Structural and Functional Insights into GID/CTLH E3 Ligase Complexes
Abstract
1. Introduction
2. The Basics: GID/CTLH Complex Composition, Characteristics, and Conservation
2.1. Composition and Conservation
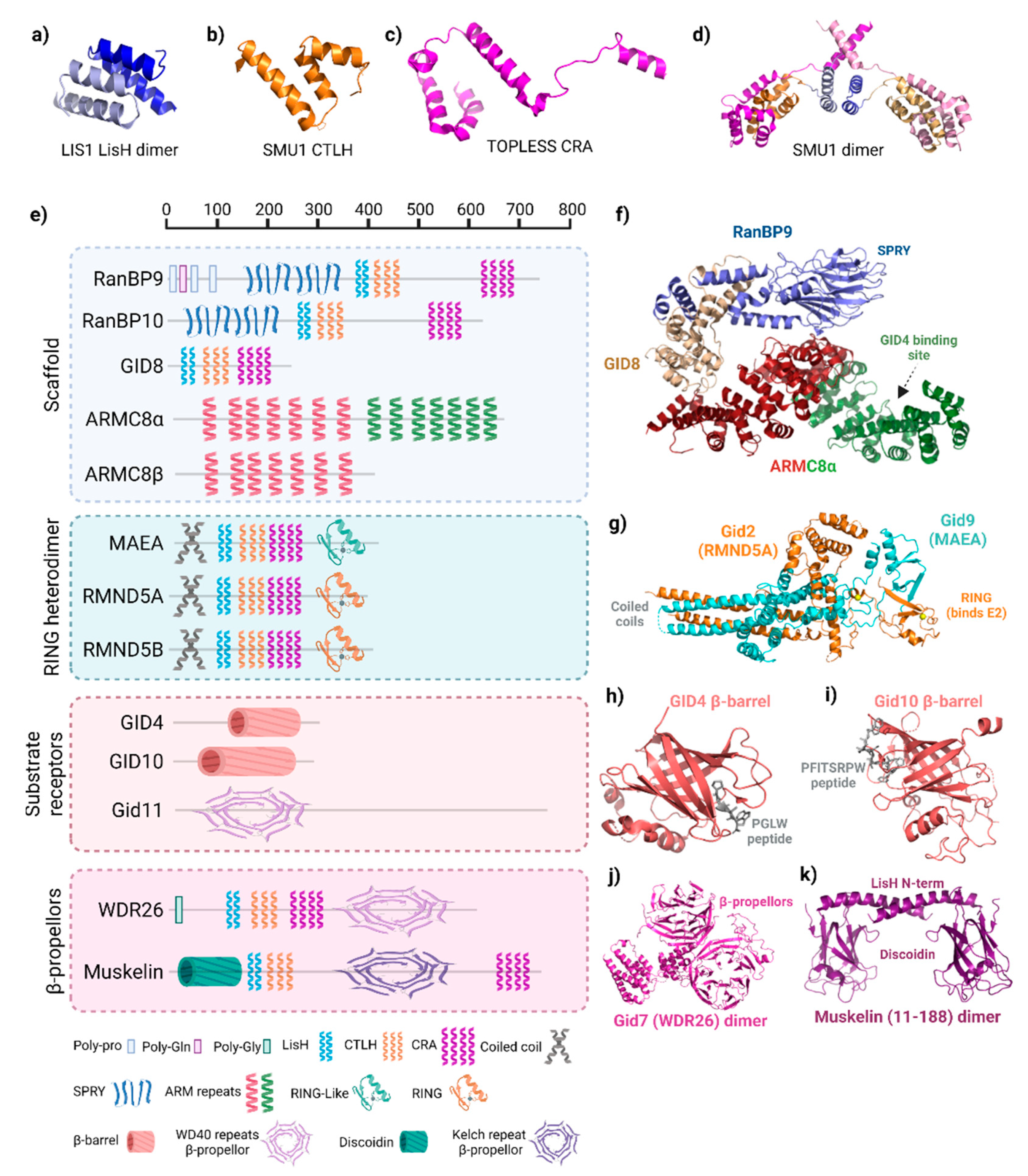
2.2. Domains in the GID/CTLH Complex: Structure and Roles in Complex Activity, Formation, or Substrate Engagement
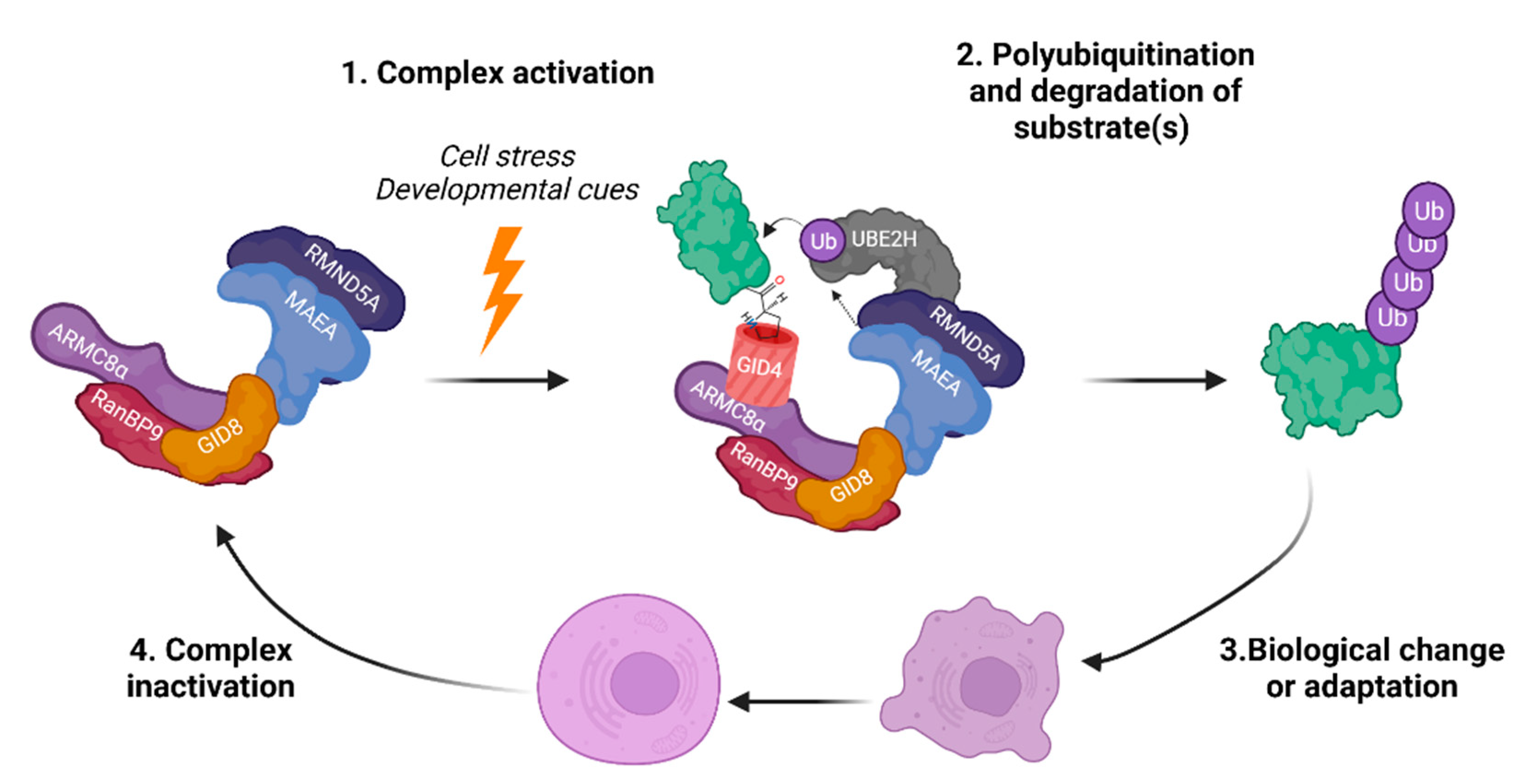
3. Complex Architecture and Activity
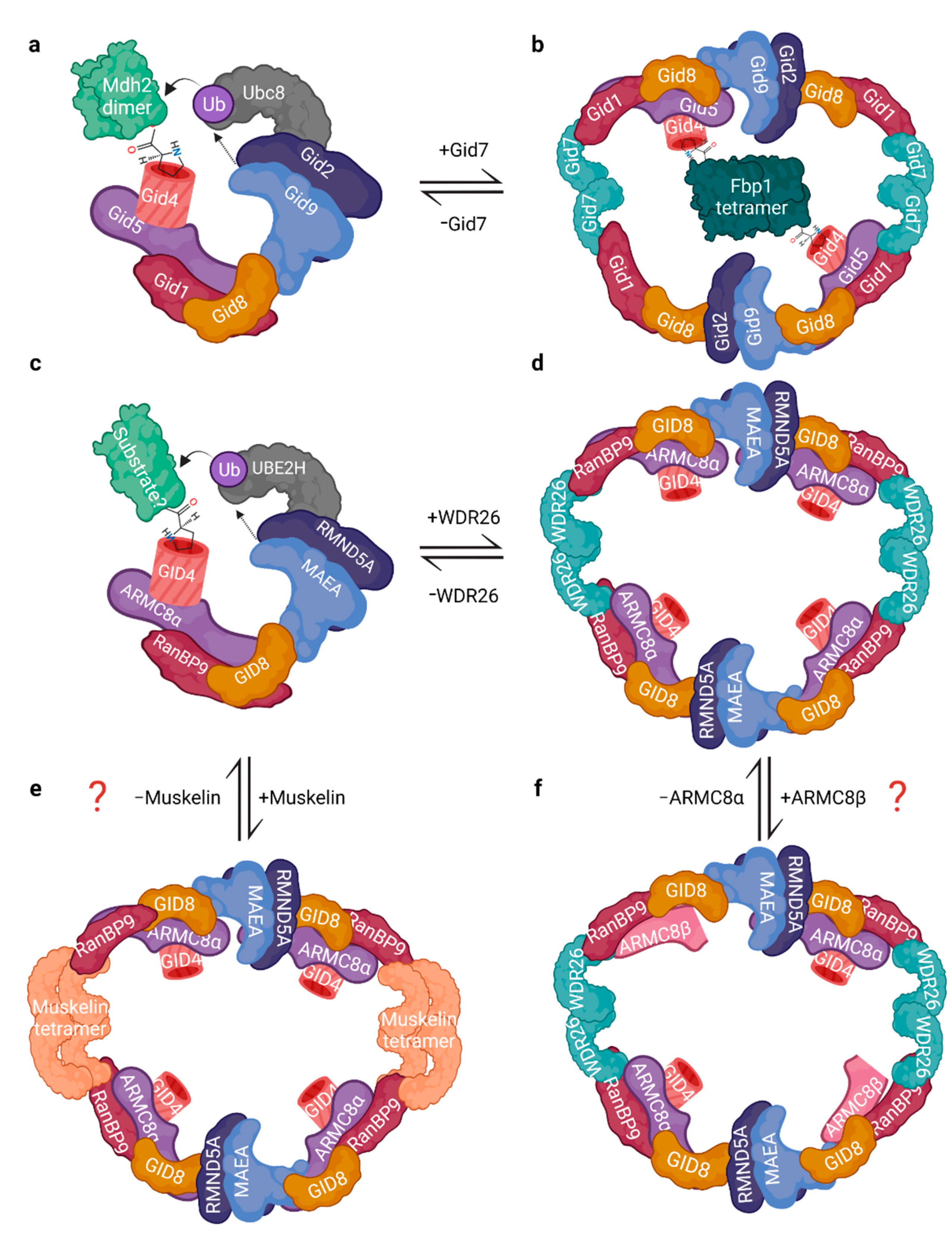
3.1. Architecture and Interactions
3.2. The RING Heterodimer: Structure and Activity
4. What Started It All: Functions of the Yeast GID Complex in Glucose Metabolism and Beyond
4.1. Catabolite Inactivation
4.2. GID-Mediated Proteasomal Degradation of Gluconeogenic Enzymes
4.3. GID-Mediated Vacuole Degradation of Gluconeogenic Enzymes
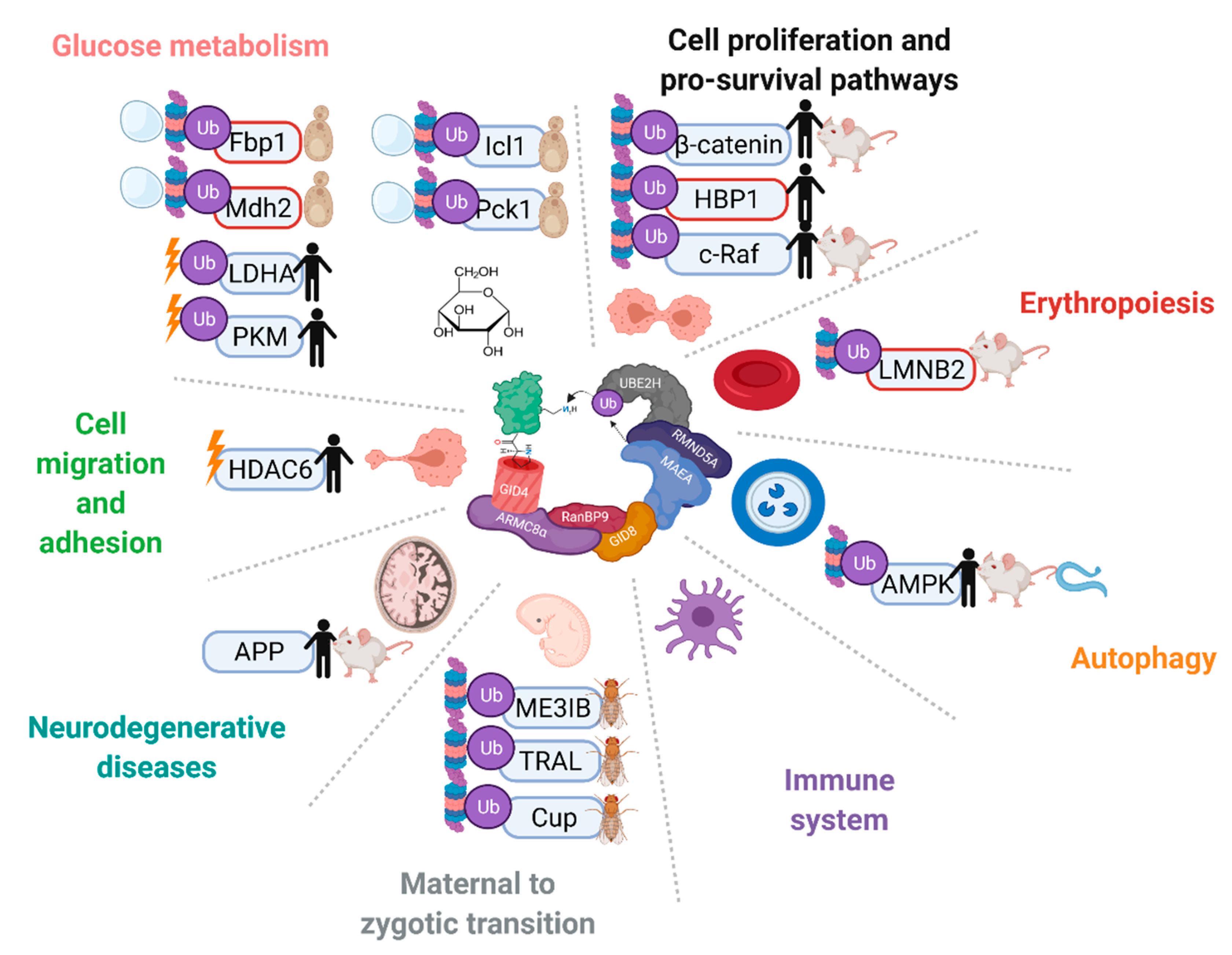
4.4. Functions and Targets beyond Catabolite Inactivation
5. Functions and Ubiquitination Targets of the CTLH Complex from Drosophila to Humans
5.1. Differentiation and Development
5.2. Cell Migration and Adhesion
5.3. Nuclear Functions
5.4. Cell Proliferation, Death, and Survival Pathways
5.5. Functions and Disease Implications in the Central Nervous System
5.6. Immune System
5.7. Endocytosis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Chaugule, V.K.; Walden, H. Specificity and Disease in the Ubiquitin System. Biochem. Soc. Trans. 2016, 44, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Sherpa, D.; Chrustowicz, J.; Schulman, B.A. How the Ends Signal the End: Regulation by E3 Ubiquitin Ligases Recognizing Protein Termini. Mol. Cell 2022, 82, 1424–1438. [Google Scholar] [CrossRef]
- Varshavsky, A. N-Degron and C-Degron Pathways of Protein Degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.M.; Scott, D.C.; Calabrese, M.F.; Zimmerman, E.S.; Zheng, N.; Schulman, B.A. Structural Regulation of Cullin-RING Ubiquitin Ligase Complexes. Curr. Opin. Struct. Biol. 2011, 21, 257–264. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and Regulation of Cullin-RING Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Schapira, M.; Calabrese, M.F.; Bullock, A.N.; Crews, C.M. Targeted Protein Degradation: Expanding the Toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. [Google Scholar] [CrossRef]
- Chen, S.J.; Wu, X.; Wadas, B.; Oh, J.H.; Varshavsky, A. An N-End Rule Pathway That Recognizes Proline and Destroys Gluconeogenic Enzymes. Science 2017, 355, eaal3655. [Google Scholar] [CrossRef]
- Santt, O.; Pfirrmann, T.; Braun, B.; Juretschke, J.; Kimmig, P.; Scheel, H.; Hofmann, K.; Thumm, M.; Wolf, D.H. The Yeast GID Complex, a Novel Ubiquitin Ligase (E3) Involved in the Regulation of Carbohydrate Metabolism. Mol. Biol. Cell 2008, 19, 3323–3333. [Google Scholar] [CrossRef]
- Francis, O.; Han, F.; Adams, J.C. Molecular Phylogeny of a RING E3 Ubiquitin Ligase, Conserved in Eukaryotic Cells and Dominated by Homologous Components, the Muskelin/RanBPM/CTLH Complex. PLoS ONE 2013, 8, e75217. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Langlois, C.R.; Chrustowicz, J.; Sherpa, D.; Karayel, O.; Hansen, F.M.; Beier, V.; von Gronau, S.; Bollschweiler, D.; Schäfer, T.; et al. Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly. Mol. Cell 2020, 77, 150–163.e9. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.Y.E.; Fischer, B.; Meurer, M.; Kats, I.; Li, Z.; Rühle, F.; Barry, J.D.; Kirrmaier, D.; Chevyreva, V.; San Luis, B.J.; et al. Timer-Based Proteomic Profiling of the Ubiquitin-Proteasome System Reveals a Substrate Receptor of the GID Ubiquitin Ligase. Mol. Cell 2021, 81, 2460–2476.e11. [Google Scholar] [CrossRef] [PubMed]
- Melnykov, A.; Chen, S.J.; Varshavsky, A. Gid10 as an Alternative N-Recognin of the Pro/N-Degron Pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 15914–15923. [Google Scholar] [CrossRef] [PubMed]
- Sherpa, D.; Chrustowicz, J.; Qiao, S.; Langlois, C.R.; Hehl, L.A.; Gottemukkala, K.V.; Hansen, F.M.; Karayel, O.; von Gronau, S.; Prabu, J.R.; et al. GID E3 Ligase Supramolecular Chelate Assembly Configures Multipronged Ubiquitin Targeting of an Oligomeric Metabolic Enzyme. Mol. Cell 2021, 81, 2445–2459.e13. [Google Scholar] [CrossRef]
- Lampert, F.; Stafa, D.; Goga, A.; Soste, M.V.; Gilberto, S.; Olieric, N.; Picotti, P.; Stoffel, M.; Peter, M. The Multi-Subunit GID/CTLH E3 Ubiquitin Ligase Promotes Cell Proliferation and Targets the Transcription Factor Hbp1 for Degradation. ELife 2018, 7, e35528. [Google Scholar] [CrossRef]
- Hosono, K.; Noda, S.; Shimizu, A.; Nakanishi, N.; Ohtsubo, M.; Shimizu, N.; Minoshima, S. YPEL5 Protein of the YPEL Gene Family Is Involved in the Cell Cycle Progression by Interacting with Two Distinct Proteins RanBPM and RanBP10. Genomics 2010, 96, 102–111. [Google Scholar] [CrossRef]
- Tomaštíková, E.; Cenklová, V.; Kohoutová, L.; Petrovská, B.; Váchová, L.; Halada, P.; Kočárová, G.; Binarová, P. Interactions of an Arabidopsis RanBPM Homologue with LisH-CTLH Domain Proteins Revealed High Conservation of CTLH Complexes in Eukaryotes. BMC Plant Biol. 2012, 12, 83. [Google Scholar] [CrossRef]
- Ho, Y.; Gruhler, A.; Heilbut, A.; Bader, G.D.; Moore, L.; Adams, S.L.; Millar, A.; Taylor, P.; Bennett, K.; Boutilier, K.; et al. Systematic Identification of Protein Complexes in Saccharomyces Cerevisiae by Mass Spectrometry. Nature 2002, 415, 180–183. [Google Scholar] [CrossRef]
- Mohamed, W.I.; Park, S.L.; Rabl, J.; Leitner, A.; Boehringer, D.; Peter, M. The Human GID Complex Engages Two Independent Modules for Substrate Recruitment. EMBO Rep. 2021, 22, e52981. [Google Scholar] [CrossRef]
