1. Introduction
The transcriptional activity of the human genome (>90% of the genome) produces long non-coding RNAs (lncRNAs) whose functions are mostly unknown. The extremities of linear chromosomes, or telomeres, actively synthesize a heterogeneous population of lncRNAs named
TERRA (telomeric repeat-containing RNA) with various sizes (from 100 bases to 10 Kb) and containing the canonical telomeric repeat sequence UUAGGG, as well as sequences unique to the subtelomeric region of each chromosome [
1,
2].
TERRA transcripts localize at telomeres and interact with the telomerase RNA subunit (
TERC) and with the telomerase catalytic subunit (
TERT) [
3,
4]. Several studies have established the role of
TERRA in telomere homeostasis and its implication in cancer. In this regard,
TERRA has been shown to sustain the activation of DNA damage response pathways at dysfunctional telomeres [
5,
6] and to facilitate completion of telomere DNA replication in cancer cells [
7]. Importantly,
TERRA depletion associates with an increase of telomerase activity, supporting the notion that
TERRA can negatively regulate telomerase in mammalian cells [
2,
3,
4]. Expression studies conducted by various technologies have revealed the dysregulation of
TERRA in various solid tumors, a frequent downregulation in specific tumor types vs. their normal tissue counterparts [
8], but also its overexpression in medulloblastoma [
9]. Moreover
TERRA expression is increased upon the treatment of human cancer cell lines with chemotherapeutic agents, suggesting that
TERRA transcripts may participate in the cellular response and/or resistance to chemotherapy [
10,
11,
12]. For hepatocellular carcinoma (HCC), it is known that telomerase activation is a hallmark of this malignant condition [
13] and that
TERRA may participate in telomerase regulation in this cancer type [
8]. HCC is the most common type of primary liver cancer and worldwide is the fifth cancer and the third cause of cancer mortality; it is a highly heterogeneous disease and it occurs most often in people with chronic liver diseases (liver hepatitis, alcohol hepatitis, viral B or C hepatitis, steatohepatitis, cirrhosis). Also for this reason, the HCC clinical management remains difficult: very often the diagnosis reveals an advanced HCC that usually is not submitted to surgical resection and a resectable HCC frequently leads to recurrent tumors [
