Development of Deep-Learning-Based Single-Molecule Localization Image Analysis
Abstract
:1. Introduction
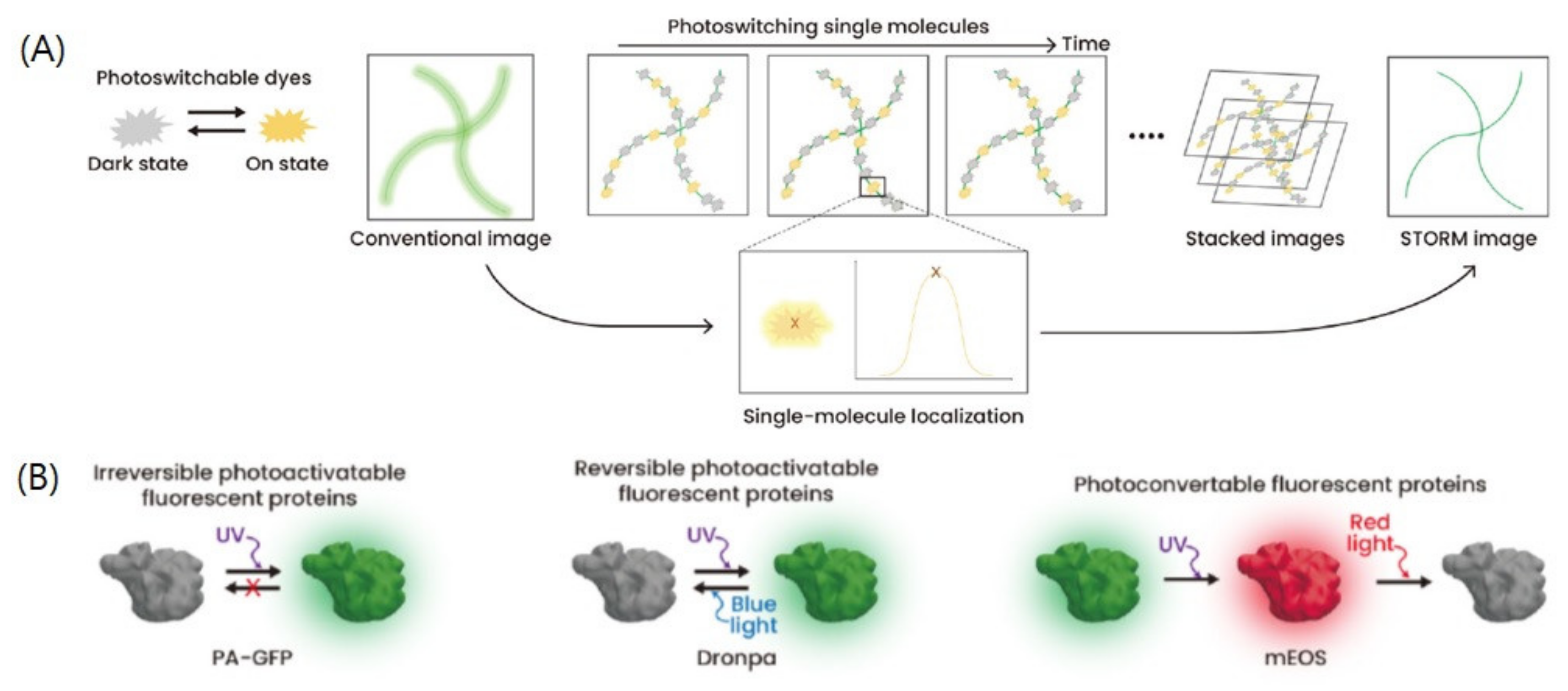
2. SMLM
2.1. Development of SMLM
2.2. Principle and Analysis of SMLM Techniques
2.3. Limitations of SMLM Analysis
3. Deep Learning in Computer Vision
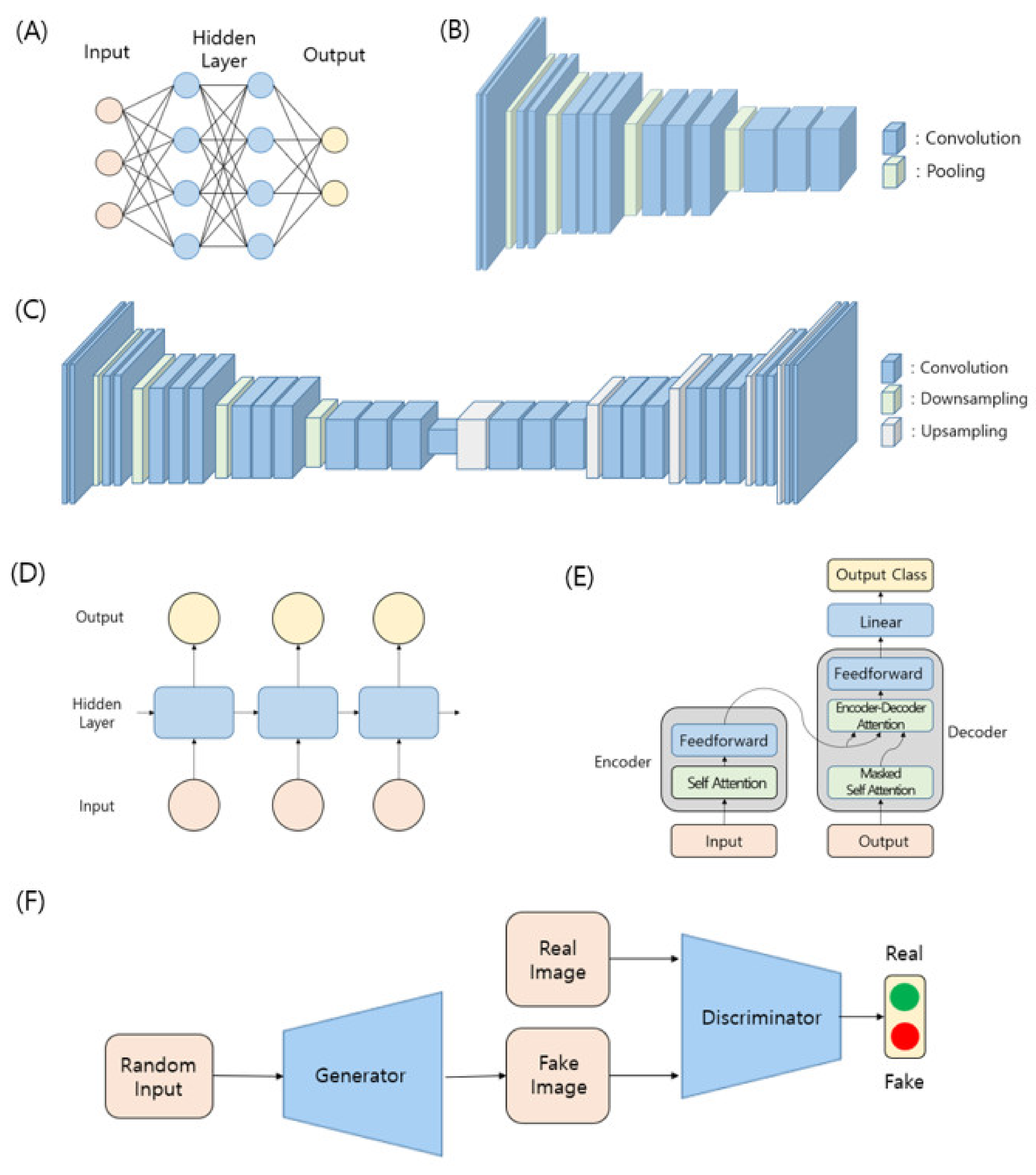
3.1. Machine Learning and Neural Network
3.2. Notable Model Architectures in Deep Learning
3.2.1. Multilayer Perceptron (MLP)
3.2.2. CNN-Based Feature Network
3.2.3. Encoder–Decoder Architecture
3.2.4. Recurrent Neural Network (RNN) and Transformer
3.2.5. Generative Models and Generative Adversarial Networks (GANs)
3.3. Algorithms in Computer Vision
3.3.1. Classification
3.3.2. Object Detection
3.3.3. Semantic Segmentation and Image Reconstruction
3.3.4. Image Generation
4. Application of Deep Learning to SMLM Image Analysis
4.1. Fast Single-Molecule Localization Using Deep Learning
4.2. Constructing High-Density Super-Resolution Image from A Low-Density Image
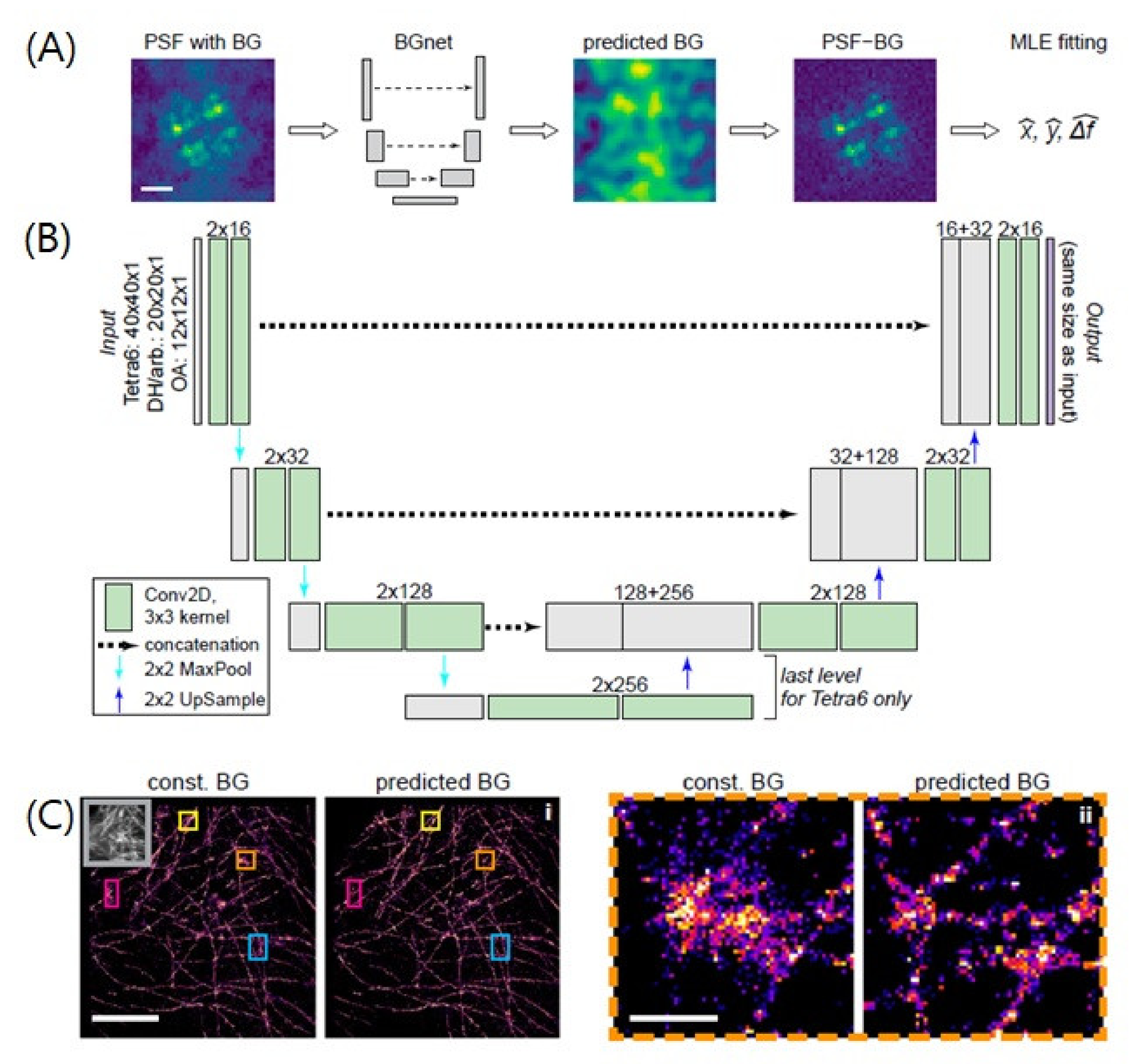
4.3. Improvement of Localization Precision
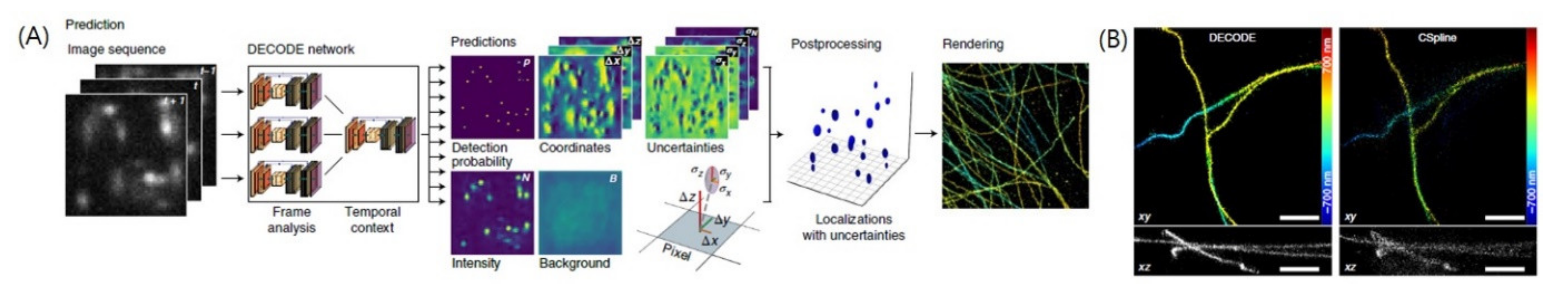
4.4. Localization of Overlapping PSFs
4.5. Extracting Additional Spectral Information from PSF
5. Perspectives on Future Deep-Learning-Based SMLM Analysis
Funding
Conflicts of Interest
Abbreviations
| SRM | super-resolution fluorescence microscopic techniques |
| SMLM | single-molecule localization microscopy |
| PSF | point spread function |
| STORM | stochastic optical reconstruction microscopy |
| PALM | photo-activated localization microscopy |
| FPALM | fluorescence photoactivation localization microscopy |
| STED | stimulated emission depletion |
| RESOLFT | reversible saturable optical fluorescence transition |
| SIM | structured-illumination microscopy |
| SSIM | saturated structured-illumination microscopy |
| EM | electron microscopy |
| ReLU | rectified linear units |
| MLP | multilayer perceptron |
| MNIST | Modified National Institute of Standards and Technology |
| CNN | convolutional neural network |
| VGG | Visual Geometry Group |
| RNN | recurrent neural network |
| LSTM | long short term memory |
| GRU | gated recurrent unit |
| GAN | generative adversarial networks |
| RCNN | region convolutional neural network |
