Performance Comparison of Five Methods for Tetrahymena Number Counting on the ImageJ Platform: Assessing the Built-in Tool and Machine-Learning-Based Extension
Abstract
:1. Introduction
2. Results
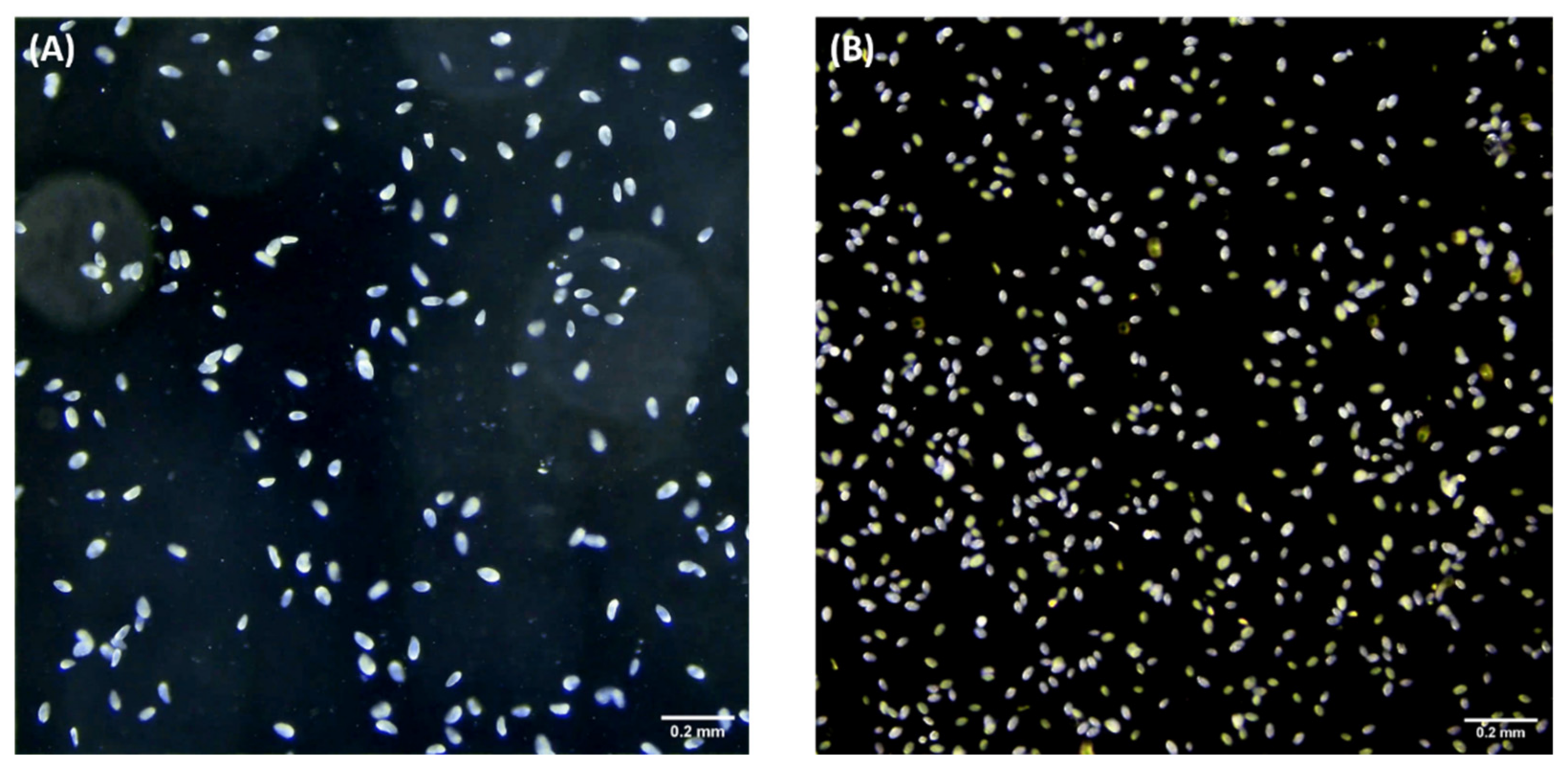
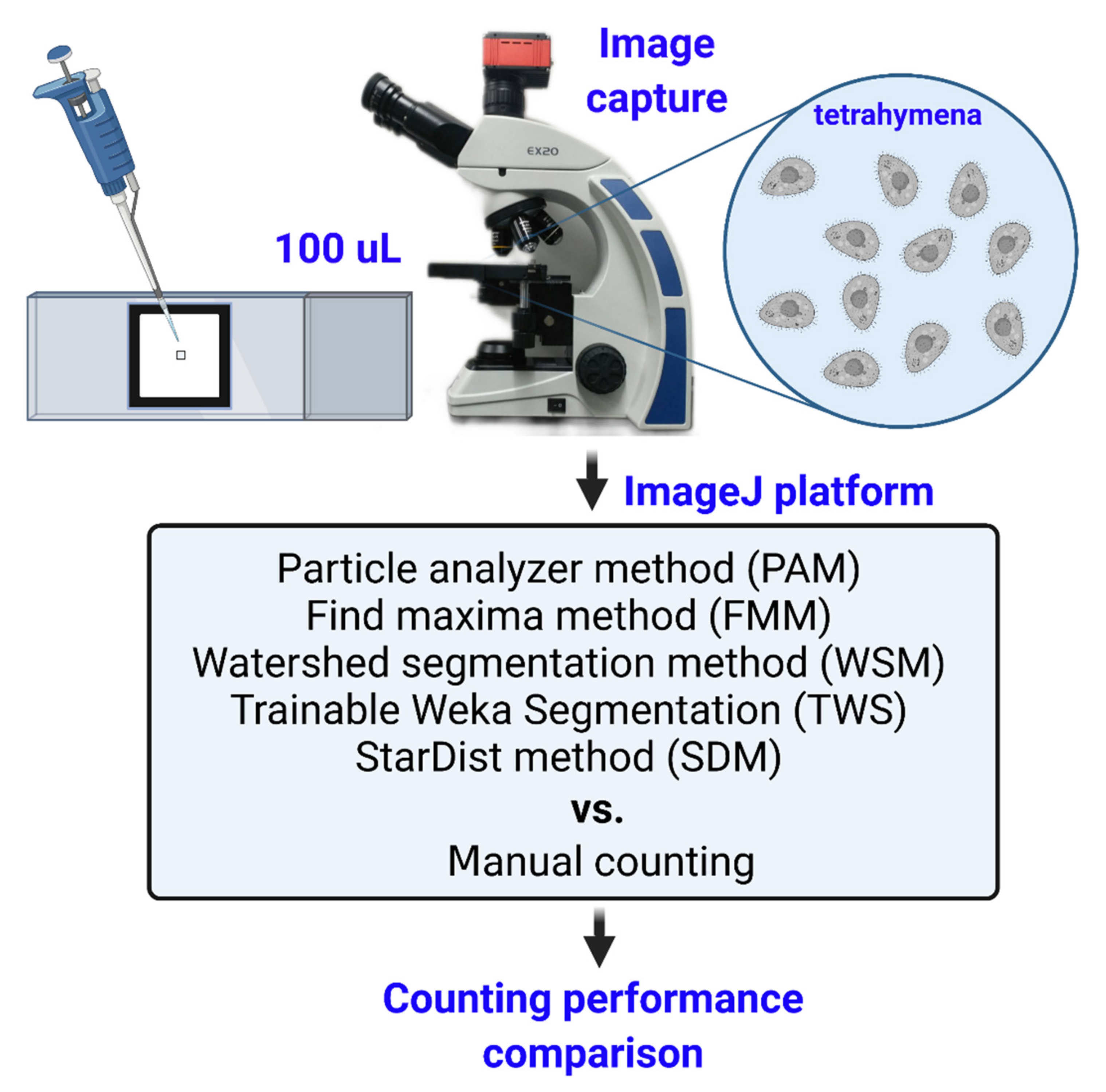
2.1. Overview of Our Setting and Analysis Pipeline for Tetrahymena Counting
2.2. Comparison of Tetrahymena Counting Performance between Known Methods
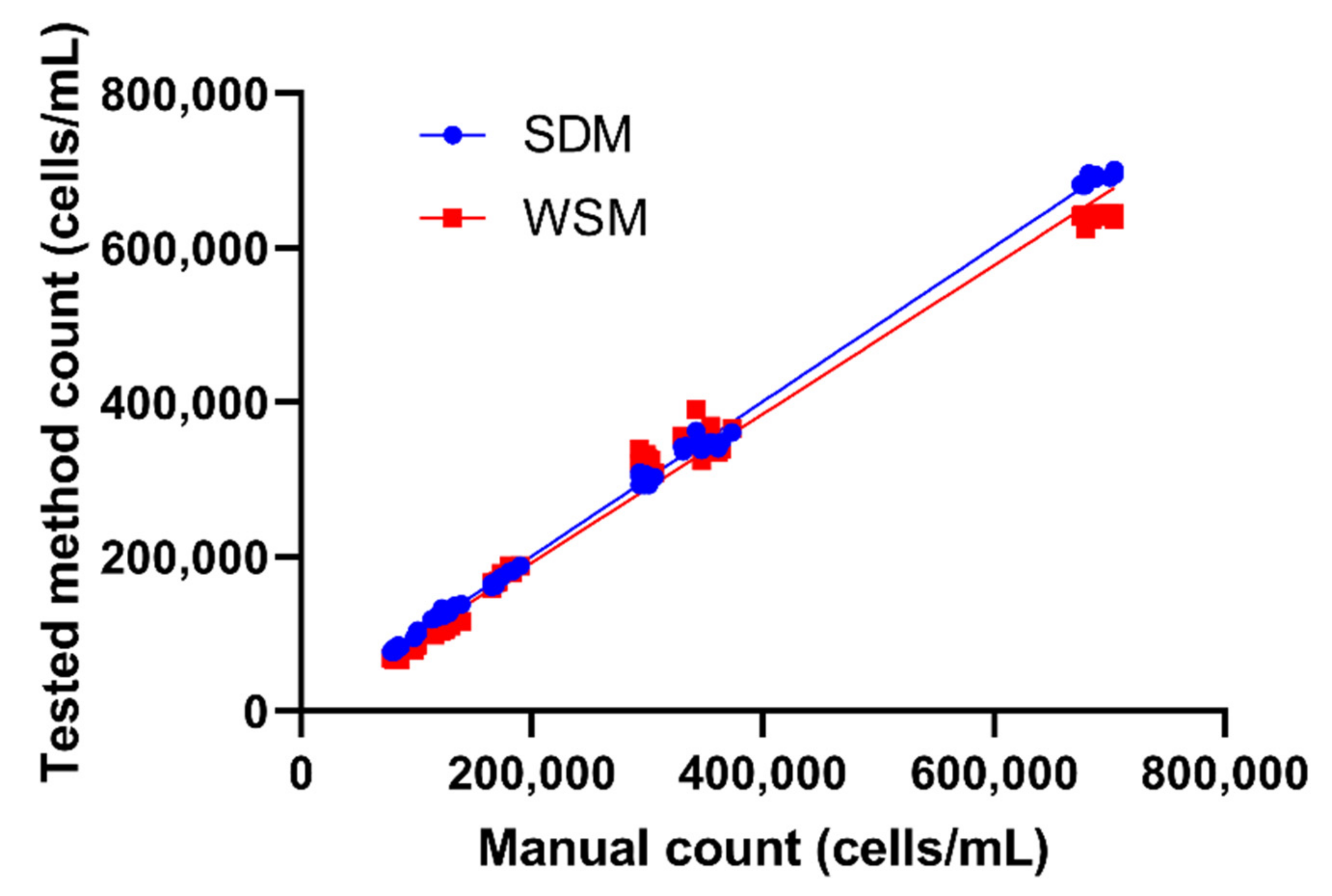
2.3. Effect of Various Tetrahymena Density on SDM and WSM Counting Performance
3. Discussion
4. Material and Methods
4.1. Tetrahymena Cell Culture and Maintenance
4.2. Tetrahymena Recording
4.3. Image Processing
4.4. Find Maxima Method (FMM)