- Gul, I.S.; Hulpiau, P.; Sanders, E.; Van Roy, F.; Van Hengel, J. Armc8 Is an Evolutionarily Conserved Armadillo Protein Involved in Cell–Cell Adhesion Complexes through Multiple Molecular Interactions. Biosci. Rep. 2019, 39, BSR20180604. [Google Scholar] [CrossRef] [PubMed]
- Maitland, M.E.R.; Onea, G.; Chiasson, C.A.; Wang, X.; Ma, J.; Moor, S.E.; Barber, K.R.; Lajoie, G.A.; Shaw, G.S.; Schild-Poulter, C. The Mammalian CTLH Complex Is an E3 Ubiquitin Ligase That Targets Its Subunit Muskelin for Degradation. Sci. Rep. 2019, 9, 9864. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Yang, J.; Ueda, A.; Suzuki, T.; Tomaru, K.; Takeno, M.; Okuda, K.; Ishigatsubo, Y. RanBPM, Muskelin, P48EMLP, P44CTLH, and the Armadillo-Repeat Proteins ARMC8α and ARMC8β Are Components of the CTLH Complex. Gene 2007, 396, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Cooper, D.R.; Oleksy, A.; Devedjiev, Y.; Derewenda, U.; Reiner, O.; Otlewski, J.; Derewenda, Z.S. The Structure of the N-Terminal Domain of the Product of the Lissencephaly Gene Lis1 and Its Functional Implications. Structure 2004, 12, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, A.K.C.; Schulz, J.F.; Kamprad, A.; Schütze, T.; Wahl, M.C. Structural Basis for the Functional Coupling of the Alternative Splicing Factors Smu1 and RED. Structure 2016, 24, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Martin-Arevalillo, R.; Nanao, M.H.; Larrieu, A.; Vinos-Poyo, T.; Mast, D.; Galvan-Ampudia, C.; Brunoud, G.; Vernoux, T.; Dumas, R.; Parcy, F. Structure of the Arabidopsis TOPLESS Corepressor Provides Insight into the Evolution of Transcriptional Repression. Proc. Natl. Acad. Sci. USA 2017, 114, 8107–8112. [Google Scholar] [CrossRef]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural Basis for Recognition of Diverse Transcriptional Repressors by the TOPLESS Family of Corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef]
- Wang, D.; Li, Z.; Schoen, S.R.; Messing, E.M.; Wu, G. A Novel MET-Interacting Protein Shares High Sequence Similarity with RanBPM, but Fails to Stimulate MET-Induced Ras/Erk Signaling. Biochem. Biophys. Res. Commun. 2004, 313, 320–326. [Google Scholar] [CrossRef]
- Hong, S.K.; Kim, K.H.; Song, E.J.; Kim, E.E.K. Structural Basis for the Interaction between the IUS-SPRY Domain of RanBPM and DDX-4 in Germ Cell Development. J. Mol. Biol. 2016, 428, 4330–4344. [Google Scholar] [CrossRef]
- Salemi, L.M.; Maitland, M.E.R.; McTavish, C.J.; Schild-Poulter, C. Cell Signalling Pathway Regulation by RanBPM: Molecular Insights and Disease Implications. Open Biol. 2017, 7, 170081. [Google Scholar] [CrossRef]
- Tewari, R.; Bailes, E.; Bunting, K.A.; Coates, J.C. Armadillo-Repeat Protein Functions: Questions for Little Creatures. Trends Cell Biol. 2010, 20, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Budhidarmo, R.; Nakatani, Y.; Day, C.L. RINGs Hold the Key to Ubiquitin Transfer. Trends Biochem. Sci. 2012, 37, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 Heterodimeric RING-RING Complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.; Pfirrmann, T.; Menssen, R.; Hofmann, K.; Scheel, H.; Wolf, D.H. Gid9, a Second RING Finger Protein Contributes to the Ubiquitin Ligase Activity of the Gid Complex Required for Catabolite Degradation. FEBS Lett. 2011, 585, 3856–3861. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zhang, H.; Li, L.; Tempel, W.; Loppnau, P.; Min, J. Molecular Basis of GID4-Mediated Recognition of Degrons for the Pro/N-End Rule Pathway Article. Nat. Chem. Biol. 2018, 14, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Park, S.H.; Kim, L.; Heo, J.; Song, H.K. Crystal Structure of Yeast Gid10 in Complex with Pro/N-Degron. Biochem. Biophys. Res. Commun. 2021, 582, 86–92. [Google Scholar] [CrossRef]
- Chrustowicz, J.; Sherpa, D.; Teyra, J.; Loke, M.S.; Popowicz, G.M.; Basquin, J.; Sattler, M.; Prabu, J.R.; Sidhu, S.S.; Schulman, B.A. Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases. J. Mol. Biol. 2022, 434, 167347. [Google Scholar] [CrossRef]
- Dong, C.; Chen, S.J.; Melnykov, A.; Weirich, S.; Sun, K.; Jeltsch, A.; Varshavsky, A.; Min, J. Recognition of Nonproline N-Terminal Residues by the Pro/N-Degron Pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 14158–14167. [Google Scholar] [CrossRef]
- Langlois, C.R.; Beier, V.; Karayel, O.; Chrustowicz, J.; Sherpa, D.; Mann, M.; Schulman, B.A. A GID E3 Ligase Assembly Ubiquitinates an Rsp5 E3 Adaptor and Regulates Plasma Membrane Transporters. EMBO Rep. 2022, e53835. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Schapira, M.; Tyers, M.; Torrent, M.; Arrowsmith, C.H. WD40 Repeat Domain Proteins: A Novel Target Class? Nat. Rev. Drug Discov. 2017, 16, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Delto, C.F.F.; Heisler, F.F.F.; Kuper, J.; Sander, B.; Kneussel, M.; Schindelin, H. The LisH Motif of Muskelin Is Crucial for Oligomerization and Governs Intracellular Localization. Structure 2015, 23, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Hong, S.K.; Hwang, K.Y.; Kim, E.E. Biological Crystallography Structure of Mouse Muskelin Discoidin Domain and Biochemical Characterization of Its Self-Association. Res. Pap. Acta Cryst. 2014, 70, 2863–2874. [Google Scholar] [CrossRef]
- Kiedzierska, A.; Smietana, K.; Czepczynska, H.; Otlewski, J. Structural Similarities and Functional Diversity of Eukaryotic Discoidin-like Domains. Biochimica Biophysica Acta (BBA)-Proteins Proteom. 2007, 1774, 1069–1078. [Google Scholar] [CrossRef]
