Biomolecules under Pressure: Phase Diagrams, Volume Changes, and High Pressure Spectroscopic Techniques
Abstract
1. Introduction
2. Pressure and Volume
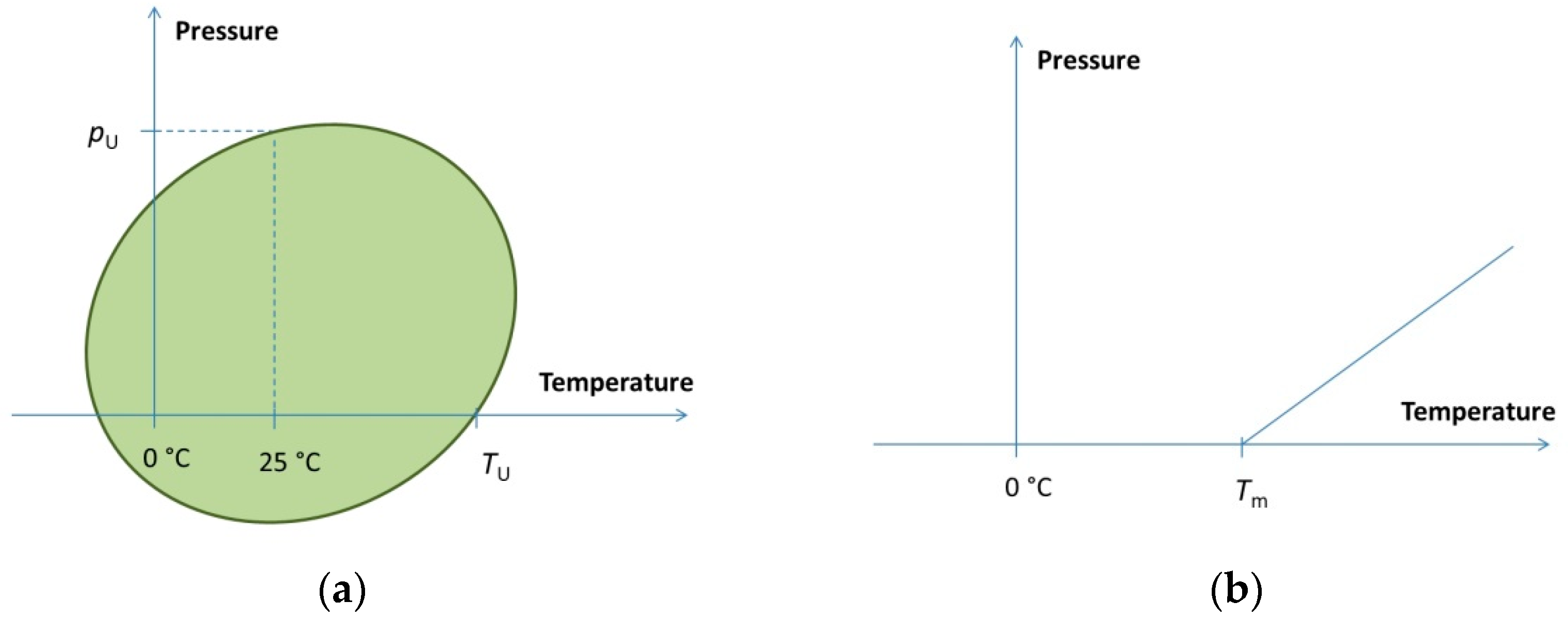
3. Pressure–Temperature Phase Diagram
4. High Pressure Techniques and Spectroscopic Methods Used in High Pressure Bioscience
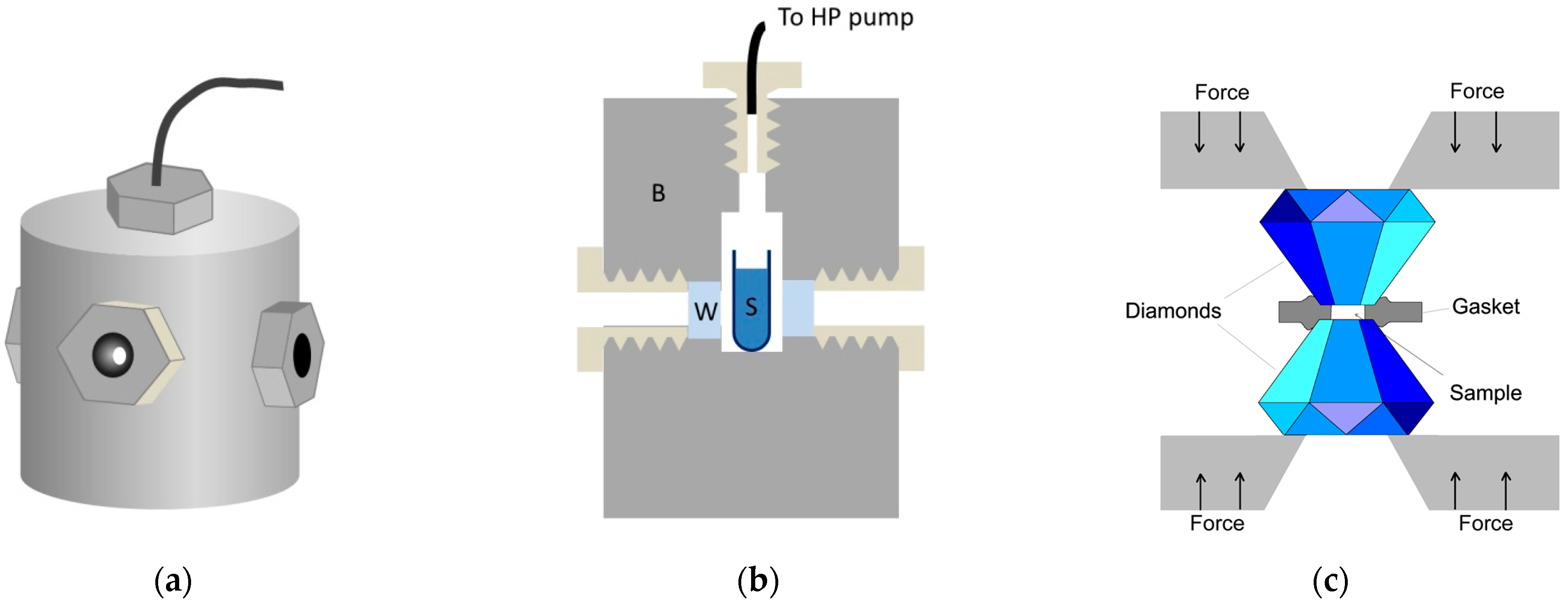
4.1. Instrumentation
4.2. Absorption Spectroscopic Techniques
4.2.1. UV-VIS Absorption Spectroscopy
4.2.2. Infrared Absorption Spectroscopy
4.3. Fluorescence Spectroscopy
4.4. Raman Spectroscopy
4.5. Nuclear Magnetic Resonance (NMR) Spectroscopy
5. Pressure Effect on Some Biomolecules
5.1. Proteins
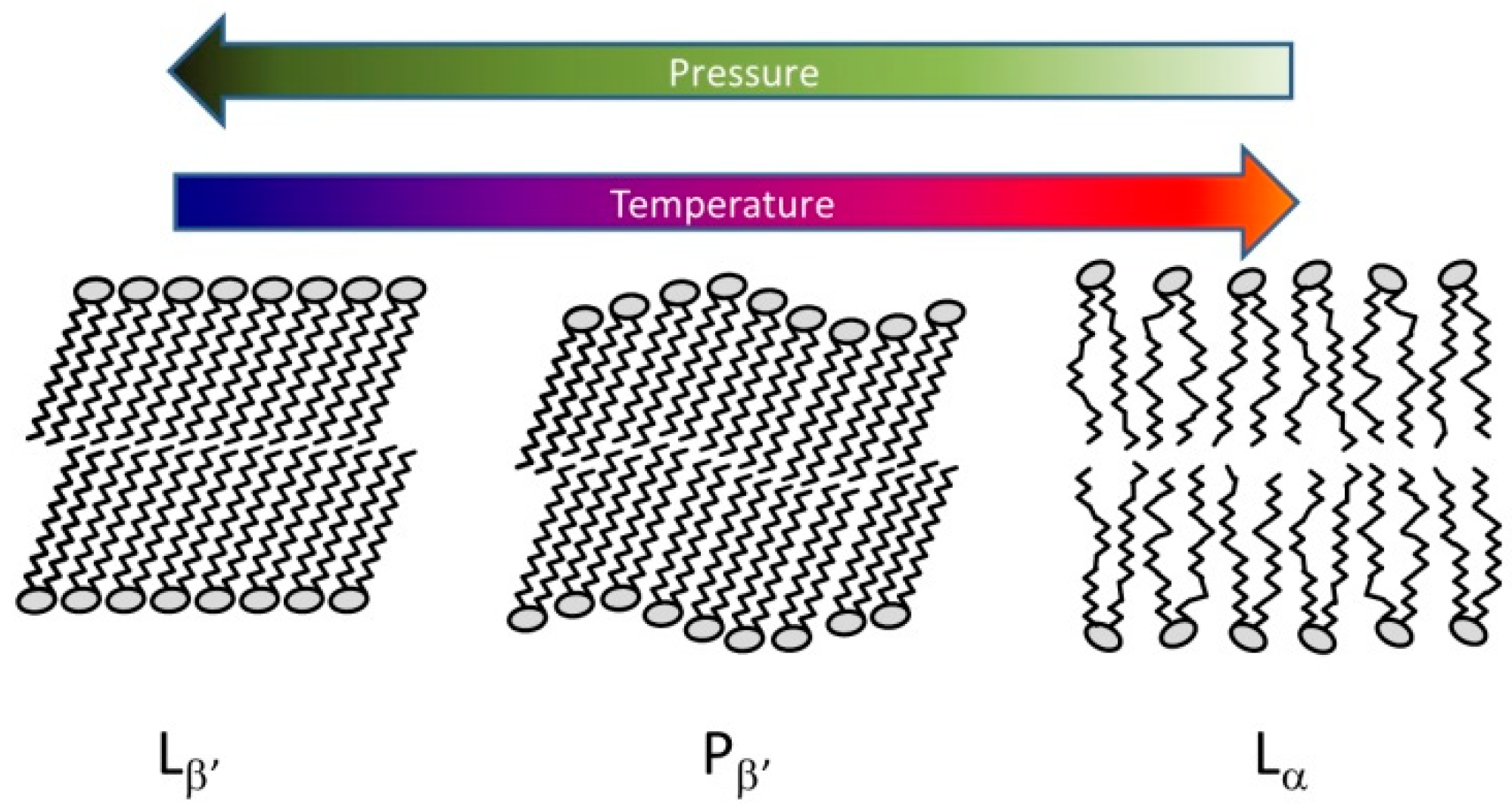
5.2. Lipids
5.3. Nucleic Acids
5.4. Supramolecular Structures
5.4.1. Viruses
5.4.2. Bacteria
6. Summary
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linley, T.D.; Gerringer, M.E.; Yancey, P.H.; Drazen, J.C.; Weinstock, C.L.; Jamieson, A.J. Fishes of the hadal zone including new species, in situ observations and depth records of Liparidae. Deep Sea Res. Part I Oceanogr. Res. Pap. 2016, 114, 99–110. [Google Scholar] [CrossRef]
- Yancey, P.H. Cellular responses in marine animals to hydrostatic pressure. J. Exp. Zool. Part A 2020, 333, 398–420. [Google Scholar] [CrossRef] [PubMed]
- Abe, F. Molecular responses to high hydrostatic pressure in eukaryotes: Genetic insights from studies on saccharomyces cerevisiae. Biology 2021, 10, 1305. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.W.; Wu, S.J.; Lu, J.K.; Shyu, Y.T.; Wang, C.Y. Current status and future trends of high-pressure processing in food industry. Food Control 2017, 72, 1–8. [Google Scholar] [CrossRef]
- Bolumar, T.; Orlien, V.; Sikes, A.; Aganovic, K.; Bak, K.H.; Guyon, C.; Stubler, A.S.; de Lamballerie, M.; Hertel, C.; Bruggemann, D.A. High-pressure processing of meat: Molecular impacts and industrial applications. Compr. Rev. Food Sci. Food Saf. 2021, 20, 332–368. [Google Scholar] [CrossRef] [PubMed]
- Nasilowska, J.; Kocot, A.; Osuchowska, P.N.; Sokolowska, B. High-pressure-induced sublethal injuries of food pathogens-microscopic assessment. Foods 2021, 10, 2940. [Google Scholar] [CrossRef]
- Rajendran, S.; Mallikarjunan, P.K.; O’Neill, E. High pressure processing for raw meat in combination with other treatments: A review. J. Food Process. Preserv. 2021, 2021, e16049. [Google Scholar] [CrossRef]
- Bridgman, P.W. The coagulation of albumen by pressure. J. Biol. Chem. 1914, 19, 511–512. [Google Scholar] [CrossRef]