14,
15,
16,
17]. To date, the treatment options are limited and advancing in the molecular characterization of HCC is a real challenge. Also, early diagnosis represents a critical step for the management of HCC and the identification of novel biomarkers of HCC will be of foremost importance.
The aim of the current study was to characterize TERRA expression in HCC, determining by qPCR its expression levels in HCC tissues and in HCC cell lines as well as its circulating amount in the plasma of HCC patients and in the cell secretomes. Further, we assessed in the same biological specimens the expression of TERC and of TERT mRNA in relation to TERRA. Our results highlight that both cellular and extracellular expressions of TERRA and TERC are dysregulated in HCC, as well as the cellular expression of TERT mRNA.
3. Discussion
The expression variations of the lncRNA
TERRA in cancer has generated a growing interest on the biology of these telomeric transcripts in various solid tumors (larynx and colon cancer, medulloblastoma, glioblastoma, head and neck cancer, cervical and endometrial carcinoma) [
2,
9,
24,
25,
26,
27,
28], but its real functional role in a given cancer type remains to be elucidated. In the present work, for the first time, we determined
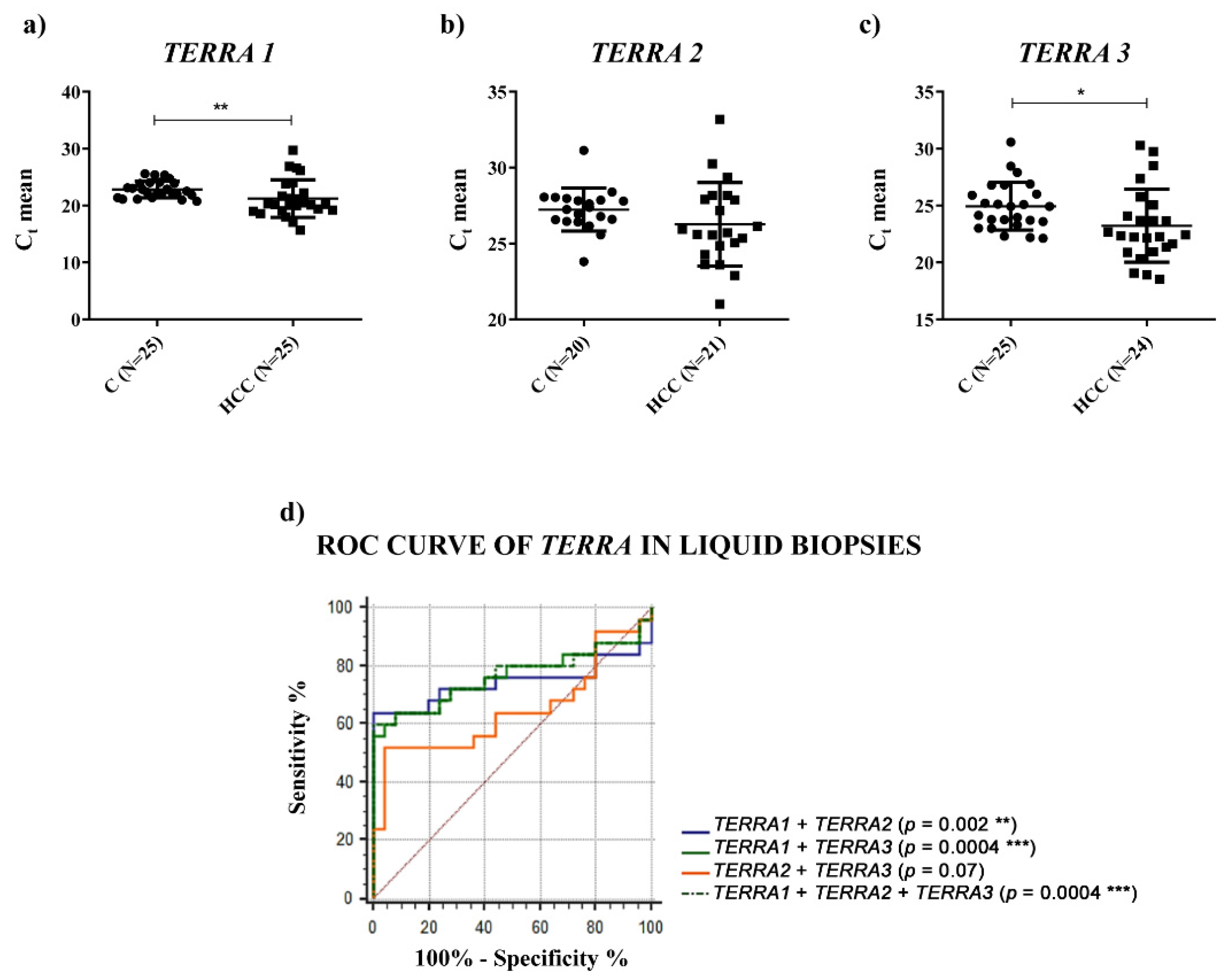
TERRA expression in HCC by qPCR, in HCC tissues and in the plasma of HCC patients. We used a qPCR assay that did not detect all
TERRA transcripts, but it detected the most abundant ones. In particular, we used three primer pairs to quantify
TERRA expression levels transcribed from seven telomeres (
TERRA1: 1_2_10_13q;
TERRA2: 15q;
TERRA3: XpYp) [
29]. Furthermore, Diman et al. [
30] demonstrated that the primer pair named
TERRA1 (1_2_10_13q) detected the transcripts expressed from the telomeres: 1_2_4_10_13_22q. Thus, according to Diman et al., our qPCR assay should recognize
TERRA transcripts expressed from nine telomeres. Furthermore, Montero et al. [
31] demonstrated that
TERRA is mainly transcribed from the 20q subtelomere in human cells and the primer pairs able to detect this transcript were not included in our qPCR assay. Moreover, considering the role of