- Zavortink, M.; Rutt, L.N.; Dzitoyeva, S.; Henriksen, J.C.; Barrington, C.; Bilodeau, D.Y.; Wang, M.; Chen, X.X.L.; Rissland, O.S. The E2 Marie Kondo and the Ctlh E3 Ligase Clear Deposited Rna Binding Proteins during the Maternal-to-Zygotic Transition. ELife 2020, 9, e53889. [Google Scholar] [CrossRef]
- Menssen, R.; Schweiggert, J.; Schreiner, J.; Kušević, D.; Reuther, J.; Braun, B.; Wolf, D.H. Exploring the Topology of the Gid Complex, the E3 Ubiquitin Ligase Involved in Catabolite-Induced Degradation of Gluconeogenic Enzymes. J. Biol. Chem. 2012, 287, 25602–25614. [Google Scholar] [CrossRef]
- Sherpa, D.; Müller, J.; Karayel, Ö.; Chrustowicz, J.; Xu, P.; Gottemukkala, K.V.; Baumann, C.; Gross, A.; Czarnezki, O.; Zhang, W.; et al. Differential UBE2H-CTLH E2-E3 Ubiquitylation Modules Regulate Erythroid Maturation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Prag, S.; de Arcangelis, A.; Georges-Labouesse, E.; Adams, J.C. Regulation of Post-Translational Modifications of Muskelin by Protein Kinase C. Int. J. Biochem. Cell Biol. 2007, 39, 366–378. [Google Scholar] [CrossRef]
- Palmieri, D.; Scarpa, M.; Tessari, A.; Uka, R.; Amari, F.; Lee, C.; Richmond, T.; Foray, C.; Sheetz, T.; Braddom, A.; et al. Ran Binding Protein 9 (RanBP9) Is a Novel Mediator of Cellular DNA Damage Response in Lung Cancer Cells. Oncotarget 2016, 7, 18371–18383. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and Recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Pfirrmann, T.; Villavicencio-Lorini, P.; Subudhi, A.K.; Menssen, R.; Wolf, D.H.; Hollemann, T. RMND5 from Xenopus Laevis Is an E3 Ubiquitin-Ligase and Functions in Early Embryonic Forebrain Development. PLoS ONE 2015, 10, e0120342. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Zhu, H.; Gou, H.; Fu, W.; Liu, L.; Chen, T.; Ke, D.; Kang, H.; Xie, Q.; Hong, Z.; et al. A Ubiquitin Ligase of Symbiosis Receptor Kinase Involved in Nodule Organogenesis. Plant Physiol. 2012, 160, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 Enzymes: More than Just Middle Men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1-BARD1 Ubiquitin Ligase Activity Counteracts Chromatin Barriers to DNA Resection. Nat. Struct. Mol. Biol. 2016, 23, 647–655. [Google Scholar] [CrossRef]
- Kaiser, P.; Seufert, W.; Höfferer, L.; Kofler, B.; Sachsenmaier, C.; Herzog, H.; Jentsch, S.; Schweiger, M.; Schneider, R. A Human Ubiquitin-Conjugating Enzyme Homologous to Yeast UBC8. J. Biol. Chem. 1994, 269, 8797–8802. [Google Scholar] [CrossRef]
- van Wijk, S.J.L.; de Vries, S.J.; Kemmeren, P.; Huang, A.; Boelens, R.; Bonvin, A.M.J.J.; Timmers, H.T.M. A Comprehensive Framework of E2–RING E3 Interactions of the Human Ubiquitin–Proteasome System. Mol. Syst. Biol. 2009, 5, 295. [Google Scholar] [CrossRef]
- Holzer, H. Catabolite Inactivation in Yeast. Trends Biochem. Sci. 1976, 1, 178–181. [Google Scholar] [CrossRef]
- Schork, S.M.; Thumm, M.; Wolf, D.H. Catabolite Inactivation of Fructose-1,6-Bisphosphatase of Saccharomyces Cerevisiae: Degradation Occurs via the Ubiquitin Pathway. J. Biol. Chem. 1995, 270, 26446–26450. [Google Scholar] [CrossRef]
- Hämmerle, M.; Bauer, J.; Rose, M.; Szallies, A.; Thumm, M.; Düsterhus, S.; Mecke, D.; Entian, K.D.; Wolf, D.H. Proteins of Newly Isolated Mutants and the Amino-Terminal Proline Are Essential for Ubiquitin-Proteasome-Catalyzed Catabolite Degradation of Fructose-1,6-Bisphosphatase of Saccharomyces Cerevisiae. J. Biol. Chem. 1998, 273, 25000–25005. [Google Scholar] [CrossRef]
- Schork, S.M.; Bee, G.; Thumm, M.; Wolf, D.H. Catabolite Inactivation of Fructose-1,6-Bisphosphatase in Yeast Is Mediated by the Proteasome. FEBS Lett. 1994, 349, 270–274. [Google Scholar] [CrossRef]
- Chiang, H.L.; Schekman, R. Regulated Import and Degradation of a Cytosolic Protein in the Yeast Vacuole. Nature 1991, 350, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Davis, C.; Broach, J.R. Efficient Transition to Growth on Fermentable Carbon Sources in Saccharomyces Cerevisiae Requires Signaling through the Ras Pathway. EMBO J. 1998, 17, 6942–6951. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.L.; Schekman, R.; Hamamoto, S. Selective Uptake of Cytosolic, Peroxisomal, and Plasma Membrane Proteins into the Yeast Lysosome for Degradation. J. Biol. Chem. 1996, 271, 9934–9941. [Google Scholar] [CrossRef]
- Regelmann, J.; Schü Le, T.; Josupeit, F.S.; Horak, J.; Rose, M.; Entian, K.-D.; Thumm, M.; Wolf, D.H. Catabolite Degradation of Fructose-1,6-Bisphosphatase in the Yeast Saccharomyces Cerevisiae: A Genome-Wide Screen Identifies Eight Novel GID Genes and Indicates the Existence of Two Degradation Pathways. Mol. Biol. Cell 2003, 14, 1652–1663. [Google Scholar] [CrossRef]
- Horak, J.; Regelmann, J.; Wolf, D.H. Two Distinct Proteolytic Systems Responsible for Glucose-Induced Degradation of Fructose-1,6-Bisphosphatase and the Gal2p Transporter in the Yeast Saccharomyces Cerevisiae Share the Same Protein Components of the Glucose Signaling Pathway. J. Biol. Chem. 2002, 277, 8248–8254. [Google Scholar] [CrossRef]
- Hung, G.C.; Brown, C.R.; Wolfe, A.B.; Liu, J.; Chiang, H.L. Degradation of the Gluconeogenic Enzymes Fructose-1,6-Bisphosphatase and Malate Dehydrogenase Is Mediated by Distinct Proteolytic Pathways and Signaling Events. J. Biol. Chem. 2004, 279, 49138–49150. [Google Scholar] [CrossRef]