- Meersman, F.; Heremans, K. High hydrostatic pressure effects in the biosphere: From molecules to microbiology. In High-Pressure Microbiology; Michiels, C., Bartlett, D.H., Aertsen, A., Eds.; American Society for Microbiology: Washington, DC, USA, 2008; pp. 1–17. [Google Scholar]
- Akasaka, K.; Maeno, A. Proteins in wonderland: The magical world of pressure. Biology 2022, 11, 6. [Google Scholar] [CrossRef]
- Wong, P.T.T.; Siminovitch, D.J.; Mantsch, H.H. Sructure and properties of model membranes—New knowledge from high-pressure vibrational spectroscopy. Biochim. Biophys. Acta 1988, 947, 139–171. [Google Scholar] [CrossRef]
- Akasaka, K.; Matsuki, H. High Pressure Bioscience: Basic Concepts, Applications and Frontiers; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, p. 730. [Google Scholar]
- Royer, C.A. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta BBA-Protein Struct. Mol. Enzymol. 2002, 1595, 201–209. [Google Scholar] [CrossRef]
- Royer, C.; Winter, R. Protein hydration and volumetric properties. Curr. Opin. Colloid Interface Sci. 2011, 16, 568–571. [Google Scholar] [CrossRef]
- Suzuki, A. High pressure-processed foods in Japan and the world. In Proceedings of the 1st International Conference on High Pressure Bioscience and Biotechnology (HPBB-2000), Kyoto, Japan, 26–30 November 2000; pp. 365–374. [Google Scholar]
- Chen, G.W.; Lin, H.T.V.; Huang, L.W.; Lin, C.H.; Lin, Y.H. Purification and identification of cholesterol micelle formation inhibitory peptides of hydrolysate from high hydrostatic pressure-assisted protease hydrolysis of fermented seabass byproduct. Int. J. Mol. Sci. 2021, 22, 5295. [Google Scholar] [CrossRef] [PubMed]
- Somkuti, J.; Smeller, L. High pressure effects on allergen food proteins. Biophys. Chem. 2013, 183, 19–29. [Google Scholar] [CrossRef]
- Somkuti, J.; Houska, M.; Smeller, L. Pressure and temperature stability of the main apple allergen Mal d1. Eur. Biophys. J. Biophys. Lett. 2011, 40, 143–151. [Google Scholar] [CrossRef]
- Somkuti, J.; Bublin, M.; Breiteneder, H.; Smeller, L. Pressure−temperature stability, Ca2+ binding, and pressure–temperature phase diagram of cod parvalbumin: Gad m 1. Biochemistry 2012, 51, 5903–5911. [Google Scholar] [CrossRef]
- Penhallurick, R.W.; Ichiye, T. Pressure adaptations in deep-sea Moritella dihydrofolate reductases: Compressibility versus stability. Biology 2021, 10, 1211. [Google Scholar] [CrossRef]