| MLE | maximum likelihood estimation |
| SR | super-resolution |
| SLM | spatial light modulator |
| SR-SMLM | super-resolution single-molecule localization microscopy |
References
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X. Nano-imaging with STORM. Nat. Photonics 2009, 3, 365–367. [Google Scholar] [CrossRef]
- Yildiz, A.; Forkey, J.N.; McKinney, S.A.; Ha, T.; Goldman, Y.E.; Selvin, P.R. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 2003, 300, 2061–2065. [Google Scholar] [CrossRef] [Green Version]
- Thompson, R.E.; Larson, D.R.; Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002, 82, 2775–2783. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.Y.; Pedley, A.M.; Kim, D.; Xia, C.; Zhuang, X.; Benkovic, S.J. Microtubule-directed transport of purine metabolons drives their cytosolic transit to mitochondria. Proc. Natl. Acad. Sci. 2018, 115, 13009–13014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, D.; Kim, D. Super—Resolution fluorescence microscopy—Based single—Molecule spectroscopy. Bull. Korean Chem. Soc. 2022, 43, 316–327. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, D. Recent developments in correlative super-resolution fluorescence microscopy and electron microscopy. Mol. Cells 2022, 45, 41. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, S.T.; Girirajan, T.P.; Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Kim, D.; Mun, J.Y. Direct visualization of actin filaments and actin-binding proteins in neuronal cells. Front. Cell Dev. Biol. 2020, 8, 1368. [Google Scholar] [CrossRef]
- Hell, S.W.; Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Eggeling, C.; Jakobs, S.; Hell, S.W. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17565–17569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsson, M.G.; Agard, D.A.; Sedat, J.W. Sevenfold improvement of axial resolution in 3D wide-field microscopy using two objective-lenses. In Proceedings of the Three-Dimensional Microscopy: Image Acquisition and Processing II, San Jose, CA, USA, 23 March 1995; pp. 147–156. [Google Scholar]
- Möckl, L.; Roy, A.R.; Moerner, W. Deep learning in single-molecule microscopy: Fundamentals, caveats, and recent developments. Biomed. Opt. Express 2020, 11, 1633–1661. [Google Scholar] [CrossRef] [PubMed]