- Brown, C.R.; Wolfe, A.B.; Cui, D.; Chiang, H.L. The Vacuolar Import and Degradation Pathway Merges with the Endocytic Pathway to Deliver Fructose-1,6-Bisphosphatase to the Vacuole for Degradation. J. Biol. Chem. 2008, 283, 26116–26127. [Google Scholar] [CrossRef]
- Hoffman, M.; Chiang, H.L. Isolation of Degradation-Deficient Mutants Defective in the Targeting of Fructose-1,6-Bisphosphatase into the Vacuole for Degradation in Saccharomyces Cerevisiae. Genetics 1996, 143, 1555–1566. [Google Scholar] [CrossRef]
- Brown, C.R.; Hung, G.C.; Dunton, D.; Chiang, H.L. The TOR Complex 1 Is Distributed in Endosomes and in Retrograde Vesicles That Form from the Vacuole Membrane and Plays an Important Role in the Vacuole Import and Degradation Pathway. J. Biol. Chem. 2010, 285, 23359–23370. [Google Scholar] [CrossRef]
- Karayel, O.; Michaelis, A.C.; Mann, M.; Schulman, B.A.; Langlois, C.R. DIA-Based Systems Biology Approach Unveils E3 Ubiquitin Ligase-Dependent Responses to a Metabolic Shift. Proc. Natl. Acad. Sci. USA 2020, 117, 32806–32815. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Kim, L.; Song, H.K.; Varshavsky, A. Aminopeptidases Trim Xaa-Pro Proteins, Initiating Their Degradation by the Pro/N-Degron Pathway. Proc. Natl. Acad. Sci. USA 2021, 118, e2115430118. [Google Scholar] [CrossRef] [PubMed]
- Menssen, R.; Bui, K.; Wolf, D.H. Regulation of the Gid Ubiquitin Ligase Recognition Subunit Gid4. FEBS Lett. 2018, 592, 3286–3294. [Google Scholar] [CrossRef] [PubMed]
- Barbin, L.; Eisele, F.; Santt, O.; Wolf, D.H. The Cdc48-Ufd1-Npl4 Complex Is Central in Ubiquitin-Proteasome Triggered Catabolite Degradation of Fructose-1,6-Bisphosphatase. Biochem. Biophys. Res. Commun. 2010, 394, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Alibhoy, A.A.; Giardina, B.J.; Dunton, D.D.; Chiang, H.L. Vps34p Is Required for the Decline of Extracellular Fructose-1,6- Bisphosphatase in the Vacuole Import and Degradation Pathway. J. Biol. Chem. 2012, 287, 33080–33093. [Google Scholar] [CrossRef]
- Giardina, B.J.; Chiang, H.-L. The Key Gluconeogenic Enzyme Fructose-1,6-Bisphosphatase Is Secreted during Prolonged Glucose Starvation and Is Internalized Following Glucose Re-Feeding via the Non-Classical Secretory and Internalizing Pathways in Saccharomyces Cerevisiae. Plant Signal. Behav. 2013, 8, e24936. [Google Scholar] [CrossRef]
- Giardina, B.J.; Stein, K.; Chiang, H.L. The Endocytosis Gene END3 Is Essential for the Glucose-Induced Rapid Decline of Small Vesicles in the Extracellular Fraction in Saccharomyces Cerevisiae. J. Extracell. Vesicles 2014, 3, 23497. [Google Scholar] [CrossRef]
- Chiang, M.C.; Chiang, H.L. Vid24p, a Novel Protein Localized to the Fructose-1,6-Bisphosphatase- Containing Vesicles, Regulates Targeting of Fructose-1,6-Bisphosphatase from the Vesicles to the Vacuole for Degradation. J. Cell Biol. 1998, 140, 1347–1356. [Google Scholar] [CrossRef]
- Alibhoy, A.A.; Giardina, B.J.; Dunton, D.D.; Chiang, H.L. Vid30 Is Required for the Association of Vid Vesicles and Actin Patches in the Vacuole Import and Degradation Pathway. Autophagy 2012, 8, 29–46. [Google Scholar] [CrossRef]
- Giardina, B.J.; Dunton, D.; Chiang, H.L. VID28 Protein Is Required for the Association of Vacuole Import and Degradation (Vid) Vesicles with Actin Patches and the Retention of Vid Vesicle Proteins in the Intracellular Fraction. J. Biol. Chem. 2013, 288, 11636–11648. [Google Scholar] [CrossRef]
- Huang, P.H.; Chiang, H.L. Identification of Novel Vesicles in the Cytosol to Vacuole Protein Degradation Pathway. J. Cell Biol. 1997, 136, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Shieh, H.L.; Chen, Y.; Brown, C.R.; Chiang, H.L. Biochemical Analysis of Fructose-1,6-Bisphosphatase Import into Vacuole Import and Degradation Vesicles Reveals a Role for UBC1 in Vesicle Biogenesis. J. Biol. Chem. 2001, 276, 10398–10406. [Google Scholar] [CrossRef] [PubMed]
- Juretschke, J.; Menssen, R.; Sickmann, A.; Wolf, D.H. The Hsp70 Chaperone Ssa1 Is Essential for Catabolite Induced Degradation of the Gluconeogenic Enzyme Fructose-1,6-Bisphosphatase. Biochem. Biophys. Res. Commun. 2010, 397, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Randell Brown, C.; McCann, J.A.; Chiang, H.L. The Heat Shock Protein Ssa2p Is Required for Import of Fructose-1,6-Bisphosphatase into Vid Vesicles. J. Cell Biol. 2000, 150, 65–76. [Google Scholar] [CrossRef]
- Lotz, S.K.; Knighton, L.E.; Nitika; Jones, G.W.; Truman, A.W. Not Quite the SSAme: Unique Roles for the Yeast Cytosolic Hsp70s. Curr. Genet. 2019, 65, 1127–1134. [Google Scholar] [CrossRef]
- Sharma, D.; Masison, D.C. Single Methyl Group Determines Prion Propagation and Protein Degradation Activities of Yeast Heat Shock Protein (Hsp)-70 Chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. USA 2011, 108, 13665–13670. [Google Scholar] [CrossRef]
- Chen, S.J.; Melnykov, A.; Varshavsky, A. Evolution of Substrates and Components of the Pro/N-Degron Pathway. Biochemistry 2020, 59, 582–593. [Google Scholar] [CrossRef]
- Sandai, D.; Yin, Z.; Selway, L.; Stead, D.; Walker, J.; Leach, M.D.; Bohovych, I.; Ene, I.V.; Kastora, S.; Budge, S.; et al. The Evolutionary Rewiring of Ubiquitination Targets Has Reprogrammed the Regulation of Carbon Assimilation in the Pathogenic Yeast Candida Albicans. mBio 2012, 3, 1–12. [Google Scholar] [CrossRef]
- Childers, D.S.; Raziunaite, I.; Mol Avelar, G.; Mackie, J.; Budge, S.; Stead, D.; Gow, N.A.R.R.; Lenardon, M.D.; Ballou, E.R.; MacCallum, D.M.; et al. The Rewiring of Ubiquitination Targets in a Pathogenic Yeast Promotes Metabolic Flexibility, Host Colonization and Virulence. PLoS Pathog. 2016, 12, e1005566. [Google Scholar] [CrossRef]