4.5. Particle Analyzer Method (PAM)
4.6. Watershed Segmentation Method (WSM)
4.7. Trainable WEKA Segmentation Method (TWS)
4.8. StarDist Method (SDM)
4.9. Manual Counting
4.10. Sensitivity Calculation
4.11. Statistics and Reproducibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Gilron, G.L.; Lynn, D.H. Ciliated protozoa as test organisms in toxicity assessments. In Microscale Testing in Aquatic Toxicology; CRC Press: Boca Raton, FL, USA, 2018; pp. 323–336. [Google Scholar]
- Maurya, R.; Pandey, A.K. Importance of protozoa Tetrahymena in toxicological studies: A review. Sci. Total Environ. 2020, 741, 140058. [Google Scholar] [CrossRef] [PubMed]
- Sauvant, N.; Pepin, D.; Piccinni, E. Tetrahymena pyriformis: A tool for toxicological studies. A review. Chemosphere 1999, 38, 1631–1669. [Google Scholar] [CrossRef]
- Nilsson, J.R. Tetrahymena in cytotoxicology: With special reference to effects of heavy metals and selected drugs. Eur. J. Protistol. 1989, 25, 2–25. [Google Scholar] [CrossRef]
- Pauli, W.; Jax, K.; Berger, S. Protozoa in wastewater treatment: Function and importance. In Biodegradation and Persistence; Springer: Berlin/Heidelberg, Germany, 2001; pp. 203–252. [Google Scholar]
- Duhra, J.K.; Fan, L.; Gill, V.S.; Lee, J.J.; Yeon, J. The effect of temperature on the population size of Tetrahymena thermophila. Expedition 2013, 3. Available online: https://ojs.library.ubc.ca/index.php/expedition/article/view/184817/ (accessed on 21 May 2022).