TERRA in the telomere function and the importance of the telomerase enzyme in the biology of HCC, in the present study, we also assessed the RNA expression of the main components of the telomerase complex, the RNA subunit
TERC and the telomerase catalytic subunit
TERT mRNA. We found a significant
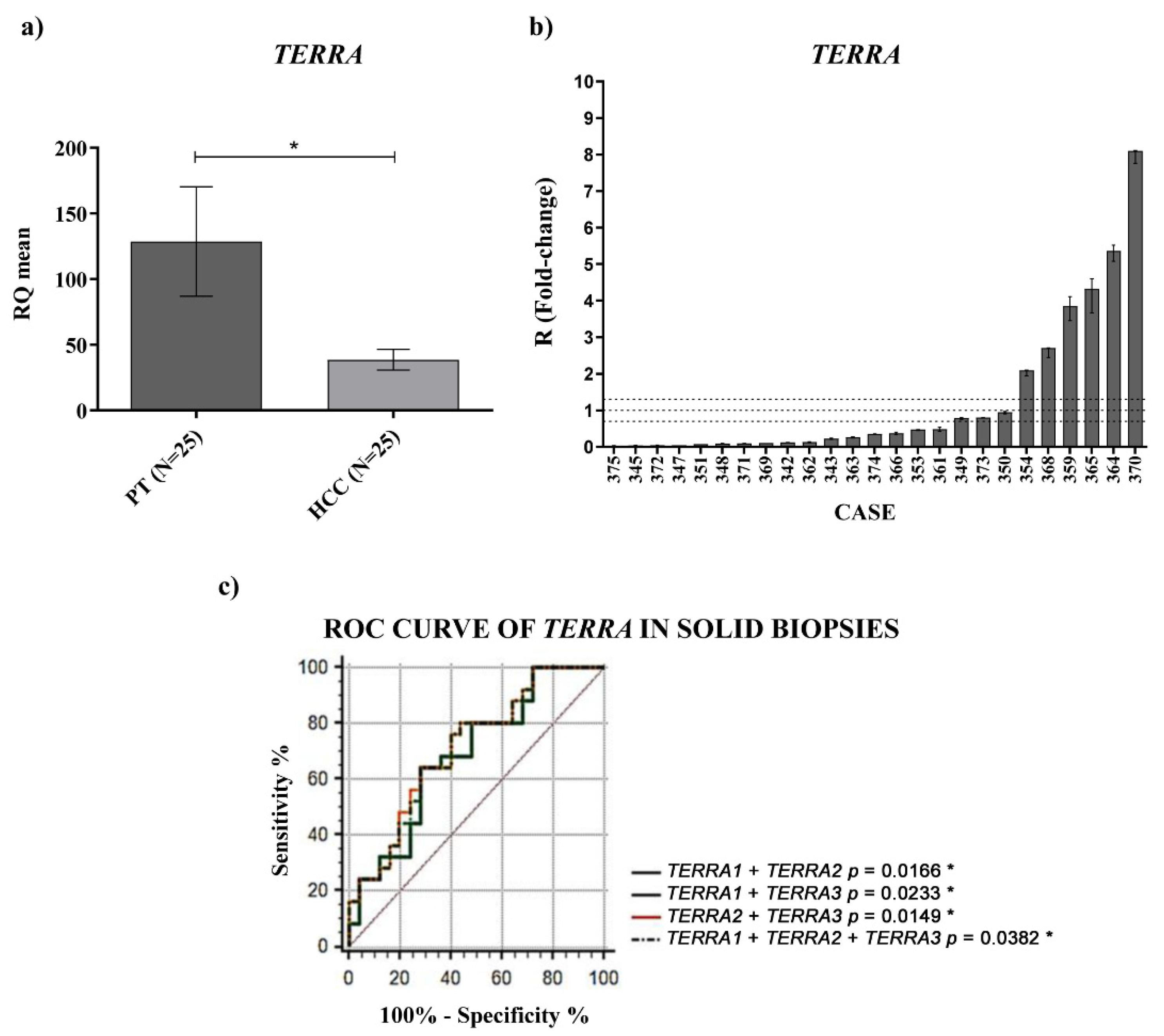
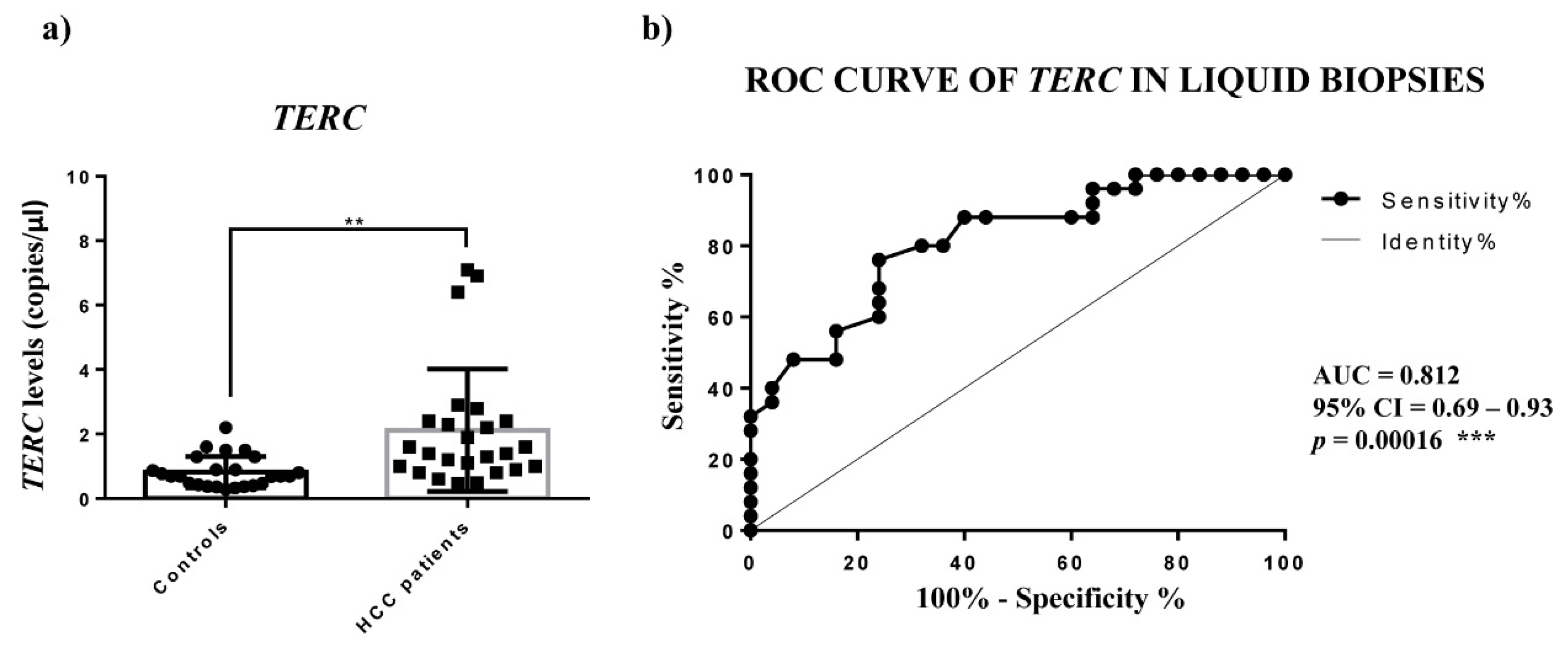
TERRA downregulation in HCC tissues vs. the corresponding PT ones and a significantly higher amount of
TERRA in the plasma of HCC patients. In both cases, for the first time, ROC curves indicated significant capabilities in distinguishing HCC tissues from PT ones and HCC patients from control subjects. Further, the qPCR determinations of
TERRA downregulation in HCC tissues, consistent with the in situ hybridization data reported by Cao et al. [
8], could be utilized as specific characterization of the malignant tissue (of HCC or other cancer types), being the qPCR technique frequently used in research laboratories.
For
TERRA transcripts in the plasma, an RNA-seq study [
18] revealed the presence of
TERRA molecules in the plasma of 2 normal subjects and of 19 patients with various cancer types (prostate, stomach, breast, colon, kidney, lung, duct, liver, melanoma, ovarian); however, to our knowledge, there are no reports on circulating
TERRA in a small or large cohort of patients with the same type of cancer, or even reports on the PCR determinations of
TERRA amounts in the plasma of cancer patients. Hence, the data reported in our study are the first concerning
TERRA expression in the plasma of a cohort of patients with the same cancer type, HCC, in comparison to healthy individuals.
For the overexpressed amount of TERRA detected in the plasma of HCC patients, we cannot know the origin of the circulating forms of TERRA, if they derive from the malignant cells or not. Despite this, our data indicate that the plasma determination of TERRA amounts can be taken into account as a possible non-invasive molecular indicator of HCC diagnosis and, in order to ascertain this hypothesis, we are planning a multicenter study to examine a larger cohort of patients.