- Snowdon, C.; Hlynialuk, C.; Van Der Merwe, G. Components of the Vid30c Are Needed for the Rapamycin-Induced Degradation of the High-Affinity Hexose Transporter Hxt7p in Saccharomyces Cerevisiae. FEMS Yeast Res. 2008, 8, 204–216. [Google Scholar] [CrossRef]
- Van der Merwe, G.K.; Cooper, T.G.; Van Vuuren, H.J.J. Ammonia Regulates VID30 Expression and Vid30p Function Shifts Nitrogen Metabolism toward Glutamate Formation Especially When Saccharomyces Cerevisiae Is Grown in Low Concentrations of Ammonia. J. Biol. Chem. 2001, 276, 28659–28666. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, C.; van der Merwe, G. Regulation of Hxt3 and Hxt7 Turnover Converges on the Vid30 Complex and Requires Inactivation of the Ras/CAMP/PKA Pathway in Saccharomyces Cerevisiae. PLoS ONE 2012, 7, e50458. [Google Scholar] [CrossRef] [PubMed]
- Zhen, R.; Moo, C.; Zhao, Z.; Chen, M.; Feng, H.; Zheng, X.; Zhang, L.; Shi, J.; Chen, C. Wdr26 Regulates Nuclear Condensation in Developing Erythroblasts. Blood 2020, 135, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ding, J.; Köhnlein, K.; Urban, N.; Ori, A.; Villavicencio-Lorini, P.; Walentek, P.; Klotz, L.O.; Hollemann, T.; Pfirrmann, T. The GID Ubiquitin Ligase Complex Is a Regulator of AMPK Activity and Organismal Lifespan. Autophagy 2020, 16, 1618–1634. [Google Scholar] [CrossRef]
- Maitland, M.E.R.; Kuljanin, M.; Wang, X.; Lajoie, G.A.; Schild-Poulter, C. Proteomic Analysis of Ubiquitination Substrates Reveals a CTLH E3 Ligase Complex-Dependent Regulation of Glycolysis. FASEB J. 2021, 35, e21825. [Google Scholar] [CrossRef]
- Cao, W.X.; Kabelitz, S.; Gupta, M.; Yeung, E.; Lin, S.; Rammelt, C.; Ihling, C.; Pekovic, F.; Low, T.C.H.; Siddiqui, N.U.; et al. Precise Temporal Regulation of Post-Transcriptional Repressors Is Required for an Orderly Drosophila Maternal-to-Zygotic Transition. Cell Rep. 2020, 31, 107783. [Google Scholar] [CrossRef]
- Goto, T.; Matsuzawa, J.; Iemura, S.I.; Natsume, T.; Shibuya, H. WDR26 Is a New Partner of Axin1 in the Canonical Wnt Signaling Pathway. FEBS Lett. 2016, 590, 1291–1303. [Google Scholar] [CrossRef]
- Wei, Q.; Boulais, P.E.; Zhang, D.; Pinho, S.; Tanaka, M.; Frenette, P.S. Maea Expressed by Macrophages, but Not Erythroblasts, Maintains Postnatal Murine Bone Marrow Erythroblastic Islands. Blood 2019, 133, 1222–1232. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef]
- Puverel, S.; Barrick, C.; Dolci, S.; Coppola, V.; Tessarollo, L. RanBPM Is Essential for Mouse Spermatogenesis and Oogenesis. Development 2011, 138, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Tang, C.; Li, J.; Zhang, Y.; Bhetwal, B.P.; Zheng, H.; Yan, W. RAN-Binding Protein 9 Is Involved in Alternative Splicing and Is Critical for Male Germ Cell Development and Male Fertility. PLoS Genet 2014, 10, 1004825. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.W.; Thiruvarangan, M.; Jeong, Y.M.; Lee, M.S.; Maddirevula, S.; Rhee, M.; Bae, Y.K.; Kim, H.G.; Kim, C.H. Mind Bomb-Binding Partner RanBP9 Plays a Contributory Role in Retinal Development. Mol. Cells 2017, 40, 271–279. [Google Scholar] [CrossRef]
- Palavicini, J.P.; Lloyd, B.N.; Hayes, C.D.; Bianchi, E.; Kang, D.E.; Dawson-Scully, K.; Lakshmana, M.K. RanBP9 Plays a Critical Role in Neonatal Brain Development in Mice. PLoS ONE 2013, 8, e66908. [Google Scholar] [CrossRef] [PubMed]
- Skraban, C.M.; Wells, C.F.; Markose, P.; Cho, M.T.; Nesbitt, A.I.; Au, P.Y.B.; Begtrup, A.; Bernat, J.A.; Bird, L.M.; Cao, K.; et al. WDR26 Haploinsufficiency Causes a Recognizable Syndrome of Intellectual Disability, Seizures, Abnormal Gait, and Distinctive Facial Features. Am. J. Hum. Genet. 2017, 101, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Bala, S.; Gwynn, B.; Sahr, K.E.; Peters, L.L.; Hanspal, M. Absence of Erythroblast Macrophage Protein (Emp) Leads to Failure of Erythroblast Nuclear Extrusion. J. Biol. Chem. 2006, 281, 20181–20189. [Google Scholar] [CrossRef] [PubMed]
- Kunert, S.; Meyer, I.; Fleischhauer, S.; Wannack, M.; Fiedler, J.; Shivdasani, R.A.; Schulze, H. The Microtubule Modulator RanBP10 Plays a Critical Role in Regulation of Platelet Discoid Shape and Degranulation. Blood 2009, 114, 5532–5540. [Google Scholar] [CrossRef]
- Meyer, I.; Kunert, S.; Schwiebert, S.; Hagedorn, I.; Italiano, J.E.; Dütting, S.; Nieswandt, B.; Bachmann, S.; Schulze, H. Altered Microtubule Equilibrium and Impaired Thrombus Stability in Mice Lacking RanBP10. Blood 2012, 120, 3594–3602. [Google Scholar] [CrossRef]
- Han, T.; Yang, C.; Chang, K.; Zhang, D.; Imam, F.B.; Rana, T.M. Identification of Novel Genes and Networks Governing Hematopoietic Stem Cell Development. EMBO Rep. 2016, 17, 1814–1828. [Google Scholar] [CrossRef]
- Woo, J.A.; Roh, S.; Lakshmana, M.K.; Kang, D.E. Pivotal Role of RanBP9 in Integrin-dependent Focal Adhesion Signaling and Assembly. FASEB J. 2012, 26, 1672–1681. [Google Scholar] [CrossRef]
- Wei, J.D.; Jang, J.H.; Kim, J.H. RanBPM Inhibits BLT2-Mediated IL-8 Production and Invasiveness in Aggressive Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2017, 483, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C.; Seed, B.; Lawler, J. Muskelin, a Novel Intracellular Mediator of Cell Adhesive and Cytoskeletal Responses to Thrombospondin-1. EMBO J. 1998, 17, 4964–4974. [Google Scholar] [CrossRef]