- Üstüntanır Dede, A.F.; Arslanyolu, M. The in vivo Tetrahymena thermophila extracellular glucose drop assay for characterization of mammalian insulin activity. Eur. J. Protistol. 2021, 79, 125803. [Google Scholar] [CrossRef] [PubMed]
- Vembadi, A.; Menachery, A.; Qasaimeh, M.A. Cell cytometry: Review and perspective on biotechnological advances. Front. Bioeng. Biotechnol. 2019, 7, 147. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Standridge, J.H.; Prieve, A.F.; Cucunato, J.C.; Bernhardt, M. Using flow cytometry to detect protozoa. J. Am. Water Work. Assoc. 1997, 89, 104–111. [Google Scholar] [CrossRef]
- Grishagin, I.V. Automatic cell counting with imagej. Anal. Biochem. 2015, 473, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Arena, E.T.; Rueden, C.T.; Hiner, M.C.; Wang, S.; Yuan, M.; Eliceiri, K.W. Quantitating the cell: Turning images into numbers with imagej. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e260. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.; Vijayalakshmi, A. Affordable image analysis using nih image/imagej. Indian J. Cancer 2004, 41, 47. [Google Scholar] [PubMed]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable weka segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Lormand, C.; Zellmer, G.F.; Németh, K.; Kilgour, G.; Mead, S.; Palmer, A.S.; Sakamoto, N.; Yurimoto, H.; Moebis, A. Weka trainable segmentation plugin in imagej: A semi-automatic tool applied to crystal size distributions of microlites in volcanic rocks. Microsc. Microanal. 2018, 24, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The imagej ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortada, M.; Sauteur, L.; Lanz, M.; Levano, S.; Bodmer, D. A deep learning approach to quantify auditory hair cells. Hear. Res. 2021, 409, 108317. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Gene, M. Cell detection with star-convex polygons. In Proceedings of the 21st International Conference on Medical Image Computing and Computer Assisted Intervention {MICCAI} 2018, Granada, Spain, 16–20 September 2018; pp. 265–273. [Google Scholar]
- Yonghua, Y.; Jian, Y.A.O.; Xiaomei, H.U.A. Effect of pesticide pollution against functional microbial diversity in soil. Wei Sheng Wu Xue Za Zhi 2000, 20, 23–25. [Google Scholar]