- Möckl, L.; Moerner, W. Super-resolution microscopy with single molecules in biology and beyond–essentials, current trends, and future challenges. J. Am. Chem. Soc. 2020, 142, 17828–17844. [Google Scholar] [CrossRef] [PubMed]
- Tianjie, Y.; Yaoru, L.; Wei, J.; Ge, Y. Advancing biological super-resolution microscopy through deep learning: A brief review. Biophys. Rep. 2021, 7, 253–266. [Google Scholar]
- Speiser, A.; Müller, L.-R.; Hoess, P.; Matti, U.; Obara, C.J.; Legant, W.R.; Kreshuk, A.; Macke, J.H.; Ries, J.; Turaga, S.C. Deep learning enables fast and dense single-molecule localization with high accuracy. Nat. Methods 2021, 18, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Jeong, D.; Kim, G.-H.; Go, S.; Song, J.; Moon, E.; Huh, Y.H.; Kim, D. Super-resolution imaging of platelet-activation process and its quantitative analysis. Sci. Rep. 2021, 11, 10511. [Google Scholar] [CrossRef] [PubMed]
- Go, S.; Jeong, D.; Chung, J.; Kim, G.-H.; Song, J.; Moon, E.; Huh, Y.H.; Kim, D. Super-resolution imaging reveals cytoskeleton-dependent organelle rearrangement within platelets at intermediate stages of maturation. Structure 2021, 29, 810–822.e3. [Google Scholar] [CrossRef]
- Xu, K.; Babcock, H.P.; Zhuang, X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 2012, 9, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wang, W.; Bates, M.; Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 2008, 319, 810–813. [Google Scholar] [CrossRef] [Green Version]
- Pavani, S.R.P.; Thompson, M.A.; Biteen, J.S.; Lord, S.J.; Liu, N.; Twieg, R.J.; Piestun, R.; Moerner, W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 2009, 106, 2995–2999. [Google Scholar] [CrossRef] [Green Version]
- Bates, M.; Huang, B.; Dempsey, G.T.; Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 2007, 317, 1749–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Kenny, S.J.; Hauser, M.; Li, W.; Xu, K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods 2015, 12, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Chung, J.; Park, H.; Kim, D. Single-Molecule Sensing by Grating-based Spectrally Resolved Super-Resolution Microscopy. Bull. Korean Chem. Soc. 2021, 42, 270–278. [Google Scholar] [CrossRef]
