Abstract
Altered protein phosphorylation is a major pathologic modification in tauopathies and Alzheimer’s disease (AD) linked to abnormal tau fibrillar deposits in neurofibrillary tangles (NFTs) and pre-tangles and β-amyloid deposits in AD. hTau transgenic mice, which express 3R and less 4R human tau with no mutations in a murine knock-out background, show increased tau deposition in neurons but not NFTs and pre-tangles at the age of nine months. Label-free (phospho)proteomics and SWATH-MS identified 2065 proteins in hTau and wild-type (WT) mice. Only six proteins showed increased levels in hTau; no proteins were down-regulated. Increased tau phosphorylation in hTau was detected at Ser199, Ser202, Ser214, Ser396, Ser400, Thr403, Ser404, Ser413, Ser416, Ser422, Ser491, and Ser494, in addition to Thr181, Thr231, Ser396/Ser404, but not at Ser202/Thr205. In addition, 4578 phosphopeptides (corresponding to 1622 phosphoproteins) were identified in hTau and WT mice; 64 proteins were differentially phosphorylated in hTau. Sixty proteins were grouped into components of membranes, membrane signaling, synapses, vesicles, cytoskeleton, DNA/RNA/protein metabolism, ubiquitin/proteasome system, cholesterol and lipid metabolism, and cell signaling. These results showed that over-expression of human tau without pre-tangle and NFT formation preferentially triggers an imbalance in the phosphorylation profile of specific proteins involved in the cytoskeletal–membrane-signaling axis.
1. Introduction
Protein phosphorylation is one of the most common and essential reversible post-translational modifications. A phosphorylated amino acid can bind molecules able to interact with other proteins, and activate or inhibit protein function [1]. Phosphorylation also affects protein conformation, protein–protein interactions, and protein signaling [2]. Protein phosphorylation usually occurs in several phosphorylation sites, thus enabling refined molecular functions [3,4]. The balance between protein phosphorylation and dephosphorylation is modulated by the dynamic combined action of specific kinases and phosphatases [5,6,7]. Conversely, abnormal or dysregulated protein phosphorylation may lead to abnormal cellular function involving a variety of vital molecular pathways [8].
Widespread dysregulated protein phosphorylation, in addition to tau hyper-phosphorylation, via extensive kinase activation occurs in Alzheimer’s disease (AD), thus contributing to abnormal function of cell membranes, cytoskeleton, synapses, neurotransmitter receptors, energy metabolism, RNA processing and splicing, protein synthesis, and cell signaling [9,10,11,12,13]. Aberrant protein phosphorylation is already detected at the first stages (stages I–III of Braak) of neurofibrillary tangle (NFT) pathology in regions such as the frontal cortex devoid of NFTs and β-amyloid deposits at these stages [12].
Abnormal protein phosphorylation also occurs in transgenic mouse models of AD that develop β-amyloid deposits in brain [14,15,16,17]. Dysregulated protein phosphorylation in these mice occurs at first stages of the disease and particularly affects synaptic proteins [18,19]. These models are very useful to learn about the core of altered phosphoproteins linked to β-amyloid pathology, but they do not permit the identification of altered proteins due to abnormal tau in AD.
Primary tauopathies are characterized by abundant 3R, 4R or 3R + 4R tau filamentous tau inclusions in neurons and glial cells not associated with extracellular amyloids [20,21]. Some of them are linked to mutations in MAPT, the tau gene, but the majority are sporadic. Widespread abnormal protein phosphorylation has been described in a few human tauopathies including aging-related tau astrogliopathy (ARTAG), a common old-age 4Rtauopathy involving astrocytes [22], and globular glial tauopathy (GGT) a rare 4Rtauopathy with characteristic inclusions in neurons, astrocytes, and oligodendrocytes [23].
Several animal models are generated to recapitulate hyper-phosphorylation of tau and the formation of NFTs as key aspects of tauopathies [24,25,26]. P301S transgenic mice express the T34 isoform of microtubule-associated protein tau bearing the human P301S mutation [27]. A recent phosphoproteomics study has been carried out on these mice [28]. The majority of dysregulated phosphoproteins, most of them hyper-phosphorylated, were proteins of the cytoskeleton, membranes, synapses, membrane trafficking, vesicles linked to endo- and exocytosis, cytoplasmic vesicles, and kinases. In contrast, proteins linked to DNA, RNA metabolism, RNA splicing, and protein synthesis were hypo-phosphorylated. Other pathways modulating energy metabolism, cell signaling, Golgi apparatus, carbohydrates, and lipids were also targets of abnormal protein phosphorylation in P301S mice [28].
In addition to tauopathies, a recent classification of diseases with accumulation of tau protein includes a miscellaneous group of various conditions with tau immunoreactivity or tau pathology, but without filamentous tau inclusions [29]. Tau immunoreactivities or tau pathologies can represent, in some cases, a preclinical stage and/or an early cytopathological phase of fibril formation [30].