- Manisegaran, M.; Bornemann, S.; Kiesel, I.; Winter, R. Effects of the deep-sea osmolyte TMAO on the temperature and pressure dependent structure and phase behavior of lipid membranes. Phys. Chem. Chem. Phys. 2019, 21, 18533–18540. [Google Scholar] [CrossRef]
- Mukherjee, S.K.; Knop, J.M.; Oliva, R.; Mobitz, S.; Winter, R. Untangling the interaction of alpha-synuclein with DNA i-motifs and hairpins by volume-sensitive single-molecule FRET spectroscopy. RSC Chem. Biol. 2021, 2, 1196–1200. [Google Scholar] [CrossRef]
- Heremans, K.; Smeller, L. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1998, 1386, 353–370. [Google Scholar] [CrossRef]
- Likhodi, O.; Chalikian, T.V. Partial molar volumes and adiabatic compressibilities of a series of aliphatic amino acids and oligoglycines in D2O. J. Am. Chem. Soc. 1999, 121, 1156–1163. [Google Scholar] [CrossRef]
- Fan, H.Y.; Shek, Y.L.; Amiri, A.; Dubins, D.N.; Heerklotz, H.; Macgregor, R.B.; Chalikian, T.V. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011, 133, 4518–4526. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Caro, J.A.; Norberto, D.R.; Barthe, P.; Roumestand, C.; Schlessman, J.L.; Garcia, A.E.; Garcia-Moreno, E.B.; Royer, C.A. Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 6945–6950. [Google Scholar] [CrossRef] [PubMed]
- Kamatari, Y.O.; Smith, L.J.; Dobson, C.M.; Akasaka, K. Cavity hydration as a gateway to unfolding: An NMR study of hen lysozyme at high pressure and low temperature. Biophys. Chem. 2011, 156, 24–30. [Google Scholar] [CrossRef]
- Xue, M.; Wakamoto, T.; Kejlberg, C.; Yoshimura, Y.; Nielsen, T.A.; Risor, M.W.; Sanggaard, K.W.; Kitahara, R.; Mulder, F.A.A. How internal cavities destabilize a protein. Proc. Natl. Acad. Sci. USA 2019, 116, 21031–21036. [Google Scholar] [CrossRef]
- Heremans, K. Protein dynamics: Hydration and cavities. Braz. J. Med. Biol. Res. 2005, 38, 1157–1165. [Google Scholar] [CrossRef]
- Smeller, L. Pressure–temperature phase diagrams of biomolecules. Biochim. Biophys. Acta-Protein Struct. Molec. Enzymol. 2002, 1595, 11–29. [Google Scholar] [CrossRef]
- Tolgyesi, F.; Bode, C.S.; Smeller, L.; Kim, D.R.; Kim, K.K.; Heremans, K.; Fidy, J. Pressure activation of the chaperone function of small heat shock proteins. Cell. Mol. Biol. 2004, 50, 361–369. [Google Scholar]
- Hawley, S.A. Reversible pressure–temperature denaturation of chymotrypsinogen. Biochemistry 1971, 10, 2436–2442. [Google Scholar] [CrossRef]
- Kapoor, S.; Werkmuller, A.; Denter, C.; Zhai, Y.; Markgraf, J.; Weise, K.; Opitz, N.; Winter, R. Temperature-pressure phase diagram of a heterogeneous anionic model biomembrane system: Results from a combined calorimetry, spectroscopy and microscopy study. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 1187–1195. [Google Scholar] [CrossRef]