- Chung, J.; Jeong, U.; Jeong, D.; Go, S.; Kim, D. Development of a New Approach for Low-Laser-Power Super-Resolution Fluorescence Imaging. Anal. Chem. 2021, 94, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Shim, S.-H.; He, J.; Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 2011, 8, 499–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shroff, H.; Galbraith, C.G.; Galbraith, J.A.; Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 2008, 5, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehme, E.; Weiss, L.E.; Michaeli, T.; Shechtman, Y. Deep-STORM: Super-resolution single-molecule microscopy by deep learning. Optica 2018, 5, 458–464. [Google Scholar] [CrossRef]
- Boyd, N.; Jonas, E.; Babcock, H.; Recht, B. DeepLoco: Fast 3D localization microscopy using neural networks. BioRxiv 2018, 267096, preprint. [Google Scholar]
- Zelger, P.; Kaser, K.; Rossboth, B.; Velas, L.; Schütz, G.; Jesacher, A. Three-dimensional localization microscopy using deep learning. Opt. Express 2018, 26, 33166–33179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jia, S.; Zhang, H.F.; Kim, D.; Babcock, H.; Zhuang, X.; Ying, L. Blind sparse inpainting reveals cytoskeletal filaments with sub-Nyquist localization. Optica 2017, 4, 1277–1284. [Google Scholar] [CrossRef]
- Ouyang, W.; Aristov, A.; Lelek, M.; Hao, X.; Zimmer, C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 2018, 36, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Gaire, S.K.; Zhang, Y.; Li, H.; Yu, R.; Zhang, H.F.; Ying, L. Accelerating multicolor spectroscopic single-molecule localization microscopy using deep learning. Biomed. Opt. Express 2020, 11, 2705–2721. [Google Scholar] [CrossRef]
- Möckl, L.; Roy, A.R.; Petrov, P.N.; Moerner, W. Accurate and rapid background estimation in single-molecule localization microscopy using the deep neural network BGnet. Proc. Natl. Acad. Sci. USA 2020, 117, 60–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.; Moon, S.; Xu, K. Information-rich localization microscopy through machine learning. Nat. Commun. 2019, 10, 1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Liu, S.; Chaurasia, A.; Ma, D.; Mlodzianoski, M.J.; Culurciello, E.; Huang, F. Analyzing complex single-molecule emission patterns with deep learning. Nat. Methods 2018, 15, 913–916. [Google Scholar] [CrossRef]