An interesting model of increased human tau expression and accumulation in the absence of MAPT mutations, in a knock-out background for murine tau protein, is hTau [31,32]. hTau transgenic mice express high levels of human 3Rtau and low levels of 4Rtau. hTau mice can also be considered as a model of the human tau expression in the mouse lacking murine tau. Whether these mice can be used as a model of “tau pathology” in humans is open to discussion. However, abnormal deposits in subpopulations of neurons in the cerebral cortex and hippocampus are identified with the antibodies PHF1 and MC1, but tau deposits are not stained with AT8, tau-PSer422, and Tau C-3 antibodies [31,32,33]. Moreover, no abnormal fibrils are observed in mice aged 9 months [31]. Thus, hTau fits with the characteristics of the miscellaneous group in the recent classification of tauopathies and tau pathology/altered tau immunoreactivity [29]. We chose this model to learn about protein modifications, particularly protein phosphorylation linked to tau pathology in the absence of tauopathy.
For this purpose, we used immunohistochemistry and western blotting, label-free (phospho)proteomics, and SWATH-MS to identify differential protein phosphorylation in hTau compared with wild-type mice.
2. Results
2.1. Immunohistochemical Characterization of hTau Mice
The cerebral cortex of WT mice showed weak 4Rtau immunoreactivity in the neuropil (Figure 1A), and negative 3Rtau staining (Figure 1C); the neuropil and a few neurons showed weak positive PHF1 immunostaining (Figure 1E), but they were negative with MC1 antibodies (Figure 1G). The antibody Tau 100 weakly stained the nuclei of neurons (Figure 1I). In contrast, the cerebral cortex of hTau mice showed weak 4Rtau immunoreactivy in subpopulations of neurons, and moderate 3Rtau immunostaining in the totality of cortical neurons (Figure 1B,D). PHF1 antibodies showed moderate and diffuse immunoreactivity mainly in the neuropil (Figure 1F), and the antibody MC1 immunostained the cytoplasm of a subpopulation of neurons (Figure 1H). The Tau 100 antibody showed more marked nuclear staining in hTau than in WT mice (Figure 1J). No cytoplasmic deposits were detected with the AT8 antibody; truncated tau C-3 immmunostaining was negative. Cytoplasmic deposits were homogeneous without the appearance of pre-tangles or neurofibrillary tangles. Threads were absent. Ubiquitin deposits were not found. Finally, immunostaining with the antibody tau-N Tyr29 was negative in WT and hTau mice. No differences were seen regarding GFAP-positive astrocytes and Iba1-immunoreactive microglia between WT and hTau.
Gallyas staining was negative.
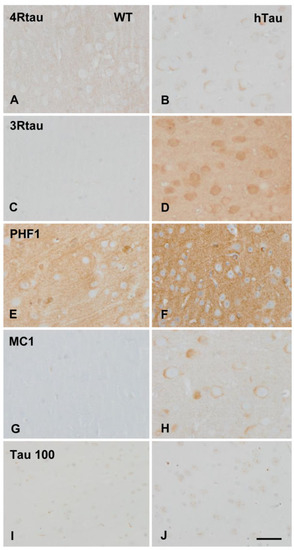
Figure 1.
Immunohistochemistry of the cerebral cortex in wild-type (WT) and hTau transgenic mice aged nine months. Weak 4Rtau immunoreacitvity decorates the neuropil in WT (A) but weak cytoplasmic neuronal deposits in hTau (B). 3Rtau immunostaining is almost absent in WT (C) but 3Rtau immunoreactivity is stronger in the neuropil and neuronal cytoplasm in hTau (D). PHF1 immunoreactivity is moderate, and MC1 staining is negative in WT (E,G respectively). In contrast, PHF1 immunostaining is stronger in hTau (F), and MC1 antibodies stain the cytoplasm of neurons in hTau (H). The antibody Tau 100 weakly stains the nuclei of neurons in WT (I), whereas more marked nuclear Tau 100 immunostaining is seen in hTau (J). Paraffin sections, lightly counterstained with hematoxylin, bar = 40 µm.
2.2. Tau Protein Expression Profile in hTau Brains
Gel electrophoresis and western blotting of total brain homogenates of 3WT and 4hTau mice were assessed in parallel. In WT mice, 3Rtau immunoreactivity was negative. Two bands of about 64 kDa and 60–50 kDa and a weak band of slightly higher molecular weight were recognized in hTau mice. WT mice showed a weak 4Rtau-immunoreactive band of about 60–50 kDa. Two bands of 68 kDa and 64 kDa were identified in hTau mice. The phospho-tau antibody tau-P Thr181 was negative in WT, but two bands of 68 kDa and 64 kDa were detected in hTau transgenic mice. The phospho-tau antibody tau-P Thr231 recognized a band of 60–50 kDa and a smear of higher molecular weight in WT, and three strong bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau mice. PHF1 antibodies exposed a unique band of about 64 kDa in WT mice but three bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau. Finally, MC1 antibodies revealed no bands in WT mice, and several bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau (Figure 2). Densitometric assessment further revealed significant differences in tau levels between WT and hTau mice (Figure 2).
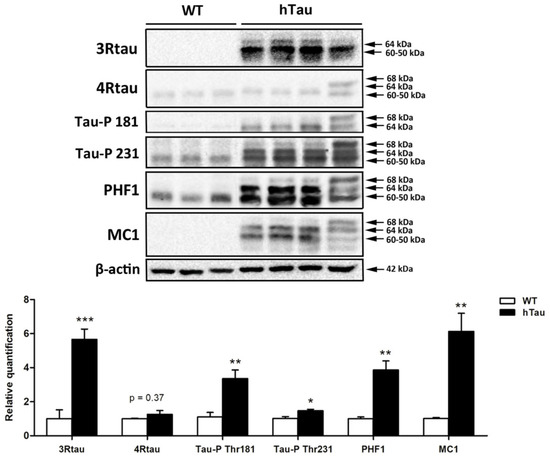
Figure 2.