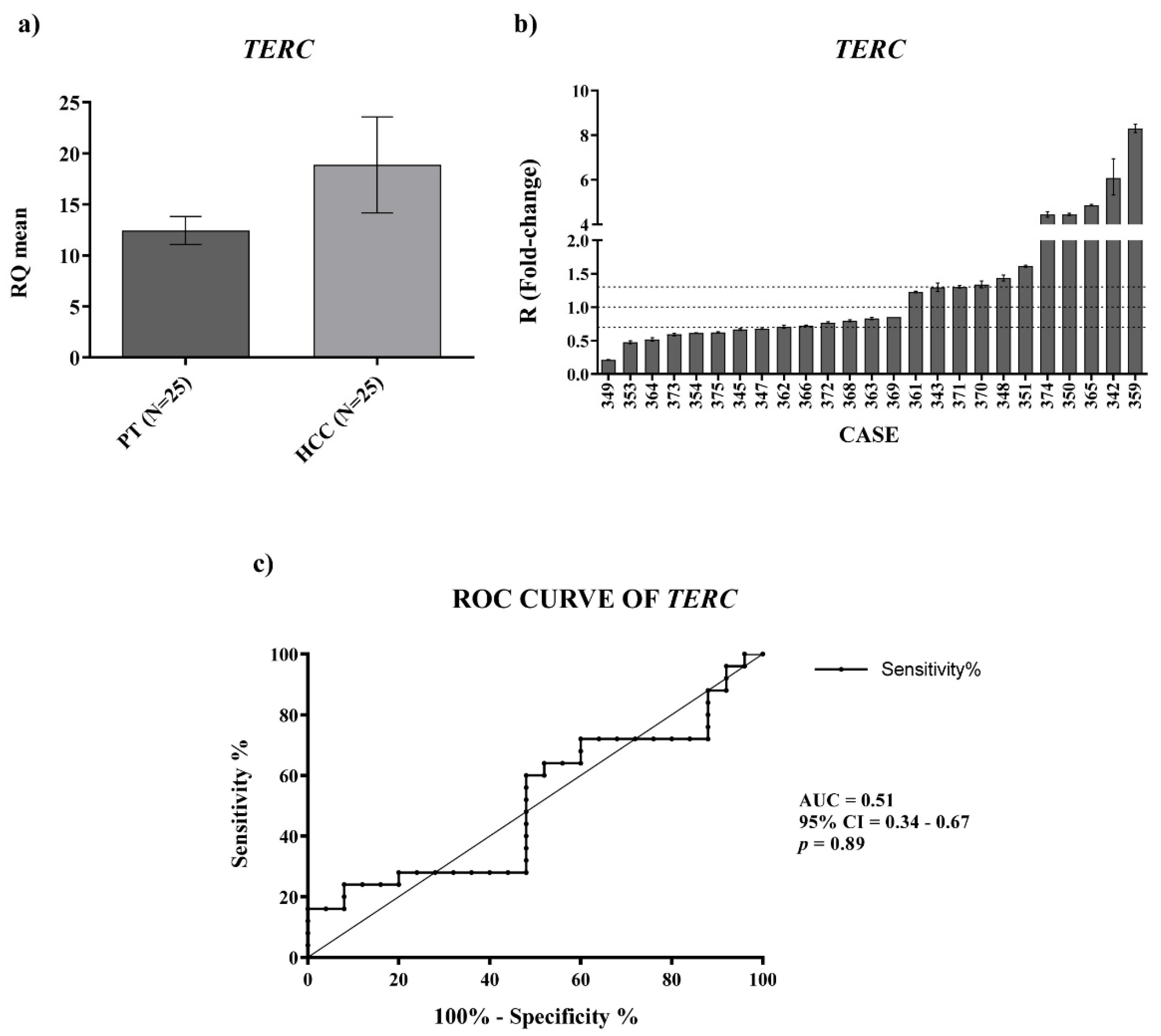
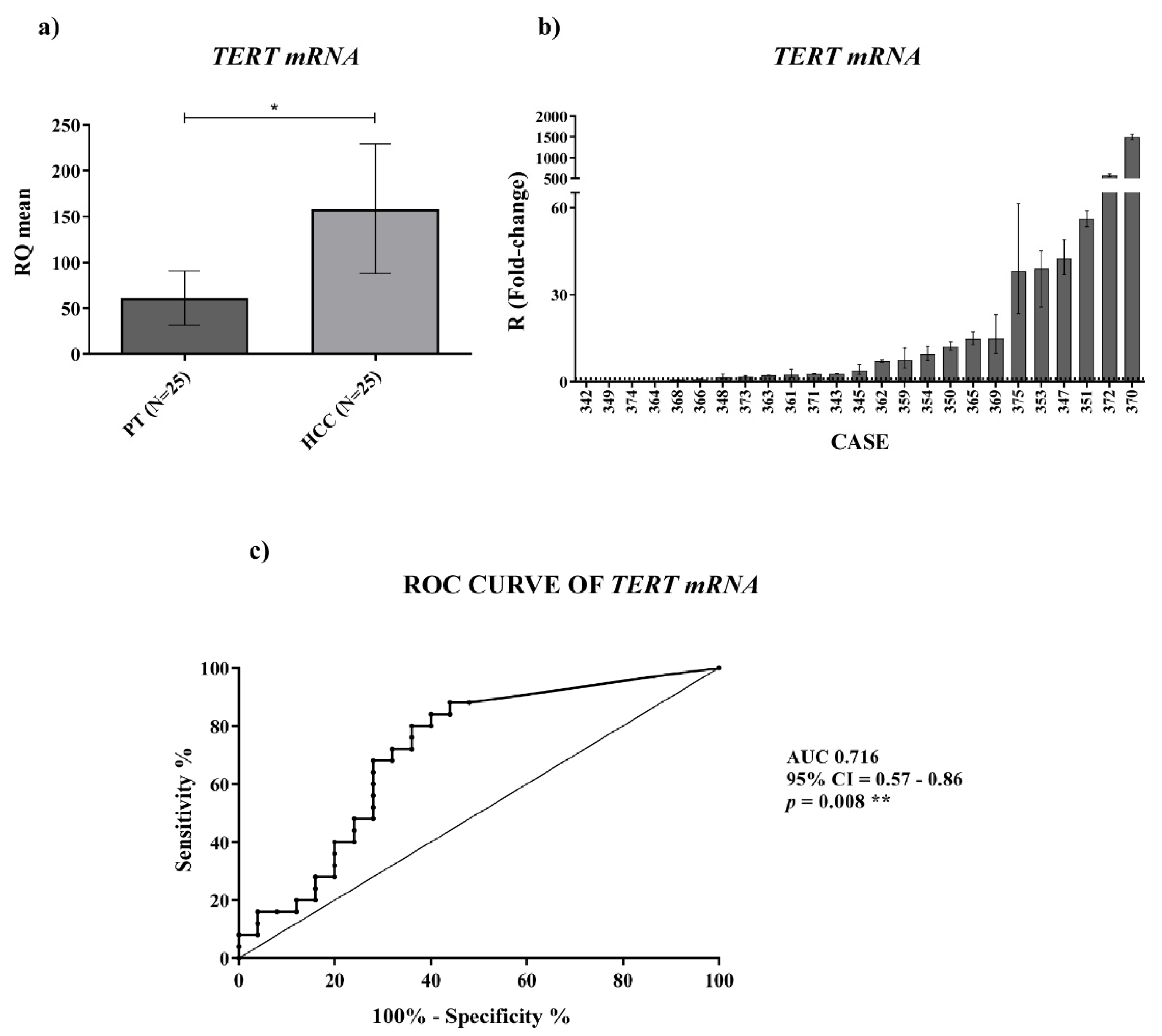
For
TERC and
TERT mRNA expression we found an upregulation in HCC tissues, which was significant for the
TERT mRNA. For measuring the level of
TERC and
TERT mRNA in plasma, we used ddPCR, the best technology to assess absolute quantifications of transcripts, especially the circulating ones. The
TERC amounts detected in the plasma of HCC patients were significantly higher, while
TERT mRNA was not detectable, thus indicating that in HCC patients the cells do not secrete the detectable extracellular form of
TERT mRNA. The ROC analysis of circulating
TERC expression revealed a significant ability in discriminating HCC patients from controls. Concerning
TERC in cancer, it is known from in situ and fluorescence hybridization data (ISH, FISH) that is overexpressed in lung, oral, prostate and cervix carcinoma [
10]; further, only Novakovic et al., to our knowledge, reported nested-PCR data that revealed the presence of
TERC in the plasma of healthy controls and of patients with breast cancer, melanoma and thyroid cancer, but did not allow the
TERC amount to be considered as a tumor marker [
29]. Hence, our findings would indicate the determination of circulating
TERRA and
TERC in HCC patients as specific non-invasive molecular indicators of this malignant condition.
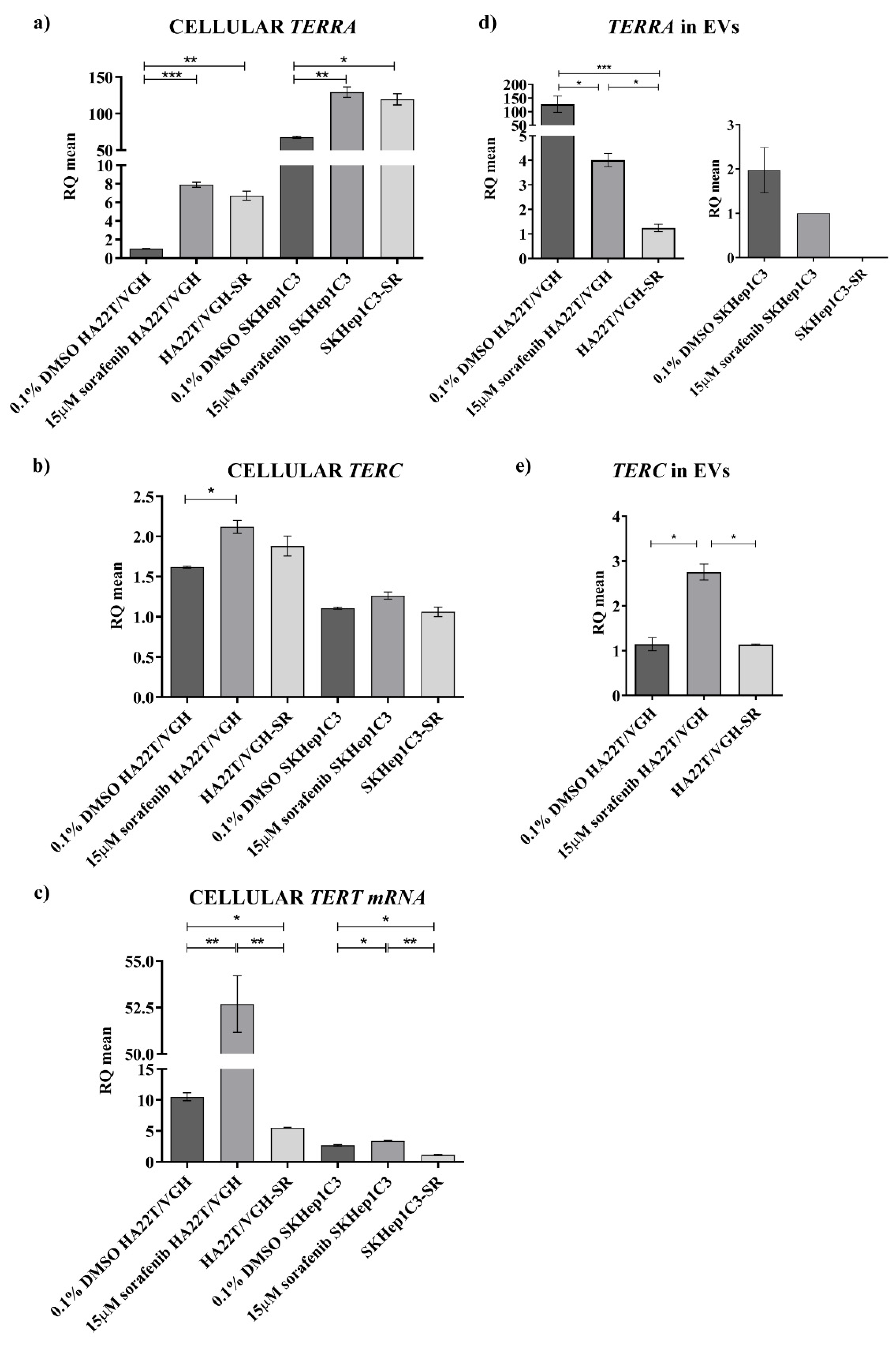
Subsequently, in the present work, to address the problem of the dysregulation of
TERRA,
TERC and
TERT mRNA in HCC from a biological point of view, we characterized two HCC cell lines for the expression of these transcripts and, for the first time, we report data on their expression both at the cellular and extracellular levels, indicating that the two HCC cell lines may represent the heterogeneity of HCC. In particular, the HA22T/VGH and SKHep1C3 cells, at low and very high
TERRA expressions, respectively, may represent the HCC cases with
TERRA downregulation and upregulation, respectively. HA22T/VGH cells producing low
TERRA amounts expressed high levels of
TERC and
TERT mRNA, and this result is consistent with previous evidence indicating that
TERRA depletion associates with increased levels of
TERC [
3]. Both HCC cell lines secrete
TERRA molecules into the secretome-derived EVs. The HA22T/VGH cells secreted very high amounts of
TERRA and low amounts of
TERC. As expected,
TERT mRNA was not detected as an extracellular form in any condition. Further, concerning the effect of an anticancer drug on
TERRA expression, both cell lines similarly responded to sorafenib treatment enhancing the expression amount of
TERRA,
TERC and
TERT mRNA at the cellular level. HA22T/VGH reacted to sf increasing the cellular expression of