- Hershko, E.; Weiss, L.E.; Michaeli, T.; Shechtman, Y. Multicolor localization microscopy and point-spread-function engineering by deep learning. Opt. Express 2019, 27, 6158–6183. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, Y.; Ying, L.; Sun, C.; Zhang, H.F. Machine-learning based spectral classification for spectroscopic single-molecule localization microscopy. Opt. Lett. 2019, 44, 5864–5867. [Google Scholar] [CrossRef]
- Nehme, E.; Freedman, D.; Gordon, R.; Ferdman, B.; Weiss, L.E.; Alalouf, O.; Naor, T.; Orange, R.; Michaeli, T.; Shechtman, Y. DeepSTORM3D: Dense 3D localization microscopy and PSF design by deep learning. Nat. Methods 2020, 17, 734–740. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the NIPS 2012, Lake Tahoe, NV, USA, 3–8 December 2012. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June –1 July 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In International Conference on Machine Learning; PMLR: New York, NY, USA, 2019; pp. 6105–6114. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the NIPS 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the NIPS 2014, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Noh, H.; Hong, S.; Han, B. Learning deconvolution network for semantic segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1520–1528. [Google Scholar]
- Newell, A.; Yang, K.; Deng, J. Stacked hourglass networks for human pose estimation. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 483–499. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Van den Oord, A.; Kalchbrenner, N.; Espeholt, L.; Vinyals, O.; Graves, A. Conditional image generation with pixelcnn decoders. In Proceedings of the NIPS 2016, Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- van den Oord, A.; Kalchbrenner, N. Pixel Recurrent Neural Networks. Proceeding of the International Conference on Machine Learning, New York City, NY, USA, 19–24 June 2016; pp. 1747–1756. [Google Scholar]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, online, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, online, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L. Swin Transformer V2: Scaling Up Capacity and Resolution. arXiv 2021, arXiv:2111.09883. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Uijlings, J.R.; Van De Sande, K.E.; Gevers, T.; Smeulders, A.W. Selective search for object recognition. Int. J. Comput. Vis. 2013, 104, 154–171. [Google Scholar] [CrossRef] [Green Version]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Boston, MA, USA, 7–12 June 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the NIPS 2015, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Tian, Z.; Shen, C.; Chen, H.; He, T. Fcos: Fully convolutional one-stage object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 9627–9636. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 213–229. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive growing of gans for improved quality, stability, and variation. arXiv 2017, arXiv:1710.10196, preprint. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 4401–4410. [Google Scholar]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]



| Type | Name | Architecture | Algorithm | Input | Output | Training Data | Reference |
|---|---|---|---|---|---|---|---|
| Acceleration of single-molecule localization | DeepSTORM | Decoder-Encoder | Image Reconstruction | Camera Images with multiple PSFs | SR image in 2D | -Simulated images of emitters and microtubules -Experimental images of microtubules | [31] |
| smNET | ResNet-like CNN | Regression | Individual Image of PSFs | 3D coordinates of PSFs, orientation, etc. | -Simulated images of emitters -Experimental images of mitochondria and bead | [39] | |
| -- | CNN | Regression | Individual Image of PSFs | 3D coordinates of PSFs | -Simulated and experimental images of beads | [33] | |
| DeepLOCO | CNN + FC with Residual Connection | Regression | Camera Images with PSFs | 3D coordinates of PSFs | -Simulated and contest data | [32] | |
| Constructing high-density super-resolution image | ANNA-PALM | U-Net, GAN | Image Generation | Widefield image, Image Sequences with multiple PSFs | Super-resolved 2D image | -Simulated image of microtubules -Experimental images of microtubules, nuclear pores and mitochondria. | [35] |
| -- | CNN | Image Reconstruction | Image of individual PSFs | Super-resolved 2D image | -Experimental images of microtubules, mitochondria, and peroxisome. | [36] | |