Gel electrophoresis and western blotting of total brain homogenates of 3WT and 4hTau mice processed in parallel. In WT mice, 3Rtau is negative, but two bands of about 64 kDa and 60–50 kDa and a weak band of slightly higher molecular weight is identified in hTau. A weak 4Rtau-immunoreactive band of about 60–50 kDa is identified in WT, and two bands of 68 kDa and 64 kDa in hTau mice. The phospho-tau antibody tau-P Thr181 (Tau-P 181) is negative in WT, but two bands of 68 kDa and 64 kDa are detected in hTau. The phospho-tau antibody tau-P Thr231 (Tau-P 231) recognizes a band of 60–50 kDa and a smear of higher molecular weight in WT, but three strong bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau. PHF1 antibodies detect a unique band of about 64kDa in WT, and three bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau. Finally, MC1 antibodies reveal no bands in WT mice, and several bands of 68 kDa, 64 kDa, and 60–50 kDa in hTau. The slightly different band pattern in hTau mice on the right reflects a higher amount of 4Rtau than in the other hTau mice. Densitometric analysis of western blots show significant differences between WT and hTau regarding the expression levels of 3Rtau, Tau-P Thr181, Tau-P Thr231, PHF1, and MC1. Unpaired student’s t-test; significance level set at * p < 0.05 ** p < 0.01, *** p < 0.001. 4Rtau are not significantly increased in hTau (p = 0.37) (modified from ref. [33]).
2.3. hTau Expression Induces Phosphoproteomic Changes at Membrane and Synaptic Levels
A total of 2065 proteins were detected in hTau and WT mice (Supplementary Table S1). In addition to protein tau, only five proteins showed increased levels in hTau compared with WT; no proteins were down-regulated in hTau mice. Four proteins were associated with the cytoskeleton: tubulin beta 2B class IIb (Tubb2b), an structural protein of the cytoskeleton and main component of the tubulin complex; transgelin 2 (Tagln2), localized in the cytoskeleton, vesicles, and endosomes which functions as a cadherin-binding protein; glyoxalase domain-containing 4 (Glod4), a cadherin-binding protein localized in mitochondria and extracellular exosomes; and NCK interacting protein with SH3 domain (Nckipsd), a cytoskeletal binding protein implicated in the regulation of actin polymerization and cell adhesion. The fifth up-regulated protein was acetyl-Coenzyme A acyltransferase 1A (Acaa1a), a protein localized in peroxisomes that participates in fatty acid metabolic pathways and unsaturated fatty acid biosynthesis.
Mass-spectrometry revealed increased tau phosphorylation at Ser199, Ser202, Ser214, Ser396, Ser400, Thr403, Ser404, Ser413, Ser416, Ser422, Ser491, and Ser494 in hTau compared with WT mice (Supplementary Table S1).
In addition, 4578 phosphopeptides (corresponding to 1622 phosphoproteins) were identified in hTau and WT mice (Supplementary Table S1). Seventy phosphosites were identified corresponding to 64 differentially phosphorylated proteins in hTau. Sixty proteins were grouped into components of membranes, membrane signaling, synapses, vesicles, cytoskeleton, DNA/RNA/protein metabolism, ubiquitin/proteasome system, cholesterol and lipid metabolism, and cell signaling. The remaining proteins (n = 4) cannot be included in one of these groups or their function in the nervous system was not known.
The largest set of differentially phosphorylated proteins (n = 37) were linked to membranes, membrane signaling, synapses, vesicles, and cytoskeleton (Table 1). Twenty-four were hyper-phosphorylated and twelve hypo-phosphorylated.

Table 1.
Dysregulated phosphorylation of proteins involved in membranes, membrane signaling, synapses, vesicles, and cytoskeleton in hTau compared with WT mice. ↑: up-regulation; ↓: down-regulation.
Seven proteins were linked to DNA, RNA, or protein synthesis; four were hyper-phosphorylated, and three hypo-phosphorylated. Six proteins were associated with the UPS; three were hyper- and three hypo-phosphorylated. Four proteins were involved in cholesterol transport and lipid metabolism; two were hypo- and two hyper-phosphorylated. Six proteins had roles in different aspects of cell signaling, five of them hyper-phosphorylated and one hypo-phosphorylated (Table 2).

Table 2.
Dysregulated phosphorylation of proteins linked to DNA, RNA, and protein metabolism; ubiquitin/proteasome system, cholesterol and lipid metabolism, and cell signaling in hTau compared with WT mice.
2.4. Interactomes
A functional interactome was constructed based on the differential phosphoprotein dataset including tau protein as external input. As shown in Figure 3, 42 out of 64 differential phosphoproteins (65%) constituted a highly related functional interactome where most of nodes were involved in cytoskeleton organization (FDR: 0.00032), synapse (FDR: 6.55 × 10−5), neuron projection (FDR: 0.0337), and neurogenesis (FDR: 0.00089). Interestingly, 12 nodes (Tubb4b, Camk2a, Syp, Sptbn2, Synj1, Pcbp1, Gsk3a, Map1a, Clasp2, Dpysl2, Dpysl3, Tuba1a) are considered functional interactors of Tau according to the STRING database.