TERRA,
TERC and
TERT mRNA and decreasing the secretion of
TERRA and
TERC; SKHep1C3 cells reacted to sf increasing the cellular expression of
TERRA, but much less
TERC and
TERT mRNA, and decreasing the secretion of
TERRA and by not secreting both
TERC and
TERT transcripts. Sorafenib is a multikinase inhibitor exerting its antitumor activity mainly affecting the proliferation and apoptosis of cancer cells, known to influence gene expression at the level of mRNAs, ncRNAs and genome methylation with mechanisms that have not been elucidated [
22,
23,
32]. Hence, further effort is needed to understand the intracellular events that led to an increase of
TERRA in cells treated with sf. From a general point of view, it is known that
TERRA is a heterogeneous group of ncRNAs generated from multiple different telomeres and with various size (from 100 to 10 Kb); it has been proposed that its upregulation can be regulated by stress inducing the processing of
TERRA molecules into smaller fragments then secreted into the EVs. It seems that
TERRA containing EVs are relatively stable and circulate in extracellular spaces, including plasma, but it is not known how the smaller extracellular forms are generated. As reported by Wang et al. [
19], the extracellular molecules of
TERRA can be breakdown products of full-length cellular
TERRA or an alternative form that is encapsulated into the EVs and secreted in extracellular spaces. It is likely that
TERRA containing EVs can transfer signaling information to surrounding cells impacting the extracellular microenvironment. The complex combination of factors that comprise EVs and the type of recipient cells sensing the EVs may determine the nature of the signaling and response.
Hence, this part of the present study is the first effort to characterize the cellular and extracellular expression of TERRA, TERC and TERT mRNA in HCC cell lines demonstrating the capability of these cells to secrete TERRA and TERC transcripts into the EVs and thus opening a new scenario in the biology of HCC cells. Novel issues will be addressed to understand the role of TERRA and TERC containing EVs in transferring signaling to surrounding cells in vitro and in an HCC tumor xenograft.
In conclusion, in the current work we have assessed the cellular and extracellular expression of TERRA, TERC and TERT mRNA studying HCC tissues, the plasma of HCC patients and the cells and secretomes of two HCC cell lines. Our novel findings obtained by qPCR show that HCC tissue is significantly characterized by TERRA downregulation and by TERT mRNA upregulation; the plasma of HCC patients revealed an overexpression of TERRA and TERC, thus indicating novel non-invasive indicators of HCC