| Improvement of localization precision | BGnet | U-Net | Image Reconstruction | Images of Individual PSFs | Background and intensity of PSFs | -Experimental images of microtubules | [37] |
| Localization of overlapping PSFs | DECODE | U-Nets | Image Reconstruction | Image Sequences with multiple PSFs | 3D coordinates of PSFs, Intensity, Background, Uncertainty | -Contest data -Experimental images of microtubules, golgi, and nuclear pore complex. | [19] |
| Extracting additional spectral information from PSF | -- | FC | Classification, Regression | Individual Image of PSFs | Axial Position, Color | -Simulated images of emitters -Experimental images of beads, microtubules and mitochondria. | [38] |
| -- | CNN, Encoder–Decoder | (1) Classification, (2) Image Reconstruction | (1) Image of individual PSFs (2) Camera Images with multiple PSFs | (1) Color channel (2) Color channel, 2D coordinates of PSFs | -Simulated images of emitters -Experimental images of Qdots, microtubules, and mitochondria. | [40] | |
| -- | FC | Classification | Full Spectra | Color Channel | -Experimental images of microtubules and mitochondria. | [41] | |
| DeepSTORM3D | CNN with skipped connection | Image Reconstruction | (1) Simulated point sources (2) Camera Images with multiple PSFs | (1) PSFs (2) 3D coordinates of PSFs | -Simulated images of emitters -Experimental images of telomeres | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyun, Y.; Kim, D. Development of Deep-Learning-Based Single-Molecule Localization Image Analysis. Int. J. Mol. Sci. 2022, 23, 6896. https://doi.org/10.3390/ijms23136896
Hyun Y, Kim D. Development of Deep-Learning-Based Single-Molecule Localization Image Analysis. International Journal of Molecular Sciences. 2022; 23(13):6896. https://doi.org/10.3390/ijms23136896
Chicago/Turabian StyleHyun, Yoonsuk, and Doory Kim. 2022. "Development of Deep-Learning-Based Single-Molecule Localization Image Analysis" International Journal of Molecular Sciences 23, no. 13: 6896. https://doi.org/10.3390/ijms23136896
APA StyleHyun, Y., & Kim, D. (2022). Development of Deep-Learning-Based Single-Molecule Localization Image Analysis. International Journal of Molecular Sciences, 23(13), 6896. https://doi.org/10.3390/ijms23136896