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Singapore, 2017; pp. 43–84. [Google Scholar]
- Omana-Zapata, I.; Mutschmann, C.; Schmitz, J.; Gibson, S.; Judge, K.; Aruda Indig, M.; Lu, B.; Taufman, D.; Sanfilippo, A.M.; Shallenberger, W. Accurate and reproducible enumeration of t-, b-, and nk lymphocytes using the bd facslyric 10-color system: A multisite clinical evaluation. PLoS ONE 2019, 14, e0211207. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.; Crichton, S.O.J.; Kulig, B.; Kiesel, B.; Hensel, O.; Sturm, B. Comparative analysis of methods and model prediction performance evaluation for continuous online non-invasive quality assessment during drying of apples from two cultivars. Therm. Sci. Eng. Prog. 2020, 18, 100461. [Google Scholar] [CrossRef]
- William, W.; Ware, A.; Basaza-Ejiri, A.H.; Obungoloch, J. A pap-smear analysis tool (pat) for detection of cervical cancer from pap-smear images. BioMedical Eng. OnLine 2019, 18, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigert, M.; Schmidt, U.; Haase, R.; Sugawara, K.; Myers, G. Star-Convex Polyhedra for 3d Object Detection and Segmentation in Microscopy. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020. [Google Scholar]
- Cassidy-Hanley, D.M. Tetrahymena in the laboratory: Strain resources, methods for culture, maintenance, and storage. Methods Cell Biol. 2012, 109, 237–276. [Google Scholar] [PubMed] [Green Version]

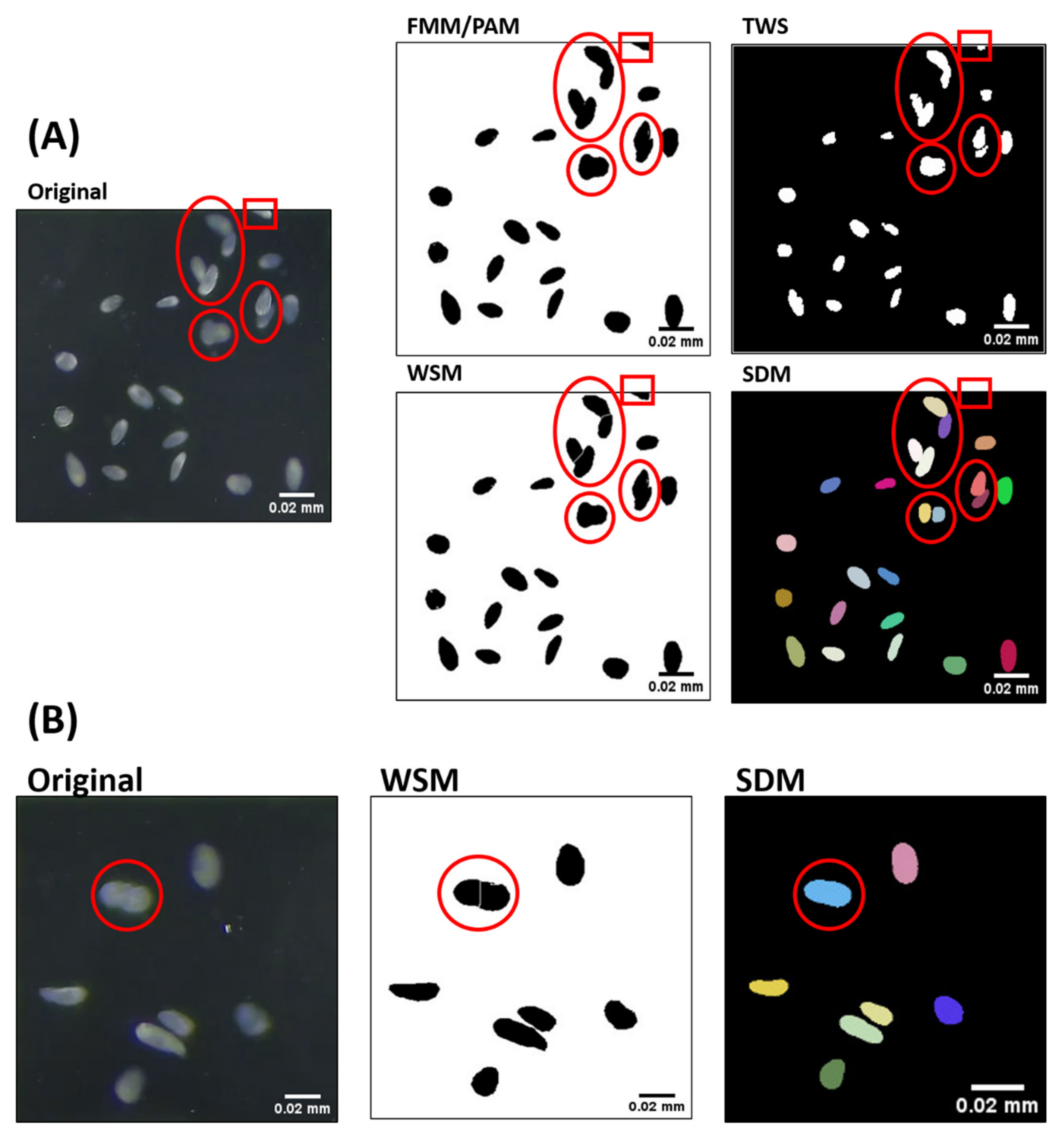
| Method | Cell Count ± SD (Cells/μL) | False Negative | Count Sensitivity | Average Cell Size ± SD (Pixel) | Total Area ± SD (Pixel) |
|---|---|---|---|---|---|
| Manual | 173.2 ± 8.0 | - | - | 1027.76 ± 85.3 | 182,583.7 ± 18,556.0 |
| FMM | 156.4 ± 8.0 | 16.8 | 90.3 ± 2.7% | Not available | Not available |
| PAM | 156.4 ± 8.5 | 16.8 | 90.3 ± 2.7% | 1145.0 ± 105.4 | 179,006.8 ± 18,148.7 |
| WSM | 172.4 ± 9.6 | 0.8 | 99.5 ± 2.3% | 1097.1 ± 105.5 | 187,745.3 ± 19,083.8 |
| TWS | 157.5 ± 8.3 | 15.7 | 91.0 ± 2.9% | 1194.4 ± 81.6 | 187,965.4 ± 14,320.5 |
| SDM | 171.3 ± 8.6 | 1.9 | 98.9 ± 1.1% | 987.9 ± 37.1 | 169,264.4 ± 11,169.2 |
| Group | SDM | WSM | TWS | PAM | FMM |
|---|---|---|---|---|---|
| Slope | 1.073 | 1.226 | 1.041 | 1.071 | 1.071 |
| 95% Lower CL # | 0.8499 | 0.5038 | 0.1019 | 0.1850 | 0.1850 |
| 95% Upper CL | 1.296 | 1.947 | 1.981 | 1.956 | 1.956 |