- Molnar, O.R.; Somkuti, J.; Smeller, L. Negative volume changes of human G-quadruplexes at unfolding. Heliyon 2020, 6, e05702. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.F.; Stadtmuller, A.A. Experimental Techniques in High-Pressure Research; Wiley: New York, NY, USA, 1987; pp. 10, 471. [Google Scholar]
- Taniguchi, H.; Takeda, S.; Satoh, R.; Taniguchi, A.; Komatsu, H.; Satoh, K. Short piston-cylinder pressure cells based on Ni-Cr-Al cylinders and their application to fragile materials. Rev. Sci. Instrum. 2010, 81, 033903. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, A. Diamond anvil cell and high-pressure physical investigations. Rev. Mod. Phys. 1983, 55, 65–108. [Google Scholar] [CrossRef]
- Wong, P.T.T.; Moffat, D.J. A new internal pressure calibrant for high-pressure infrared spectroscopy of aquesous systems. Appl. Spectrosc. 1989, 43, 1279–1281. [Google Scholar] [CrossRef]
- Wong, P.T.T.; Moffatt, D.J.; Baudais, F.L. Crystalline quartz as an internal-pressure calibrant for high-pressure infrared-spectroscopy. Appl. Spectrosc. 1985, 39, 733–735. [Google Scholar] [CrossRef]
- Forman, R.A.; Piermarini, G.J.; Dean, B.J.; Stanely, B. Pressure measurement made by the utilization of ruby sharp-line luminescence. Science 1972, 176, 284–285. [Google Scholar] [CrossRef] [PubMed]
- Syassen, K. Ruby under pressure. High Press. Res. 2008, 28, 75–126. [Google Scholar] [CrossRef]
- Bassett, W.A. Diamond anvil cell, 50th birthday. High Press. Res. 2009, 29, 163–186. [Google Scholar] [CrossRef]
- Minic, S.; Annighofer, B.; Helary, A.; Hamdane, D.; Hoa, G.H.B.; Loupiac, C.; Brulet, A.; Combet, S. Effect of ligands on HP-induced unfolding and oligomerization of beta-lactoglobulin. Biophys. J. 2020, 119, 2262–2274. [Google Scholar] [CrossRef]
- Kangur, L.; Jones, M.R.; Freiberg, A. Hydrogen bonds in the vicinity of the special pair of the bacterial reaction center probed by hydrostatic high-pressure absorption spectroscopy. Biophys. Chem. 2017, 231, 27–33. [Google Scholar] [CrossRef]
- Lange, R.; Frank, J.; Saldana, J.L.; Balny, C. Fourth derivative UV-spectroscopy of proteins under high pressure. 1. Factors affecting the fourth derivative spectrum of the aromatic amino acids. Eur. Biophys. J. Biophys. Lett. 1996, 24, 277–283. [Google Scholar] [CrossRef]