whose clinical diagnostic significance will be investigated further in a larger cohort of patients. It is clear that among HCC tissues there are two sets, one with TERRA down-modulation, the other with TERRA upregulation. Moreover, the characterization of two HCC cell lines, representative of this feature of HCC heterogeneity, has clearly shown the ability of HCC cells to produce and secrete TERRA and TERC into the EVs; the ability to produce TERT mRNA that is not secreted; and the ability to respond to sorafenib treatment increasing TERRA expression. From a general point of view, our findings contribute to the basic knowledge of the cellular and extracellular forms of TERRA and TERC produced by cultured cancer cell lines and those detectable in HCC tissues and in the plasma of HCC patients.
4. Materials and Methods
4.1. Tissue Samples and Liquid Biopsies Collection
All human HCC tissues as well as the corresponding peritumoral (PT) (resected 1–2 cm from the malignant tumor) and the peripheral blood were obtained from 25 HCC patients who underwent HCC resection at Spedali Civili, Surgical Clinic of Brescia, Italy (
Table S3). The peripheral blood of healthy volunteers (
n = 25) was collected at the Immunohematology and Transfusion Medicine Service at ASST Civili Hospital in Brescia, Italy. HCC tissues and the PT ones were subsequently preserved in RNA-Later (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for further investigation. The subjects consisted of 20 men and 5 women, ranging from 60 to 82 years of age. The subjects did not have any apparent distant metastases, and none of them had been previously treated for HCC. Each individual was screened for the presence of the hepatitis B virus (HBV) or hepatitis C virus (HCV): 3 patients were positive for HBV, 15 were positive for HCV and 8 were found to be negative for both HBV and HCV. Each biopsy specimen underwent a pathological examination as previously described [
22] that revealed the presence of different background diseases: 3 steatosis, 14 cirrhosis, 6 chronic hepatitis, 1 von Meyenburg complex and 1 no apparent background pathology (normal hepatic parenchyma); for 1 sample, no information was available. Liquid biopsies were collected from the peripheral blood of each patient before HCC surgical resection in a VACUETTE
® (Greiner Bio-One, S.r.l., Kremsmünster, Austria) EDTA-coated blood collection tubes. Plasma samples were obtained from 1 mL of peripheral blood with a first centrifugation step at 3000 rpm for 10 min at 4 °C followed by a second centrifugation step at 4000 rpm for 20 min at 4 °C. Samples showing hemolysis were excluded. The plasma was transferred to a new tube and stored at −80 °C until RNA extraction.