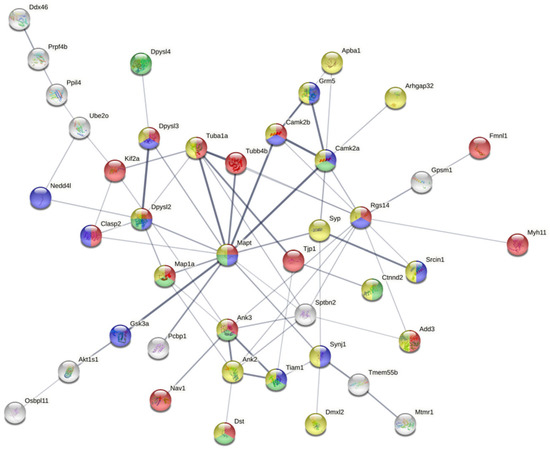
Figure 3.
Functional interactome constituted by differential phosphoproteins detected in hTau transgenic compared with WT mice aged nine months; 42 out of 64 differential phosphoproteins (65%) constitute a highly-related functional interactome where most of nodes are involved in cytoskeleton organization, synapse, neuron projection and neurogenesis. Moreover, 12 nodes are considered functional interactors of Tau according to STRING database.
3. Discussion
Increased levels of 3Rtau together with higher tau phosphorylation are the main differences between WT and hTau transgenic mice. Western blotting revealed tau hyperphosphorylation at residues Thr181, Thr231, Ser396/Ser404, and mass spectrometry at residues Ser199, Ser202, Ser214, Ser396, Ser400, Thr403, Ser404, Ser413, Ser416, Ser422, Ser491, and Ser494. However, tau was not phosphorylated at Ser202/Thr205 evinced by the lack of staining with the AT8 antibody; this is not a false negative result as tau inclusions in human and mouse tauopathies show strong AT8 immunoreactivity. Tau in hTau mice had an abnormal conformation, as shown with the antibody MC1 in immunohistochemistry and western blotting. L-ack of immunostaining with the tau C-3 antibody revealed no tau truncation at aspartic acid 421. Gallyas staining was negative. Tau truncation at Asp421 is an early change preceding neurofibrillary tangle formation [34]. Tau ubiquitinitation, which is absent in hTau, is a late tau post-translational event in NFT formation [35]. hTau mice did not contain tau nitrated at Tyr29 which is in contrast with the presence of Tau-N Tyr29 in NFTs, dystrophic neurites, and neuropil threads in AD, and NFTs in corticobasal degeneration and progressive supranuclear palsy [36], and P301S transgenic mice [28].
Tau alterations in hTau here observed were similar to those reported in animals aged 9 months in the original descriptions of hTau although less pronounced with PHF1 and MC1 antibodies [31]. Paired helical filaments, as illustrated in hTau mice aged 13 months [31] were not assessed in our study. Considering morphological and molecular characteristics, hTau transgenic mice largely differ from AD, and whichever of the described human sporadic and familial tauopathies [20,21,29,37,38,39]. Moreover, transgenic mice carrying human tau mutations, as the P301S construct, also differ from hTau mice [27,28,40,41]. Therefore, the present model at the age of nine months is suitable for the study of altered protein expression and dysregulated protein phosphorylation in murine tau knockout mice expressing human 3Rtau and less 4Rtau, and accompanied by tau phosphorylation at selected sites, in the absence of NFTs and pre-tangles. A limitation of the current scenario lies on the lack of control over the modifications due to the knockout of murine tau on which hTau is generated. Curiously, we identified increased levels of only five proteins, in addition to tau, four of them being cytoskeletal proteins, and one a peroxisomal protein that participates in fatty acid metabolism and unsaturated fatty acid biosynthesis.
Regarding the study of phosphorylated proteins, we used a similar equipment and methodological approach as the employed to analyze (phospho)proteomics in AD [12], and primary tauopathies ARTAG [22], GGT [23], and P301S transgenic mice [28].
We identified 230 dysregulated phosphoproteins in the entorhinal cortex and 82 in the frontal cortex at different stages of AD [12].
In ARTAG, 109 proteins were hyper-phosphorylated and 31 hypo-phosphorylated compared with controls. In addition to GFAP that was phosphorylated at positions 8, 14, 82, and 424, many neuronal and glial proteins were differentially phosphorylated in ARTAG. These included proteins of the cytoskeleton, kinases, proteins linked to calcium/calmodulin signalling, cAMP signaling and DNA repair, nuclear and nucleolar regulators, proteins linked to tight junctions, proteins linked to proteolysis, and synaptic proteins. [22]. The study revealed that altered protein phosphorylation in ARTAG was not restricted to astrocytes.
In GGT, phosphoproteome profiling revealed 74 dysregulated proteins in the frontal cortex and 15 in the white matter. Cytoskeleton, axon guidance, exocytosis, chaperone-mediated protein folding, and myelination were part of the significantly over-represented dysregulated biological processes in the frontal cortex. Interestingly, dysregulated specific protein clusters were related to microtubule polymerization, and pre- and post-synaptic transmission [23].