- Tripathi, B.K.; Lowy, D.R.; Zelenka, P.S. The Cdk5 Activator P39 Specifically Links Muskelin to Myosin II and Regulates Stress Fiber Formation and Actin Organization in Lens. Exp. Cell Res. 2015, 330, 186–198. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valiyaveettil, M.; Bentley, A.A.; Gursahaney, P.; Hussien, R.; Chakravarti, R.; Kureishy, N.; Prag, S.; Adams, J.C. Novel Role of the Muskelin–RanBP9 Complex as a Nucleocytoplasmic Mediator of Cell Morphology Regulation. J. Cell Biol. 2008, 182, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Tang, X.; Sun, Z.; Chen, S. Upregulated WDR26 Serves as a Scaffold to Coordinate PI3K/AKT Pathway-Driven Breast Cancer Cell Growth, Migration, and Invasion. Oncotarget 2016, 7, 17854–17869. [Google Scholar] [CrossRef]
- Hasegawa, M.; Parkos, C.A.; Nusrat, A. WD40 Repeat Protein 26 Negatively Regulates Formyl Peptide Receptor-1 Mediated Wound Healing in Intestinal Epithelial Cells. Am. J. Pathol. 2020, 190, 2029–2038. [Google Scholar] [CrossRef]
- Salemi, L.M.; Maitland, M.E.R.; Yefet, E.R.; Schild-Poulter, C. Inhibition of HDAC6 Activity through Interaction with RanBPM and Its Associated CTLH Complex. BMC Cancer 2017, 17, 460. [Google Scholar] [CrossRef]
- Cox, J.L.; Wilder, P.J.; Gilmore, J.M.; Wuebben, E.L.; Washburn, M.P.; Rizzino, A. The SOX2-Interactome in Brain Cancer Cells Identifies the Requirement of MSI2 and USP9X for the Growth of Brain Tumor Cells. PLoS ONE 2013, 8, e62857. [Google Scholar] [CrossRef]
- Kim, J.S.; He, X.; Liu, J.; Duan, Z.; Kim, T.; Gerard, J.; Kim, B.; Pillai, M.M.; Lane, W.S.; Noble, W.S.; et al. Systematic Proteomics of Endogenous Human Cohesin Reveals an Interaction with Diverse Splicing Factors and RNA-Binding Proteins Required for Mitotic Progression. J. Biol. Chem. 2019, 294, 8760–8772. [Google Scholar] [CrossRef]
- Yatim, A.; Benne, C.; Sobhian, B.; Laurent-Chabalier, S.; Deas, O.; Judde, J.G.; Lelievre, J.D.; Levy, Y.; Benkirane, M. NOTCH1 Nuclear Interactome Reveals Key Regulators of Its Transcriptional Activity and Oncogenic Function. Mol. Cell 2012, 48, 445–458. [Google Scholar] [CrossRef]
- Chen, X.; Lu, D.; Gao, J.; Zhu, H.; Zhou, Y.; Gao, D.; Zhou, H. Identification of a USP9X Substrate NFX1-123 by SILAC-Based Quantitative Proteomics. J. Proteome Res. 2019, 18, 2654–2665. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, S.Y.; Gong, F.; Battenhouse, A.M.; Boutz, D.R.; Bashyal, A.; Refvik, S.T.; Chiang, C.M.; Xhemalce, B.; Paull, T.T.; et al. Systematic Bromodomain Protein Screens Identify Homologous Recombination and R-Loop Suppression Pathways Involved in Genome Integrity. Genes Dev. 2019, 33, 1751–1774. [Google Scholar] [CrossRef] [PubMed]
- Woods, N.T.; Mesquita, R.D.; Sweet, M.; Carvalho, M.A.; Li, X.; Liu, Y.; Nguyen, H.; Thomas, C.E.; Iversen, E.S.; Marsillac, S.; et al. Charting the Landscape of Tandem BRCT Domain-Mediated Protein Interactions. Sci. Signal. 2012, 5, rs6. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P.; Mandl, S.; Schweiger, M.; Schneider, R. Characterization of Functionally Independent Domains in the Human Ubiquitin Conjugating Enzyme UbcH2. FEBS Lett. 1995, 377, 193–196. [Google Scholar] [CrossRef]
- Jones, J.M.; Bhattacharyya, A.; Simkus, C.; Vallieres, B.; Veenstra, T.D.; Zhou, M. The RAG1 V(D)J Recombinase/Ubiquitin Ligase Promotes Ubiquitylation of Acetylated, Phosphorylated Histone 3.3. Immunol. Lett. 2011, 136, 156–162. [Google Scholar] [CrossRef]
- Atabakhsh, E.; Wang, J.H.; Wang, X.; Carter, D.E.; Schild-Poulter, C. RanBPM Expression Regulates Transcriptional Pathways Involved in Development and Tumorigenesis. Am. J. Cancer Res. 2012, 2, 549–565. [Google Scholar]
- Poirier, M.B.; Laflamme, L.; Langlois, M.F. Identification and Characterization of RanBPM, a Novel Coactivator of Thyroid Hormone Receptors. J. Mol. Endocrinol. 2006, 36, 313–325. [Google Scholar] [CrossRef]
- Harada, N.; Yokoyama, T.; Yamaji, R.; Nakano, Y.; Inui, H. RanBP10 Acts as a Novel Coactivator for the Androgen Receptor. Biochem. Biophys. Res. Commun. 2008, 368, 121–125. [Google Scholar] [CrossRef]
- Rao, M.A.; Cheng, H.; Quayle, A.N.; Nishitani, H.; Nelson, C.C.; Rennie, P.S. RanBPM, a Nuclear Protein That Interacts with and Regulates Transcriptional Activity of Androgen Receptor and Glucocorticoid Receptor. J. Biol. Chem. 2002, 277, 48020–48027. [Google Scholar] [CrossRef]
- Yang, Y.C.; Feng, T.H.; Chen, T.Y.; Huang, H.H.; Hung, C.C.; Liu, S.T.; Chang, L.K. RanBPM Regulates Zta-Mediated Transcriptional Activity in Epstein-Barr Virus. J. Gen. Virol. 2015, 96, 2336–2348. [Google Scholar] [CrossRef]
- Chang, L.K.; Liu, S.T.; Kuo, C.W.; Wang, W.H.; Chuang, J.Y.; Bianchi, E.; Hong, Y.R. Enhancement of Transactivation Activity of Rta of Epstein-Barr Virus by RanBPM. J. Mol. Biol. 2008, 379, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Atabakhsh, E.; Schild-Poulter, C. RanBPM Is an Inhibitor of ERK Signaling. PLoS ONE 2012, 7, e47803. [Google Scholar] [CrossRef] [PubMed]
- McTavish, C.J.; Bérubé-Janzen, W.; Wang, X.; Maitland, M.E.R.; Salemi, L.M.; Hess, D.A.; Schild-Poulter, C. Regulation of C-Raf Stability through the CTLH Complex. Int. J. Mol. Sci. 2019, 20, 934. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Okamoto, M.; Terada, T.; Sakurai, F.; Mizuguchi, H.; Tomita, K.; Nishinaka, T. Adenovirus Vector-Mediated Macrophage Erythroblast Attacher (MAEA) Overexpression in Primary Mouse Hepatocytes Attenuates Hepatic Gluconeogenesis. Biochem. Biophys. Rep. 2017, 10, 192–197. [Google Scholar] [CrossRef]