This study was approved by the ethical committee of ASST Civili Hospital of Brescia on 2 October 2012 (NP1230). Informed consent was obtained from all the subjects enrolled in this study. All methods were performed in accordance with the relevant guidelines and regulations.
4.2. Cell Cultures, Treatment with Sorafenib and EV Isolation
In the present study, human tumor cell lines derived from HCC (HA22T/VGH and SKHep1C3) were used. HA22T⁄VGH undifferentiated cells, kindly provided by N. D’Alessandro (University of Palermo, Palermo, Italy), were maintained in RPMI-1640 (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 100 nM sodium pyruvate (Thermo Fisher Scientific, Inc., Waltham, MA, USA). SKHep1Clone3 (SKHep1C3), selected from human HCC-derived cells (SKHep1: ATCC HTB-52), was maintained in Earle’s MEM (Thermo Fisher Scientific, Inc., Waltham, MA, USA). All culture media were supplemented with 10% fetal bovine serum (FBS; Euroclone, S.p.a., Pero, Italy) and 10,000 U/mL penicillin/streptomycin (Thermo Fisher Scientific, Inc., Waltham, MA, USA). HA22T/VGH and SKHep1C3 sorafenib resistant cells (HA22T/VGH-SR and SKHep1C3-SR), produced in our lab, were maintained as previously described [
22]. Cells were cultured at 37 °C with 5% CO
2 in 10 cm Ø plates up to almost 90% of confluence. Sorafenib [N-(3-trifluoromethyl-4-chlorophenyl)-N-(4-(2-methylcarbamoylpyridin-4-yl) oxyphenyl) urea] was synthesized and provided by Bayer Corporation (West Haven, CT, USA). The compound was dissolved in 100% dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck, Inc., Darmstadt, Germany) and diluted with RPMI-1640 or MEM to the required concentration (10 µM, 15 µM). In in vitro experiments, 0.1% DMSO was added to cultures as a negative solvent control. In order to get extracellular vesicles (EVs)-containing media (named in this text as secretome), HA22T/VGH and SKHep1C3 cells were seeded in a 10 cm Ø plate (4 replicates/each condition) at a density of 5 × 10
6 cells and maintained in complete media until they reached 75% of confluence. Then, after gentle washes with PBS, cells were subsequently supplied with serum-free medium for 24 h and treated with sorafenib at the specific concentrations (15 µM).
Before starting the nickel-based isolation (NBI) of EV, collected media were centrifuged at 2800×
g for 10 min to remove cell debris and mixed to a nickel-based matrix of beads in order to capture EVs, which were subsequently analyzed by Q-NANO instrument (IZON Science, Ltd., Christchurch, New Zealand), as previously reported [
33]. We recovered a particle concentration range between 1.63 × 10
8 and 2.09 × 10
8 EV/mL for HA22T/VGH cells, from 3.08 × 10
8 to 11.01 × 10
8 EV/mL for SKHep1C3 cells according to different cell densities and the amount of medium processed to further optimize the RNA recovery. The EV showed various sizes of the mean diameter, from 185 to 252 nm for HA22T/VGH cells and from 250 to 317 nm for the SKHep1C3 ones.
4.3. RNA Isolation
Total RNA was obtained from 10 cm Ø cells plates and by grinding ~1 cm
3 of tumor and peritumor tissue with a TissueRupture (Qiagen, Inc., Hilden, Germany) in 1 mL of TRIzol Lysis Reagent (Qiagen, Inc., Hilden, Germany) according to the Chomczynski–Sacchi method [
34]. RNA purity and concentration were assessed and measured by a NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA), while the integrity was evaluated on 0.8% agarose gel stained with ethidium bromide. Extracellular total RNA was extracted from 200 µL of isolated EVs and 200 µL of plasma, using a miRNeasy Mini Kit (Qiagen, Inc., Hilden, Germany), according to the manufacturer’s instructions as previously reported [
20].
4.4. Northern Blotting
The secretome RNA samples were extracted as described above, the control total RNA from HeLa (ATCC: CCL-2) and U2OS (ATCC: HTB-96) cell lines was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturers’ protocol.
U2OS (human osteosarcoma cell line) and HeLa (human cervical adenocarcinoma cell line) cells were obtained from ATCC (Manassas, VA, USA). The cells were grown in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 2 mM L-glutamine, 1% penicillin-streptomycin and 10% fetal bovine serum at 37 °C and 5% CO2 to 50–70% confluency before harvesting for RNA extraction.
RNA purity and concentrations were assessed as stated in
Section 4.3. RNA was mixed with RNA gel loading dye (Thermo Fisher Scientific, Inc., Waltham, MA, USA), denatured for 10 min at 65 °C and separated for 2 h at 9.5 V/cm in 1% agarose MOPS gel with 1.9% formaldehyde, then blotted onto the Amersham Hybond-N+ hybridization membrane (Cytiva) with the semi-dry Power Blotter System (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and cross-linked in the CL-1000 Ultraviolet Crosslinker (Fisher Scientific, Inc., Hampton, NH, USA) at the pre-set conditions. A probe consisting of five telomeric repeats (5′-CCCTAA-3′)5 was labeled with the DIG Oligonucleotide 3′-End Labeling Kit (F. Hoffmann-La Roche, Ltd.; Basel, Switzerland) according to the manufacturers’ instructions and denatured for 5 min at 95 °C before hybridization. The hybridization was performed at 50 °C overnight, the membrane was washed, blocked, incubated with anti-digoxygenin-AP (F. Hoffmann-La Roche, Ltd.; Basel, Switzerland; dilution 1:1500), visualized by incubation with CDP-Star Merck KGaA, Darmstadt, Germany and captured on the Chemidoc XRS+ (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
4.5. RT-qPCR and Gene Expression Analysis
cDNA for
TERC and
TERT mRNA studies was synthesized from 1 μg of tissues and cells total RNA using M-MLV reverse transcriptase (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with random hexamer (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer’s instructions. The reverse transcription (RT) reaction was performed at 37 °C for 60 min, followed by inactivation at 95 °C for 5 min in a T100 Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
TERRA first-strand cDNA was synthesized following Feretzaki’s protocol [
35]. Briefly, 1 μg of tissues and cells total RNA was first partially denatured at 65 °C before RT. Then, a
TERRA-cDNA-pool was synthesized in a final volume of 20 µL with RT-specific primers: lncRNA
TERRA 5′-(CCCTAA)5-3′ and β-actin 5′-AGTCCGCCTAGAAGCATTTG-3′. cDNA for circulating lncRNA
TERRA was synthesized from 2.5 µL of plasma total RNA using the iScript Explore One-Step RT and PreAmp Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) in a final reaction volume of 25 µL, according to the manufacturer’s instructions. The pre-amplification step consisted of 14 cycles of temperature variation at 95 °C for 15 s and 60 °C for 4 min.