In a parallel study, we identified 328 phosphoproteins differentially regulated in the telencephalon of P301S transgenic mice at the age of 9 months when compared with age-matched WT mice; 179 hyper-phosphorylated, 134 hypo-phosphorylated, and 16 both hyper- and hypo-phosphorylated at different phosphorylation sites [28]. These numbers are quite different from those found in hTau when compared with WT mice in the present study. Seventy proteins were differentially phosphorylated in hTau. Of 60 proteins with known functions in the nervous system, proteins showed an increment in specific phosphosites whereas 19 proteins evidenced a reduction in phosphoprotein levels. Although less marked than in AD, and tauopathies, altered tau expression in hTau mice alters phosphorylation of determined proteins linked mainly with the cytoskeleton, membranes, membrane signaling, synapses, and vesicles; kinases; DNA/RNA/protein synthesis, UPS; and cell signaling.
These targets match with the recently validated principal tau interacting partners (n = 153), among total 245 reported tau interacting partners [42], which include proteins involved in phosphorylation and dephosphorylation (n = 65), cytoskeleton and intracellular transport (n = 24), ubiquitin system and chaperones (n = 27), signal transduction (n = 11), cleavage and truncation (n = 11), and DNA/RNA/protein synthesis (n = 4), among others [42].
In addition, we identified four dysregulated phosphoproteins involved in cholesterol transport and lipid metabolism in hTau. This observation is relevant because there is a close functional relationship between cytoskeletal proteins and cell membranes through protein–protein interactions, electrostatic interactions with lipid membranes, and lipid tails. Lipid rafts are microdomains of cell membranes which favor multiple cell signaling interactions at the cell membrane [43]. The exoplasmic leaflet is enriched with glycosphingolipids and sphingomyelin, the cytoplasmic leaflet is enriched with glycerolipids; cholesterol is present in both. The limited number of dysregulated phosphoproteins in hTau transgenic mice aged 9 months is in contrast with the large number of identified proteins forming the phosphorylated tau interactome in the human AD brain [44]. Of 542 proteins identified in microdissected NFTs, 75 interacted with PHF-1-immunoreactive phosphorylated tau, some of them previously associated with total tau [45] but not yet linked to phosphorylated tau [44]. Although we did not perform co-immunoprecipitation studies, it can be assumed that differences between AD and hTau are largely dependent on the limited recruitment of abnormal proteins in hTau neuronal deposits when compared with the abundance of abnormal phosphorylated and non-phosphorylated proteins in NFTs in AD. Moreover, we cannot rule out the possibility that dysregulated phosphoproteins are not trapped by tau deposits in our model. Finally, other abnormally phosphorylated proteins do not apparently interact with tau in AD, human tauopathies, and murine tau models [12,22,23,28].
4. Material and Methods
4.1. Animals
The experiments were carried out on 9-month-old C57BL/6 WT mice and heterozygous mice expressing human isoforms of tau protein (hTau; B6.Cg- (GFP)Klt Tg(MAPT)8cPdav/J) in a C57BL/6 background (Jackson laboratories; reference: RRID:IMSR_JAX:005491; Bar Harbor, ME, USA) [31,32]. This mouse model can only be hemizygous to the transgene as homozygous animals are not viable. The number of mice was eight per group with equal numbers of males and females per group. Transgenic mice were identified by genotyping genomic DNA isolated from tail clips using the polymerase chain reaction conditions indicated by Jackson laboratories. All animal procedures were carried out following the guidelines of the European Communities Council Directive 2010/63/EU and with the approval of the local ethical committee (C.E.E.A: Comitè Ètic d’Experimentació Animal; University of Barcelona, Spain; reference number: 426/18). Animals were killed by cervical dislocation and their brains were then rapidly removed and processed for study. The left cerebral hemisphere was dissected on ice, immediately frozen, and stored at −80 °C until use for biochemical studies. The rest of the brain was fixed in 4% paraformaldehyde, cut in coronal sections, and embedded in paraffin. De-waxed sections were stained with hematoxylin and eosin, Gallyas staining, or processed for immunohistochemistry.
4.2. Immunohistochemistry
De-waxed sections, 4 microns thick, were processed for immunohistochemistry as detailed elsewhere [23]. The sections were incubated at 4 °C overnight with one of the primary antibodies listed in Table 3. Control of immunostaining included omission of the primary antibody; no signal was obtained following incubation with only the secondary antibody.

Table 3.
Antibodies used for immunohistochemistry and western blotting.
4.3. Gel Electrophoresis and Western Blotting
Frozen samples of the posterior part of the left hemisphere from 3 WT and 4 hTau mice were homogenized in RIPA lysis buffer and phosphatase inhibitor cocktail (Roche Molecular Systems, San Jose, CA, USA). The homogenates were centrifuged for 20 min at 12,000 rpm. Equal amounts of protein (12 μg) for each sample were loaded and separated by electrophoresis on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto nitrocellulose membranes (Amersham, Freiburg, Germany). Incubation in 3% albumin in PBS was used to block non-specific bindings. After washing, membranes were incubated overnight at 4 °C with antibodies detailed in Table 3. β-actin (1:30,000, Sigma, Barcelona, Spain) was used as a control of protein loading. Membranes were incubated with HRP-conjugated secondary antibodies (1:3000, Dako, Barcelona, Spain); the immunoreaction was visualized with a chemiluminescence reagent (ECL, Amersham, UK). Results were analyzed with SPSS 19.0 (SPSS Inc., USA; https://www.ibm.com) software and GraphPad PRISM (GraphPad Software; https://www.graphpad.com) software. Data were presented as mean ± standard error of the mean (SEM). The unpaired student’s t-test was used to compare groups. Significance level was set at * p < 0.05, ** p < 0.01, *** p < 0.001.