- Leal-Esteban, L.C.; Rothé, B.; Fortier, S.; Isenschmid, M.; Constam, D.B. Role of Bicaudal C1 in Renal Gluconeogenesis and Its Novel Interaction with the CTLH Complex. PLoS Genet. 2018, 14, e1007487. [Google Scholar] [CrossRef]
- De Berardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Feng, Y.; Zhao, J.; Hou, H.; Zhang, H.; Jiao, Y.; Wang, J.; Wang, Y.; Sun, Y. WDR26 Promotes Mitophagy of Cardiomyocytes Induced by Hypoxia through Parkin Translocation. Acta Biochim. Biophys. Sin. 2016, 48, 1075–1084. [Google Scholar] [CrossRef]
- Sato, A.; Shimizu, M.; Goto, T.; Masuno, H.; Kagechika, H.; Tanaka, N.; Shibuya, H. WNK Regulates Wnt Signalling and β-Catenin Levels by Interfering with the Interaction between β-Catenin and GID. Commun. Biol. 2020, 3, 166. [Google Scholar] [CrossRef]
- Anvarian, Z.; Nojima, H.; van Kappel, E.C.; Madl, T.; Spit, M.; Viertler, M.; Jordens, I.; Low, T.Y.; van Scherpenzeel, R.C.; Kuper, I.; et al. Axin Cancer Mutants Form Nanoaggregates to Rewire the Wnt Signaling Network. Nat. Struct. Mol. Biol. 2016, 23, 324–332. [Google Scholar] [CrossRef]
- Hilger, M.; Mann, M. Triple SILAC to Determine Stimulus Specific Interactions in the Wnt Pathway. J. Proteome Res. 2012, 11, 982–984. [Google Scholar] [CrossRef]
- Dansereau, D.A.; Lasko, P. RanBPM Regulates Cell Shape, Arrangement, and Capacity of the Female Germline Stem Cell Niche in Drosophila Melanogaster. J. Cell Biol. 2008, 182, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Atabakhsh, E.; Bryce, D.M.; Lefebvre, K.J.; Schild-Poulter, C. RanBPM Has Proapoptotic Activities That Regulate Cell Death Pathways in Response to Dna Damage. Mol. Cancer Res. 2009, 7, 1962–1972. [Google Scholar] [CrossRef] [PubMed]
- Salemi, L.M.; Almawi, A.W.; Lefebvre, K.J.; Schild-Poulter, C. Aggresome Formation Is Regulated by RanBPM through an Interaction with HDAC6. Biol. Open 2014, 3, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Tessari, A.; Parbhoo, K.; Pawlikowski, M.; Fassan, M.; Rulli, E.; Foray, C.; Fabbri, A.; Embrione, V.; Ganzinelli, M.; Capece, M.; et al. RANBP9 Affects Cancer Cells Response to Genotoxic Stress and Its Overexpression Is Associated with Worse Response to Platinum in NSCLC Patients. Oncogene 2018, 37, 6463–6476. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.A.; Jung, A.R.; Lakshmana, M.K.; Bedrossian, A.; Lim, Y.; Bu, J.H.; Park, S.A.; Koo, E.H.; Mook-Jung, I.; Kang, D.E. Pivotal Role of the RanBP9-Cofilin Pathway in AΒ-Induced Apoptosis and Neurodegeneration. Cell Death Differ. 2012, 19, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Roh, S.E.; Woo, J.A.; Ryu, H.; Kang, D.E. Cooperative Role of RanBP9 and P73 in Mitochondria-Mediated Apoptosis. Cell Death Dis. 2013, 4, e476. [Google Scholar] [CrossRef]
- Dai, H.; Lv, Y.F.; Yan, G.N.; Meng, G.; Zhang, X.; Guo, Q.N. RanBP9/TSSC3 Complex Cooperates to Suppress Anoikis Resistance and Metastasis via Inhibiting Src-Mediated Akt Signaling in Osteosarcoma. Cell Death Dis. 2016, 7, e2572. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, X.; Yuan, Y.; Wang, Y.; Zhou, M.; Li, X.; Zhen, P. MiR-664 Protects Against UVB Radiation-Induced HaCaT Cell Damage via Downregulating ARMC8. Dose-Response 2020, 18. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, C.; Luo, Q.; Wei, X.; Jiang, B.; Zhu, H.; Zhang, L.; Jiang, L.; Liu, M.; Xiao, X. A Novel WD-Repeat Protein, WDR26, Inhibits Apoptosis of Cardiomyocytes Induced by Oxidative Stress. Free. Radic. Res. 2012, 46, 777–784. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Y.; Wei, X.; Yuan, C.; Yuan, X.; Xiao, X. A Novel WD-40 Repeat Protein WDR26 Suppresses H2O2-Induced Cell Death in Neural Cells. Neurosci. Lett. 2009, 460, 66–71. [Google Scholar] [CrossRef]
- Li, H.; Wang, W.; Liu, X.; Paulson, K.E.; Yee, A.S.; Zhang, X. Transcriptional Factor HBP1 Targets P16(INK4A), Upregulating Its Expression and Consequently Is Involved in Ras-Induced Premature Senescence. Oncogene 2010, 29, 5083–5094. [Google Scholar] [CrossRef] [PubMed]
- Huffman, N.; Palmieri, D.; Coppola, V. The CTLH Complex in Cancer Cell Plasticity. J. Oncol. 2019, 2019, 4216750. [Google Scholar] [CrossRef] [PubMed]
- Heisler, F.F.; Loebrich, S.; Pechmann, Y.; Maier, N.; Zivkovic, A.R.; Tokito, M.; Hausrat, T.J.; Schweizer, M.; Bähring, R.; Holzbaur, E.L.F.; et al. Muskelin Regulates Actin Filament- and Microtubule-Based GABAA Receptor Transport in Neurons. Neuron 2011, 70, 66–81. [Google Scholar] [CrossRef]
- Heisler, F.F.; Pechmann, Y.; Wieser, I.; Altmeppen, H.C.; Veenendaal, L.; Muhia, M.; Schweizer, M.; Glatzel, M.; Krasemann, S.; Kneussel, M. Muskelin Coordinates PrPC Lysosome versus Exosome Targeting and Impacts Prion Disease Progression. Neuron 2018, 99, 1155–1169.e9. [Google Scholar] [CrossRef] [PubMed]
- Lakshmana, M.K.; Yoon, I.S.; Chen, E.; Bianchi, E.; Koo, E.H.; Kang, D.E. Novel Role of RanBP9 in BACE1 Processing of Amyloid Precursor Protein and Amyloid β Peptide Generation. J. Biol. Chem. 2009, 284, 11863–11872. [Google Scholar] [CrossRef]
- Lakshmana, M.K.; Hayes, C.D.; Bennett, S.P.; Bianchi, E.; Reddy, K.M.; Koo, E.H.; Kang, D.E. Role of RanBP9 on Amyloidogenic Processing of APP and Synaptic Protein Levels in the Mouse Brain. FASEB J. 2012, 26, 2072–2083. [Google Scholar] [CrossRef]
- Woo, J.A.; Liu, T.; Zhao, X.; Trotter, C.; Yrigoin, K.; Cazzaro, S.; De Narvaez, E.; Khan, H.; Witas, R.; Bukhari, A.; et al. Enhanced Tau Pathology via RanBP9 and Hsp90/Hsc70 Chaperone Complexes. Hum. Mol. Genet. 2017, 26, 3973–3988. [Google Scholar] [CrossRef]