qPCR for
TERC and
TERT mRNA were carried out using TaqMan Universal PCR Master Mix 2X (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the appropriate qPCR assays 20X, following the manufacturer’s instructions:
TERC (assay ID: ID Hs03454202_s1; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
TERT (assay ID: Hs00972650_m1; Thermo Fisher Scientific, Inc., Waltham, MA, USA). GAPDH was used as endogenous control (assay ID: Hs99999905_m1; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The thermocycling conditions were: initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s then 60 °C for 1 min. qPCR for lncRNA
TERRA was performed mixing 2 µL of
TERRA-cDNA-pool (1:2 from tissues or cells) or 3 µL of
TERRA-cDNA-pool (1:10 of pre-amplified product) and the specific forward/reverse primers with Power SYBR Green PCR Master Mix 2X (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a final volume of 20 µL, according to the manufacturer’s instructions.
TERRA qPCR was followed by the dissociation stage for melting curve analysis for each primer-pair used at 95 °C for 25 min, 60 °C for 1 min and 95 °C for 15 min. The sequence of the specific primers (Integrated DNA Technologies, Inc.) was:
TERRA 1 (hTel_1_2_10_13q) 5′-GAATCCTGCGCACCGAGAT-3′ (forward) 5′-CTGCACTTGAACCCTGCAATAC-3′ (reverse);
TERRA 2 (hTel_15q) 5′-CAGCGAGATTCTCCCAAGCTAAG-3′ (forward) 5′-AACCCTAACCACATGAGCAACG-3′ (reverse);
TERRA 3 (hTel_XpYp) 5′-GCAAAGAGTGAAAGAACGAAGCTT-3′ (forward) 5′-CCCTCTGAAAGTGGACCAATCA-3′ (reverse). β-actin was used as endogenous control 5′-CCTCGCCTTTGCCGATCC-3′ (forward), 5′-GGATCTTCATGAGGTAGTCAGT-3′ (reverse). Each reaction was performed in triplicate. Data analysis was performed by the 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). In tissues and cells, gene expression was measured with the Livak–Schmittgen (2-ΔΔCq) method [
36] and the
TERRA expression was obtained as the mean of the relative quantification (RQ) values for
TERRA 1,
TERRA 2 and
TERRA 3. When all 25 patients were analyzed, the plates were virtually merged by the 7500 Applied Biosystems software in order to determine the same calibrator per target (
TERRA1,
TERRA1,
TERRA3) that was the sample with the lowest
TERRA expression (in our cohort the sample HCC347). For all these reasons, RQ values of the different samples can be compared.
Due to the lack of a known reference gene that would allow the normalization of circulating TERRA, qPCR data were reported as the average raw cycle thresholds (Ct) in plasma samples.
4.6. ddPCR and Gene Expression Analysis
Total RNA from plasma was reverse-transcribed into cDNA by random hexamers using M-MLV reverse transcriptase (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Then, 9 μL of the resulting cDNA was prepared for amplification in a 20 μL reaction volume containing 10 μL 2X ddPCR Supermix for probes (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and 1 μL 20X TaqMan (Thermo Fisher Scientific, Inc., Waltham, MA, USA) PCR probe assay (TERT: assay ID Hs00972650_m1; TERC: assay ID Hs03454202_s1). Each ddPCR assay mixture (20 μL) was loaded into a disposable droplet generator cartridge (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Then, 70 μL of droplet generation oil for probes (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was loaded into each of the eight oil wells. The cartridge was then placed inside the QX200 droplet generator (Bio-Rad Laboratories, Inc., Hercules, CA, USA). When droplet generation was completed, the droplets were transferred to a 96-well PCR plate (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using a multichannel pipette. The plate was heat-sealed with foil and placed in a conventional thermal cycler. Thermal cycling conditions were: 95 °C for 10 min, then 40 cycles of 94 °C for 30 s and 60 °C for 1 min (ramping rate reduced to 2%), with a final step at 98 °C for 10 min and a 4 °C indefinite hold. A no template control (NTC) and a negative control for each reverse transcription reaction (RT-neg) were included in every assay.
4.7. Statistical Analysis
Statistical analysis was carried out using GraphPad Prism v7.0 (GraphPad Software, Inc., San Diego, CA, USA) software. The Wilcoxon test was used to compare the expression levels of selected transcripts between peritumoral and tumoral tissues. An unpaired t-test was used to test the significant differences in the plasma levels of selected transcripts between HCC patients and healthy subjects and for in vitro results. Diagnostic performance was evaluated using receiver operating characteristic (ROC) curve analysis. To assess the diagnostic values of multi-assays, the logistic regression method was applied. The diagnostic performance of individual as well as combinations of classifiers was computed using MedCalc v19.1.7 (MedCalc Software, Ltd., Ostend, Belgium) software. To evaluate the correlation between the clinical pathological characteristics and TERRA levels in tissues or in plasma samples, the patients were divided into 2 groups with high and low TERRA levels by using R (RQHCC/RQPT) = 0.7 for tissue expression or Ct = 22.81 (the median value) for plasma levels. Fisher’s exact test was applied. Data were considered statistically significant when p-value ≤ 0.05.