4.4. Phosphoproteomics
Anterior left hemisphere from WT and hTau mice were subjected to phosphoproteome analysis as previously described [12]. The (phospho)proteomes and proteomes were independently analyzed by conventional label-free phosphoproteomics [46] and SWATH-MS (sequential window acquisition of all theoretical fragment ion spectra mass spectrometry) [47], respectively. The High-Select™ TiO2 Phosphopeptide Enrichment Kit (Thermo Scientific, Barcelona, Spain) was used to obtain the phosphorylated peptide fractions, according to the manufacturer’s instructions. Protein extraction, in-solution digestion, peptide purification, and reconstitution prior to mass spectrometric analysis were performed as previously reported [12]. Library generation, nanochromatography, mass-spectrometry settings and database searching parameters were as previously described [12]. MS/MS spectra associated to phosphoproteomic data were processed using MaxQuant (1.6.7.0). Database searching was performed using the Uniprot reference for Mus musculus (ID: UP000000_10090, 29 July 2021). FDR filter less than 1% was applied at protein, peptide, and PTM level. In the case of SWATH proteome data, MS/MS data acquisition was performed using AnalystTF 1.7 (Sciex, Vaughan, ON, Canada), and spectra files were processed through ProteinPilot v5.0 search engine (Sciex) using the ParagonTM Algorithm (v.4.0.0.0) (https://www.paragon-sofware.com) [48]. A non-lineal fitting method [49] was used to determine the FDR, reporting results with a 1% global FDR or better. Quantitative data were analyzed using the Perseus software (https://maxquant.net) (1.6.14.0) for statistical analysis and data visualization. MS data and search result files are found in Supplementary Table S1.
4.5. Bioinformatics
Metascape was used to characterize the significantly dysregulated metabolic pathways based on proteomic profiles [50]. Network analysis was performed using STRING (Search Tool for the Retrieval of Interacting Genes) software (http://stringdb.org/, accessed on 1 April 2022) [51]. All edges were supported by at least one reference from the literature or from canonical information stored in the STRING database. False positives as well as false negatives were minimized using only interactions tagged as “medium confidence” (>0.4) in STRING.
5. Conclusions
To sum up, this study showed that over-expression of human tau without pre-tangle and NFT formation in hTau transgenic mice preferentially triggers an imbalance in the phosphorylation profile of specific protein sets involved in the cytoskeletal–membrane-signaling axis rather than a generalized proteome variation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23126427/s1.
Author Contributions
I.F., P.A.-B. and E.S. designed the study; P.A.-B. prepared the colonies and carried out the western blots; J.F.-I., K.A. and E.S. performed the proteomic analysis; I.F., P.A.-B., J.A.d.R., K.A., P.C.-C., M.L.-M., J.F.-I. and E.S. analyzed and interpreted the data; I.F. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The project leading to these results received funding from the “la Caixa” Foundation (ID 100010434) under the agreement LCF/PR/HR19/52160007, HR18-00452 to I.F. We thank CERCA Programme/Generalitat de Catalunya for institutional support. The Proteomics Platform of Navarrabiomed is a member of Proteored (PRB3-ISCIII), and is supported by grant PT17/0019/009 to J.F.I., of the PE I+D+I 2013–2016 funded by ISCIII and FEDER. Part of this work was funded by a grant from the Spanish Ministry of Science Innovation and Universities (Ref. PID2019-110356RB-I00) to J.F.I. and E.S., and the Department of Economic and Business Development of the Government of Navarra (Ref. 0011-1411-2020-000028) to ES.
Institutional Review Board Statement
All animal procedures were carried out following the guidelines of the European Communities Council Directive 2010/63/EU and with the approval of the local ethical committee (C.E.E.A: Comitè Ètic d’Experimentació Animal; University of Barcelona, Spain; reference number: 426/18).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We wish to thank Tom Yohannan for the final language revision of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, C.; Höfer, T. Multisite protein phosphorylation—From molecular mechanisms to kinetic models. FEBS J. 2009, 276, 3177–3198. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 2002, 3, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Barik, D.; Sananes, M.R.; Tyson, J.J.; Novák, B. Bistability by multiple phosphorylation of regulatory proteins. Prog. Biophys. Mol. Biol. 2009, 100, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P. The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 2000, 25, 596–601. [Google Scholar] [CrossRef]
- Johnson, L.N.; Lewis, R.J. Structural basis for control by phosphorylation. Chem. Rev. 2001, 101, 2209–2242. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [Green Version]
