossRef]
- Meda, P. Connexines, canaux jonctionnels et communications cellulaires. Médecine/Science 1996, 12, 909–920. [Google Scholar] [CrossRef]
- Sánchez, H.A.; Verselis, V.K. Aberrant Cx26 hemichannels and keratitis-ichthyosis-deafness syndrome: Insights into syndromic hearing loss. Front. Cell. Neurosci. 2014, 8, 354. [Google Scholar] [CrossRef] [Green Version]
- Jagger, D.J.; Forge, A. Connexins and gap junctions in the inner ear--it’s not just about K+ recycling. Cell Tissue Res. 2015, 360, 633–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carson, J.L.; Reed, W.; Moats-Staats, B.M.; Brighton, L.E.; Gambling, T.M.; Hu, S.-C.; Collier, A.M. Connexin 26 expression in human and ferret airways and lung during development. Am. J. Respir. Cell Mol. Biol. 1998, 18, 111–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moisan, S.; Le Nabec, A.; Quillévéré, A.; Le Maréchal, C.; Férec, C. Characterization of GJB2 cis-regulatory elements in the DFNB1 locus. Hum. Genet. 2019, 138, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, D.; Le Maréchal, C.; Jonard, L.; Thierry, P.; Czajka, C.; Couderc, R.; Ferec, C.; Denoyelle, F.; Marlin, S.; Fellmann, F. A new large deletion in the DFNB1 locus causes nonsyndromic hearing loss. Eur. J. Med. Genet. 2009, 52, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Bliznetz, E.A.; Makienko, O.N.; Okuneva, E.G.; Markova, T.G.; Polyakov, A.V. New recurrent large deletion, encompassing both GJB2 and GB6 genes, results in isolated sensorineural hearing impairment with autosomal recessive mode of inheritance. Russ. J. Genet. 2014, 50, 415–420. [Google Scholar] [CrossRef]
- Lerer, I.; Sagi, M.; Ben-Neriah, Z.; Wang, T.; Levi, H.; Abeliovich, D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum. Mutat. 2001, 18, 460. [Google Scholar] [CrossRef]
- del Castillo, I.; Villamar, M.; Moreno-Pelayo, M.A.; del Castillo, F.J.; Álvarez, A.; Tellería, D.; Menéndez, I.; Moreno, F. A Deletion Involving the Connexin 30 Gene in Nonsyndromic Hearing Impairment. New Engl. J. Med. 2002, 346, 243–249. [Google Scholar] [CrossRef]
- Pallares-Ruiz, N.; Blanchet, P.; Mondain, M.; Claustres, M.; Roux, A.-F. A large deletion including most of GJB6 in recessive non syndromic deafness: A digenic effect? Eur. J. Hum. Genet. 2002, 10, 72–76. [Google Scholar] [CrossRef] [Green Version]
- del Castillo, F.J.; Rodriguez-Ballesteros, M.; Alvarez, A.; Hutchin, T.; Leonardi, E.; de Oliveira, C.A.; Azaiez, H.; Brownstein, Z.; Avenarius, M.R.; Marlin, S.; et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005, 42, 588–594. [Google Scholar] [CrossRef] [Green Version]
- Wilch, E.; Azaiez, H.; Fisher, R.A.; Elfenbein, J.; Murgia, A.; Birkenhäger, R.; Bolz, H.; Da Silva-Costa, S.M.; Del Castillo, I.; Haaf, T.; et al. A novel DFNB1 deletion allele supports the existence of a distant cis-regulatory region that controls GJB2 and GJB6 expression. Clin. Genet. 2010, 78, 267–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tayoun, A.N.A.; Mason-Suares, H.; Frisella, A.L.; Bowser, M.; Duffy, E.; Mahanta, L.; Funke, B.; Rehm, H.L.; Amr, S.S. Targeted Droplet-Digital PCR as a Tool for Novel Deletion Discovery at the DFNB1 Locus. Hum. Mutat. 2015, 37, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Nishio, S.-Y.; Yokota, Y.; Moteki, H.; Kumakawa, K.; Usami, S.-I. Diagnostic pitfalls for GJB2 -related hearing loss: A novel deletion detected by Array-CGH analysis in a Japanese patient with congenital profound hearing loss. Clin. Case Rep. 2018, 6, 2111–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safka Brozkova, D.; Uhrova Meszarosova, A.; Lassuthova, P.; Varga, L.; Staněk, D.; Borecká, S.; Laštůvková, J.; Čejnová, V.; Rašková, D.; Lhota, F.; et al. The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region. Genes 2021, 12, 684. [Google Scholar] [CrossRef] [PubMed]
- Common, J.E.A.; Bitner-Glindzicz, M.; O’Toole, E.A.; Barnes, M.R.; Jenkins, L.; Forge, A.; Kelsell, D.P. Specific loss of connexin 26 expression in ductal sweat gland epithelium associated with the deletion mutation del(GJB6-D13S1830). Clin. Exp. Dermatol. 2005, 30, 688–693. [Google Scholar] [CrossRef]