- Peyrano, F.; de Lamballerie, M.; Avanza, M.V.; Speroni, F. Gelation of cowpea proteins induced by high hydrostatic pressure. Food Hydrocoll. 2021, 111, 106191. [Google Scholar] [CrossRef]
- Kwak, M.I.; Jeon, B.R.; Kim, S.K.; Jang, Y.J. Binding mode of cationic porphyrin with CT-DNA: Importance of the location and the number of positively charged of periphery cationic ions of porphyrin. ACS Omega 2018, 3, 946–953. [Google Scholar] [CrossRef]
- Csik, G.; Egyeki, M.; Herenyi, L.; Majer, Z.; Toth, K. Role of structure-proteins in the porphyrin-DNA interaction. J. Photochem. Photobiol. B-Biol. 2009, 96, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Jeworrek, C.; Uelner, S.; Winter, R. Phase behavior and kinetics of pressure-jump induced phase transitions of bicellar lipid mixtures. Soft Matter 2011, 7, 2709–2719. [Google Scholar] [CrossRef]
- Somkuti, J.; Martonfalvi, Z.; Kellermayer, M.S.Z.; Smeller, L. Different pressure–temperature behavior of the structured and unstructured regions of titin. Biochim. Biophys. Acta-Proteins Proteom. 2013, 1834, 112–118. [Google Scholar] [CrossRef]
- Guzman, M.R.; Liquier, J.; Brahmachari, S.K.; Taillandier, E. Characterization of parallel and antiparallel G-tetraplex structures by vibrational spectroscopy. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2006, 64, 495–503. [Google Scholar] [CrossRef]
- Susi, H.; Byler, D.M. Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. Methods Enzymol. 1986, 130, 290–311. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Ismail, A.A.; Mantsch, H.H.; Wong, P.T.T. Aggregation of chymotripsinogen—Portrait by infrared-spectroscopy. Biochim. Biophys. Acta 1992, 1121, 183–188. [Google Scholar] [CrossRef]
- Smeller, L.; Meersman, F.; Heremans, K. Refolding studies using pressure: The folding landscape of lysozyme in the pressure–temperature plane. Biochim. Biophys. Acta-Proteins Proteom. 2006, 1764, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Metrick, M.A.; MacDonald, G. Hofmeister ion effects on the solvation and thermal stability of model proteins lysozyme and myoglobin. Colloids Surf. A-Physicochem. Eng. Asp. 2015, 469, 242–251. [Google Scholar] [CrossRef]
- Paschou, A.M.; Katsikini, M.; Christofilos, D.; Arvanitidis, J.; Ves, S. High pressure Raman study of type-I collagen. FEBS J. 2018, 285, 2641–2653. [Google Scholar] [CrossRef] [PubMed]
- Smeller, L.; Goossens, K.; Heremans, K. Determination of the secondary structure of proteins at high pressure. Vib. Spectrosc. 1995, 8, 199–203. [Google Scholar] [CrossRef]