- Tenreiro, S.; Eckermann, K.; Outeiro, T.F. Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 2014, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Dammer, E.B.; Lee, A.K.; Duong, D.M.; Gearing, M.; Lah, J.J.; Levey, A.I.; Seyfried, N. Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. Proteomics 2015, 15, 508–519. [Google Scholar] [CrossRef] [Green Version]
- Sathe, G.; Mangalaparthi, K.K.; Jain, A.; Darrow, J.; Troncoso, J.; Albert, M.; Moghekar, A.; Pandey, A. Multiplexed phosphoproteomic study of the brain in patients with Alzheimer’s disease and age-matched cognitively healthy controls. OMICS 2020, 24, 2016–2227. [Google Scholar] [CrossRef]
- Bai, B.; Wang, X.; Li, Y.; Chen, P.C.; Yu, K.; Dey, K.K.; Yarbro, J.M.; Han, X.; Lutz, B.M.; Rao, S.; et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron 2020, 105, 975–991.e7. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Andrés-Benito, P.; Ausín, K.; Pamplona, R.; Del Rio, J.A.; Fernández-Irigoyen, J.; Santamaría, E. Dysregulated protein phosphorylation: A determining condition in the continuum of brain aging and Alzheimer’s disease. Brain Pathol. 2021, 31, e12996. [Google Scholar] [CrossRef] [PubMed]
- Andres-Benito, P.; Carmona, M.; Jordán-Pirla, M.; Torrejón-Escribano, B.; del Rio, J.A.; Ferrer, I. Dysregulated protein phosphorylation as main contributor of granulovacuolar degeneration at the first stages of neurofibrillary tangle pathology. Neuroscience 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Blanchard, A.P.; Elisma, P.; Granger, M.; Xu, H.; Bennett, S.A.; Figeys, D.; Zou, H. Phosphoproteomics analysis of an early onset mouse model (TgCRND8) of Alzheimer’s disease reveals temporal changes in neuronal and glia signaling pathways. Proteomics 2013, 13, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Kempf, S.J.; Metaxas, A.; Ibanez-Vea, M.; Darvesh, S.; Finsen, B.; Larsen, M.R. An integrated proteomics approach shows synaptic plasticity changes in an APP/PS1 Alzheimer’s mouse model. Oncotarget 2016, 7, 33627–33648. [Google Scholar] [CrossRef] [Green Version]
- Lachen-Montes, M.; González-Morales, A.; de Morentin, X.M.; Pérez-Valderrama, E.; Ausín, K.; Zelaya, M.V.; Serna, A.; Aso, E.; Ferrer, I.; Fernández-Irigoyen, J.; et al. An early dysregulation of FAK and MEK/ERK signaling pathways precedes the β-amyloid deposition in the olfactory bulb of APP/PS1 mouse model of Alzheimer’s disease. J. Proteom. 2016, 148, 149–158. [Google Scholar] [CrossRef]
- Kim, D.K.; Park, J.; Han, D.; Yang, J.; Kim, A.; Woo, J.; Kim, Y.; Mook-Jung, I. Molecular and functional signatures in a novel Alzheimer’s disease mouse model assessed by quantitative proteomics. Mol. Neurodegener. 2018, 13, 2. [Google Scholar] [CrossRef] [Green Version]
- Tagawa, K.; Homma, H.; Saito, A.; Fujita, K.; Chen, X.; Imoto, S.; Oka, T.; Ito, H.; Motoki, K.; Yoshida, C.; et al. Comprehensive phosphoproteome analysis unravels the core signalling network that initiates the earliest synapse pathology in preclinical Alzheimer disease brain. Hum. Mol. Genet. 2014, 24, 540–558. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Xu, J.; Zhou, X.; Liu, S.; Zhang, Y.; Ma, S.; Fu, A.K.Y.; Ip, N.Y.; Chen, Y. Changes of protein phosphorylation are associated with synaptic functions during the early stage of Alzheimer’s disease. ACS Chem. Neurosci. 2019, 10, 3986–3996. [Google Scholar] [CrossRef]
- Kovacs, G.G. Tauopathies. In Neuropathology of Neurodegenerative Diseases: A Practical Guide; Kovacs, G.G., Ed.; Cambridge University: Cambridge, UK, 2015; pp. 109–148. [Google Scholar]
- Arendt, T.; Stieler, J.; Holzer, M. Tau and Tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef]
- Ferrer, I.; García, M.A.; González, I.L.; Lucena, D.D.; Villalonga, A.R.; Tech, M.C.; Llorens, F.; Garcia-Esparcia, P.; Martinez-Maldonado, A.; Mendez, M.F.; et al. Aging-related tau astrogliopathy (ARTAG): Not only tau phosphorylation in astrocytes. Brain Pathol. 2018, 28, 965–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer, I.; Andrés-Benito, P.; Zelaya, M.V.; Aguirre, M.E.E.; Carmona, M.; Ausín, K.; Lachén-Montes, M.; Fernández-Irigoyen, J.; Santamaría, E.; Del Rio, J.A. Familial globular glial tauopathy linked to MAPT mutations: Molecular neuropathology and seeding capacity of a prototypical mixed neuronal and glial tauopathy. Acta Neuropathol. 2020, 139, 735–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, F.M.; Camargos, E.R.; De Souza, L.C.; Teixeira, A.L. Animal models of neurodegenerative diseases. Braz. J. Psychiatry 2013, 35, S82–S91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, V.M.; Kenyon, T.K.; Trojanowski, J.Q. Transgenic animal models of tauopathies. Biochim. Biophys. Acta 2005, 1739, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Dujardin, S.; Colin, M.; Buée, L. Animal models of tauopathies and their implications for research/translation into the clinic. Neuropathol. Appl. Neurobiol. 2015, 41, 59–80. [Google Scholar] [CrossRef]