- Rodriguez-Paris, J.; Schrijver, I. The digenic hypothesis unraveled: The GJB6 del(GJB6-D13S1830) mutation causes allele-specific loss of GJB2 expression in cis. Biochem. Biophys. Res. Commun. 2009, 389, 354–359. [Google Scholar] [CrossRef]
- Rodriguez-Paris, J.; Tamayo, M.L.; Gelvez, N.; Schrijver, I. Allele-Specific Impairment of GJB2 Expression by GJB6 Deletion del(GJB6-D13S1854). PLoS ONE 2011, 6, e21665. [Google Scholar] [CrossRef]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2015, 17, 5–15. [Google Scholar] [CrossRef] [Green Version]
- Konermann, S.; Brigham, M.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.; Habib, N.; Gootenberg, J.; Nishimasu, H.; et al. Genome-Scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2014, 517, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Schulte, D.; Geerts, D. MEIS transcription factors in development and disease. Development 2019, 146, ev174706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.-K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef]
- Moisan, S.; Berlivet, S.; Ka, C.; Le Gac, G.; Férec, C.; Dostie, J. WS06.1 Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. J. Cyst. Fibros. 2016, 15, S9. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, J.; Wu, J.; Jiang, R.; Guo, C.; Tang, Y.; Wang, H.; Kong, S.; Wang, S. HOXA10 co-factor MEIS1 is required for the decidualization in human endometrial stromal cell. J. Mol. Endocrinol. 2020, 64, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, Y.; Cao, H.; Zhang, Y.; Gu, Z.; Liu, X.; Yu, A.; Kaphle, P.; Dickerson, K.E.; Ni, M.; et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Hazan, J.; Bester, A.C. CRISPR-Based Approaches for the High-Throughput Characterization of Long Non-Coding RNAs. Non-Coding RNA 2021, 7, 79. [Google Scholar] [CrossRef]
- Cirillo, L.A.; Lin, F.R.; Cuesta, I.; Friedman, D.; Jarnik, M.; Zaret, K.S. Opening of Compacted Chromatin by Early Developmental Transcription Factors HNF3 (FoxA) and GATA-4. Mol. Cell 2002, 9, 279–289. [Google Scholar] [CrossRef]
- Eeckhoute, J.; Carroll, J.S.; Geistlinger, T.R.; Torres-Arzayus, M.I.; Brown, M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006, 20, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Narita, T.; Ito, S.; Higashijima, Y.; Chu, W.K.; Neumann, K.; Walter, J.; Satpathy, S.; Liebner, T.; Hamilton, W.B.; Maskey, E.; et al. Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol. Cell 2021, 81, 2166–2182.e6. [Google Scholar] [CrossRef]
- Kubo, N.; Ishii, H.; Xiong, X.; Bianco, S.; Meitinger, F.; Hu, R.; Hocker, J.D.; Conte, M.; Gorkin, D.; Yu, M.; et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021, 28, 152–161. [Google Scholar] [CrossRef]
- Krijger, P.H.; Geeven, G.; Bianchi, V.; Hilvering, C.R.; de Laat, W. 4C-seq from beginning to end: A detailed protocol for sample preparation and data analysis. Methods 2020, 170, 17–32. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Nabec, A.; Blotas, C.; Briset, A.; Collobert, M.; Férec, C.; Moisan, S. 3D Chromatin Organization Involving MEIS1 Factor in the cis-Regulatory Landscape of GJB2. Int. J. Mol. Sci. 2022, 23, 6964. https://doi.org/10.3390/ijms23136964
Le Nabec A, Blotas C, Briset A, Collobert M, Férec C, Moisan S. 3D Chromatin Organization Involving MEIS1 Factor in the cis-Regulatory Landscape of GJB2. International Journal of Molecular Sciences. 2022; 23(13):6964. https://doi.org/10.3390/ijms23136964
Chicago/Turabian StyleLe Nabec, Anaïs, Clara Blotas, Alinéor Briset, Mégane Collobert, Claude Férec, and Stéphanie Moisan. 2022. "3D Chromatin Organization Involving MEIS1 Factor in the cis-Regulatory Landscape of GJB2" International Journal of Molecular Sciences 23, no. 13: 6964. https://doi.org/10.3390/ijms23136964
APA StyleLe Nabec, A., Blotas, C., Briset, A., Collobert, M., Férec, C., & Moisan, S. (2022). 3D Chromatin Organization Involving MEIS1 Factor in the cis-Regulatory Landscape of GJB2. International Journal of Molecular Sciences, 23(13), 6964. https://doi.org/10.3390/ijms23136964