- Smeller, L.; Goossens, K.; Heremans, K. How to minimize certain artifacts in Fourier self-deconvolution. Appl. Spectrosc. 1995, 49, 1538–1542. [Google Scholar] [CrossRef]
- Maeno, A.; Akasaka, K. High-pressure fluorescence spectroscopy. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, pp. 687–705. [Google Scholar]
- Herberhold, H.; Marchal, S.; Lange, R.; Scheyhing, C.H.; Vogel, R.F.; Winter, R. Characterization of the pressure-induced intermediate and unfolded state of red-shifted green fluorescent protein—A static and kinetic FTIR, UV/VIS and fluorescence spectroscopy study. J. Mol. Biol. 2003, 330, 1153–1164. [Google Scholar] [CrossRef]
- Scheyhing, C.H.; Meersman, F.; Ehrmann, M.A.; Heremans, K.; Vogel, R.F. Temperature-pressure stability of green fluorescent protein: A Fourier transform infrared spectroscopy study. Biopolymers 2002, 65, 244–253. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Scheyhing, C.H.; Vogel, R.F. In vitro stability and expression of green fluorescent protein under high pressure conditions. Lett. Appl. Microbiol. 2001, 32, 230–234. [Google Scholar] [CrossRef]
- Gustavsson, T.; Markovitsi, D. Fundamentals of the intrinsic DNA fluorescence. Acc. Chem. Res. 2021, 54, 1226–1235. [Google Scholar] [CrossRef]
- Molnar, O.R.; Vegh, A.; Somkuti, J.; Smeller, L. Characterization of a G-quadruplex from hepatitis B virus and its stabilization by binding TMPyP4, BRACO19 and PhenDC3. Sci. Rep. 2021, 11, 23243. [Google Scholar] [CrossRef]
- Patra, S.; Schuabb, V.; Kiesel, I.; Knop, J.M.; Oliva, R.; Winter, R. Exploring the effects of cosolutes and crowding on the volumetric and kinetic profile of the conformational dynamics of a poly dA loop DNA hairpin: A single-molecule FRET study. Nucleic Acids Res. 2019, 47, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.L.; Nesbitt, D.J. High pressure single-molecule FRET studies of the lysine riboswitch: Cationic and osmolytic effects on pressure induced denaturation. Phys. Chem. Chem. Phys. 2020, 22, 15853–15866. [Google Scholar] [CrossRef] [PubMed]
- Knop, J.M.; Patra, S.; Harish, B.; Royer, C.A.; Winter, R. The deep sea osmolyte trimethylamine N-oxide and macromolecular crowders rescue the antiparallel conformation of the human telomeric G-quadruplex from urea and pressure stress. Chem.-Eur. J. 2018, 24, 14346–14351. [Google Scholar] [CrossRef] [PubMed]