- López-González, I.; Aso, E.; Carmona, M.; Armand-Ugon, M.; Blanco, R.; Naudí, A.; Cabré, R.; Portero-Otin, M.; Pamplona, R.; Ferrer, I. Neuroinflammatory gene regulation, mitochondrial function, oxidative stress, and brain lipid modifications with disease progression in tau P301S transgenic mice as a model of frontotemporal lobar degeneration-tau. J. Neuropathol. Exp. Neurol. 2015, 74, 975–999. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, I.; Andrés-Benito, P.; Ausín, K.; Cartas-Cejudo, P.; Lachén-Montes, M.; del Rio, J.A.; Fernández-Irigoyen, J.; Santamaría, E. Dysregulated protein phosphorylation in tauopathy. J. Neuropathol. Exp. Neurol. 2022. submitted. [Google Scholar]
- Kovacs, G.G.; Ghetti, B.; Goedert, M. Classification of diseases with accumulation of Tau protein. Neuropathol. Appl. Neurobiol. 2022, 48, e12792. [Google Scholar] [CrossRef]
- Kovacs, G.G. Molecular pathology of neurodegenerative diseases: Principles and practice. J. Clin. Pathol. 2019, 72, 725–735. [Google Scholar] [CrossRef]
- Andorfer, C.; Kress, Y.; Espinoza, M.; de Silva, R.; Tucker, K.L.; Barde, Y.A.; Duff, K.; Davies, P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 2003, 86, 582–590. [Google Scholar] [CrossRef]
- Andorfer, C.; Acker, C.M.; Kress, Y.; Hof, P.R.; Duff, K.; Davies, P. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J. Neurosci. 2005, 25, 5446–5454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andres-Benito, P.; Carmona, M.; Jordán, M.; Fernández-Irigoyen, J.; Santamaria, E.; del Rio, J.A.; Ferrer, I. Host tau genotype specifically designs and regulates tau seeding and spreading and host tau transformation following intrahippocampal injection of identical tau AD inoculums. Int. J. Mol. Sci. 2022, 23, 718. [Google Scholar] [CrossRef] [PubMed]
- Guillozet-Bongaarts, A.L.; Garcia-Sierra, F.; Reynols, M.R.; Horowitz, P.M.; Fu, Y.; Wang, T.; Cahill, M.E.; Bigio, E.H.; Berry, R.W.; Binder, L.I. Tau truncation during neurofibrillary tangle evolution in Alzheimer’s disease. Neurobiol. Aging 2005, 26, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Alquezar, C.; Arya, S.; Kao, A.W. Tau post-translational modifications: Dynamic transformers of tau function, degradation, and aggregation. Front. Neurol. 2021, 11, 595532. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Reyes, J.F.; Fu, Y.; Bigio, E.H.; Guillozet-Bongaarts, A.L.; Berry, R.W.; Binder, L.I. Tau nitration occurs at tyrosine 29 in the fibrillar lesions of Alzheimer’s disease and other tauopathies. J. Neurosci. 2006, 26, 1036–1045. [Google Scholar] [CrossRef] [Green Version]
- Arnsten, A.F.T.; Datta, D.; Del Tredici, K.; Braak, H. Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimer’s Dement. 2020, 17, 115–124. [Google Scholar] [CrossRef]
- Ferrer, I.; López-González, I.; Carmona, M.; Arregui, L.; Dalfó, E.; Torrejón-Escribano, B.; Diehl, R.; Kovacs, G.G. Glial and neuronal tau pathology in tauopathies: Characterization of disease-specific phenotypes and tau pathology progression. J. Neuropathol. Exp. Neurol. 2014, 73, 81–97. [Google Scholar] [CrossRef] [Green Version]
- Chung, D.C.; Roemer, S.; Petrucelli, L.; Dickson, D.W. Cellular and pathological heterogeneity of primary tauopathies. Mol. Neurodegener. 2021, 16, 57. [Google Scholar] [CrossRef]
- Yoshiyama, Y.; Higuchi, M.; Zhang, B.; Huang, S.M.; Iwata, N.; Saido, T.C.; Maeda, J.; Suhara, T.; Trojanowski, J.Q.; Lee, V.M. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 2007, 53, 337–351. [Google Scholar] [CrossRef] [Green Version]
- Allen, B.; Ingram, E.; Takao, M.; Smith, M.J.; Jakes, R.; Virdee, K.; Yoshida, H.; Holzer, M.; Craxton, M.; Emson, P.C.; et al. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J. Neurosci. 2002, 22, 9340–9351. [Google Scholar] [CrossRef] [Green Version]
- Sinsky, J.; Pichlerova, K.; Hanes, J. Tau protein interaction partners and their roles in Alzheimer’s disease and other tauopathies. Int. J. Mol. Sci. 2021, 22, 9207. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, E.; Pires, G.; MacMurray, C.; Askenazi, M.; Nayak, S.; Bourdon, M.; Safar, J.; Ueberheide, B.; Wisniewski, T. Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain 2020, 143, 2803–2817. [Google Scholar] [CrossRef]
- Ayyadevara, S.; Balasubramaniam, M.; Parcon, P.A.; Barger, S.; Griffin, W.S.T.; Alla, R.; Tackett, A.J.; Mackintosh, S.G.; Petricoin, E.; Zhou, W.; et al. Proteins that mediate protein aggregation and cytotoxicity distinguish Alzheimer’s hippocampus from normal controls. Aging Cell 2016, 15, 924–939. [Google Scholar] [CrossRef] [PubMed]
- Montoya, A.; Beltran, L.; Casado, P.; Rodríguez-Prados, J.C.; Cutillas, P.R. Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods 2011, 54, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.C.; Hunter, C.L.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.L.; Chan, D.W.; Gibson, B.W.; Gingras, A.-C.; Held, J.M.; et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Huntyer, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.H.; Shilov, I.V.; Seymour, S.L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 2008, 7, 3661–3667. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).