- Lim, K.H.; Joo, J.Y. Predictive Potential of Circulating Ube2h MRNA as an E2 Ubiquitin-Conjugating Enzyme for Diagnosis or Treatment of Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 3398. [Google Scholar] [CrossRef]
- Li, Y.-P.; Lecker, S.H.; Chen, Y.; Waddell, I.D.; Goldberg, A.L.; Reid, M.B. TNF-α Increases Ubiquitin-conjugating Activity in Skeletal Muscle by Up-regulating UbcH2/E2 20k. FASEB J. 2003, 17, 1048–1057. [Google Scholar] [CrossRef]
- Subramanian, M.; Hayes, C.D.; Thome, J.J.; Thorp, E.; Matsushima, G.K.; Herz, J.; Farber, D.L.; Liu, K.; Lakshmana, M.; Tabas, I. An AXL/LRP-1/RANBP9 Complex Mediates DC Efferocytosis and Antigen Cross-Presentation in Vivo. J. Clin. Investig. 2014, 124, 1296–1308. [Google Scholar] [CrossRef]
- Wang, L.; Fu, C.; Cui, Y.; Xie, Y.; Yuan, Y.; Wang, X.; Chen, H.; Huang, B.R. The Ran-Binding Protein RanBPM Can Depress the NF-ΚB Pathway by Interacting with TRAF6. Mol. Cell. Biochem. 2012, 359, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Baeg, G.H.; Zhou, R.; Perrimon, N. Genome-Wide RNAi Analysis of JAK/STAT Signaling Components in Drosophila. Genes Dev. 2005, 19, 1861–1870. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.P.; Almasy, K.M.; Mcdonald, E.F.; Plate, L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect. Dis. 2020, 6, 3174. [Google Scholar] [CrossRef]
- Davis, Z.H.; Verschueren, E.; Jang, G.M.; Kleffman, K.; Johnson, J.R.; Park, J.; VonDollen, J.; Maher, M.C.; Johnson, T.; Newton, W.; et al. Global Mapping of Herpesvirus-Host Protein Complexes Reveals a Transcription Strategy for Late Genes. Mol. Cell 2015, 57, 349–360. [Google Scholar] [CrossRef]
- Reitsma, J.M.; Savaryn, J.P.; Faust, K.; Sato, H.; Halligan, B.D.; Terhune, S.S. Antiviral Inhibition Targeting the HCMV Kinase PUL97 Requires PUL27-Dependent Degradation of Tip60 Acetyltransferase and Cell-Cycle Arrest. Cell Host Microbe 2011, 9, 103–114. [Google Scholar] [CrossRef]
- Sato, Y.; Kato, A.; Maruzuru, Y.; Oyama, M.; Kozuka-Hata, H.; Arii, J.; Kawaguchi, Y. Cellular Transcriptional Coactivator RanBP10 and Herpes Simplex Virus 1 ICP0 Interact and Synergistically Promote Viral Gene Expression and Replication. J. Virol. 2016, 90, 3173–3186. [Google Scholar] [CrossRef]
- Tomaru, K.; Ueda, A.; Suzuki, T.; Kobayashi, N.; Yang, J.; Yamamoto, M.; Takeno, M.; Kaneko, T.; Ishigatsubo, Y. Armadillo Repeat Containing 8α Binds to HRS and Promotes HRS Interaction with Ubiquitinated Proteins. Open Biochem. J. 2010, 4, 1–8. [Google Scholar] [CrossRef]
- Lausen, J.; Pless, O.; Leonard, F.; Kuvardina, O.N.; Koch, B.; Leutz, A. Targets of the Tal1 Transcription Factor in Erythrocytes: E2 Ubiquitin Conjugase Regulation by Tal1. J. Biol. Chem. 2010, 285, 5338–5346. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, X.; Hao, Y.; Tang, L.; He, Z. MicroRNA-26a-5p Alleviates Neuronal Apoptosis and Brain Injury in Intracerebral Hemorrhage by Targeting RAN Binding Protein 9. Acta Histochem. 2020, 122, 1511571. [Google Scholar] [CrossRef]
- Her, L.S.; Mao, S.H.; Chang, C.Y.; Cheng, P.H.; Chang, Y.F.; Yang, H.I.; Chen, C.M.; Yang, S.H. MiR-196a Enhances Neuronal Morphology through Suppressing RANBP10 to Provide Neuroprotection in Huntington’s Disease. Theranostics 2017, 7, 2452–2462. [Google Scholar] [CrossRef] [PubMed]
- Koenig, A.B.; Barajas, J.M.; Guerrero, M.J.; Ghoshal, K. A Comprehensive Analysis of Argonaute-CLIP Data Identifies Novel, Conserved and Species-Specific Targets of MiR-21 in Human Liver and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2018, 19, 851. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Kumar, A.; Soni, S.; Sinha, S.; Hanspal, M. Emp Is a Component of the Nuclear Matrix of Mammalian Cells and Undergoes Dynamic Rearrangements during Cell Division. Biochem. Biophys. Res. Commun. 2006, 342, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Tang, X.; Lin, F.; Chen, S. The WD40 Repeat Protein WDR26 Binds Gβγ and Promotes Gβγ-Dependent Signal Transduction and Leukocyte Migration. J. Biol. Chem. 2011, 286, 43902–43912. [Google Scholar] [CrossRef] [PubMed]


| CTLH Complex Subunit | Pearson | Spearman |
|---|---|---|
| RANBP9 | 0.575 | 0.565 |
| RANBP10 | 0.480 | 0.465 |
| WDR26 | 0.473 | 0.472 |
| ARMC8 | 0.408 | 0.457 |
| GID4 | 0.344 | 0.396 |
| RMND5A | 0.328 | 0.295 |
| MKLN1 | 0.297 | 0.322 |
| RMND5B | 0.159 | 0.172 |
| GID8 | 0.105 | 0.113 |
| MAEA | 0.041 | 0.064 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maitland, M.E.R.; Lajoie, G.A.; Shaw, G.S.; Schild-Poulter, C. Structural and Functional Insights into GID/CTLH E3 Ligase Complexes. Int. J. Mol. Sci. 2022, 23, 5863. https://doi.org/10.3390/ijms23115863
Maitland MER, Lajoie GA, Shaw GS, Schild-Poulter C. Structural and Functional Insights into GID/CTLH E3 Ligase Complexes. International Journal of Molecular Sciences. 2022; 23(11):5863. https://doi.org/10.3390/ijms23115863
Chicago/Turabian StyleMaitland, Matthew E. R., Gilles A. Lajoie, Gary S. Shaw, and Caroline Schild-Poulter. 2022. "Structural and Functional Insights into GID/CTLH E3 Ligase Complexes" International Journal of Molecular Sciences 23, no. 11: 5863. https://doi.org/10.3390/ijms23115863
APA StyleMaitland, M. E. R., Lajoie, G. A., Shaw, G. S., & Schild-Poulter, C. (2022). Structural and Functional Insights into GID/CTLH E3 Ligase Complexes. International Journal of Molecular Sciences, 23(11), 5863. https://doi.org/10.3390/ijms23115863