- Doona, C.J.; Feeherry, F.E.; Setlow, B.; Wang, S.; Li, W.; Nichols, F.C.; Talukdar, P.K.; Sarker, M.R.; Li, Y.Q.; Shen, A.; et al. Effects of high-pressure treatment on spores of clostridium species. Appl. Environ. Microbiol. 2016, 82, 5287–5297. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Butler, I.S. Pressure-tuning infrared and Raman microscopy study of the DNA bases: Adenine, guanine, cytosine, and thymine. J. Biomol. Struct. Dyn. 2013, 31, 1490–1496. [Google Scholar] [CrossRef]
- Bondos, S.E.; Sligar, S.; Jonas, J. High-pressure denaturation of apomyoglobin. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 2000, 1480, 353–364. [Google Scholar] [CrossRef]
- Jonas, J. High-resolution nuclear magnetic resonance studies of proteins. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 2002, 1595, 145–159. [Google Scholar] [CrossRef]
- Akasaka, K.; Tezuka, T.; Yamada, H. Pressure-induced changes in the folded structure of lysozyme. J. Mol. Biol. 1997, 271, 671–678. [Google Scholar] [CrossRef]
- Lassalle, M.W.; Yamada, H.; Akasaka, K. The pressure–temperature free energy-landscape of staphylococcal nuclease monitored by H-1 NMR. J. Mol. Biol. 2000, 298, 293–302. [Google Scholar] [CrossRef]
- Kalbitzer, H.R. High pressure NMR methods for characterizing functional substates of proteins. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, pp. 179–197. [Google Scholar]
- Dubois, C.; Herrada, I.; Barthe, P.; Roumestand, C. Combining high-pressure perturbation with NMR spectroscopy for a structural and dynamical characterization of protein folding pathways. Molecules 2020, 25, 5551. [Google Scholar] [CrossRef]
- Akasaka, K. High pressure NMR spectroscopy. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, pp. 707–721. [Google Scholar]
- Van Deuren, V.; Yang, Y.S.; de Guillen, K.; Dubois, C.; Royer, C.A.; Roumestand, C.; Barthe, P. Comparative assessment of NMR probes for the experimental description of protein folding pathways with high-pressure NMR. Biology 2021, 10, 656. [Google Scholar] [CrossRef] [PubMed]
- Harish, B.; Gillilan, R.E.; Zou, J.J.; Wang, J.Q.; Raleigh, D.P.; Royer, C.A. Protein unfolded states populated at high and ambient pressure are similarly compact. Biophys. J. 2021, 120, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, R. High-pressure NMR spectroscopy reveals functional sub-states of ubiquitin and ubiquitin-like proteins. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, pp. 199–214. [Google Scholar]
- Erlach, M.B.; Munte, C.E.; Kremer, W.; Hartl, R.; Rochelt, D.; Niesner, D.; Kalbitzer, H.R. Ceramic cells for high pressure NMR spectroscopy of proteins. J. Magn. Reson. 2010, 204, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, R.; Hata, K.; Li, H.; Williamson, M.P.; Akasaka, K. Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 71, 35–58. [Google Scholar] [CrossRef]