| y-intercept | −14,537 | −39,866 | −22,865 | −29,011 | −29,011 |
| 95% Lower CL | −53,465 | −162,036 | −183,179 | −179,390 | −179,390 |
| 95% Upper CL | 24,392 | 82,304 | 137,448 | 121,368 | 121,368 |
| p value | <0.0001 (****) | 0.0003 (***) | 0.0054 (**) | 0.0028 (**) | 0.0028 (**) |
| Correlation coefficient (r) | 0.9784 | 0.9096 | 0.8007 | 0.8320 | 0.8320 |
| Group | SDM | WSM |
|---|---|---|
| Slope | 1.002 | 0.963 |
| 95% Lower CL # | 0.9934 | 0.9346 |
| 95% Upper CL | 1.01 | 0.9914 |
| y-intercept | 457.1 | −300 |
| 95% Lower CL | −5825 | −1135 |
| 95% Upper CL | 5225 | 2049 |
| p value | <0.0001 (****) | <0.0001 (****) |
| Correlation coefficient (r) | 0.9934 | 0.9995 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurnia, K.A.; Sampurna, B.P.; Audira, G.; Juniardi, S.; Vasquez, R.D.; Roldan, M.J.M.; Tsao, C.-C.; Hsiao, C.-D. Performance Comparison of Five Methods for Tetrahymena Number Counting on the ImageJ Platform: Assessing the Built-in Tool and Machine-Learning-Based Extension. Int. J. Mol. Sci. 2022, 23, 6009. https://doi.org/10.3390/ijms23116009
Kurnia KA, Sampurna BP, Audira G, Juniardi S, Vasquez RD, Roldan MJM, Tsao C-C, Hsiao C-D. Performance Comparison of Five Methods for Tetrahymena Number Counting on the ImageJ Platform: Assessing the Built-in Tool and Machine-Learning-Based Extension. International Journal of Molecular Sciences. 2022; 23(11):6009. https://doi.org/10.3390/ijms23116009
Chicago/Turabian StyleKurnia, Kevin Adi, Bonifasius Putera Sampurna, Gilbert Audira, Stevhen Juniardi, Ross D. Vasquez, Marri Jmelou M. Roldan, Che-Chia Tsao, and Chung-Der Hsiao. 2022. "Performance Comparison of Five Methods for Tetrahymena Number Counting on the ImageJ Platform: Assessing the Built-in Tool and Machine-Learning-Based Extension" International Journal of Molecular Sciences 23, no. 11: 6009. https://doi.org/10.3390/ijms23116009
APA StyleKurnia, K. A., Sampurna, B. P., Audira, G., Juniardi, S., Vasquez, R. D., Roldan, M. J. M., Tsao, C.-C., & Hsiao, C.-D. (2022). Performance Comparison of Five Methods for Tetrahymena Number Counting on the ImageJ Platform: Assessing the Built-in Tool and Machine-Learning-Based Extension. International Journal of Molecular Sciences, 23(11), 6009. https://doi.org/10.3390/ijms23116009