- Trajkovski, M.; Endoh, T.; Tateishi-Karimata, H.; Ohyama, T.; Tanaka, S.; Plavec, J.; Sugimoto, N. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018, 46, 4301–4315. [Google Scholar] [CrossRef]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Muller, D.; Bessi, I.; Richter, C.; Schwalbe, H. The folding landscapes of human telomeric RNA and DNA G-quadruplexes are markedly different. Angew. Chem. Int. Ed. 2021, 60, 10895–10901. [Google Scholar] [CrossRef]
- Smeller, L. Protein denaturation on p-t axes—Thermodynamics and analysis. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2015; Volume 72, pp. 19–39. [Google Scholar]
- Zipp, A.; Kauzmann, W. Pressure denaturation of metmyoglobin. Biochemistry 1973, 12, 4217–4228. [Google Scholar] [CrossRef]
- Spinozzi, F.; Mariani, P.; Saturni, L.; Carsughi, F.; Bernstorff, S.; Cinelli, S.; Onori, G. Met-myoglobin association in dilute solution during pressure-induced denaturation: An analysis at pH 4.5 by high-pressure small-angle X-ray scattering. J. Phys. Chem. B 2007, 111, 3822–3830. [Google Scholar] [CrossRef]
- Meersman, F.; Smeller, L.; Heremans, K. Extending the pressure–temperature state diagram of myoglobin. Helv. Chim. Acta 2005, 88, 546–556. [Google Scholar] [CrossRef]
- Panick, G.; Vidugiris, G.J.A.; Malessa, R.; Rapp, G.; Winter, R.; Royer, C.A. Exploring the temperature-pressure phase diagram of staphylococcal nuclease. Biochemistry 1999, 38, 4157–4164. [Google Scholar] [CrossRef] [PubMed]
- Meersman, F.; Smeller, L.; Heremans, K. Comparative Fourier transform infrared spectroscopy study of cold-, pressure-, and heat-induced unfolding and aggregation of myoglobin. Biophys. J. 2002, 82, 2635–2644. [Google Scholar] [CrossRef]
- Smeller, L.; Rubens, P.; Heremans, K. Pressure effect on the temperature-induced unfolding and tendency to aggregate of myoglobin. Biochemistry 1999, 38, 3816–3820. [Google Scholar] [CrossRef] [PubMed]
- Smeller, L.; Meersman, F.; Heremans, K. Stable misfolded states of human serum albumin revealed by high-pressure infrared spectroscopic studies. Eur. Biophys. J. Biophys. Lett. 2008, 37, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef]
- Foguel, B.; Silva, J.L. New insights into the mechanisms of protein misfolding and aggregation in amyloidogenic diseases derived from pressure studies. Biochemistry 2004, 43, 11361–11370. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Gidalevitz, T.; Kikis, E.A.; Morimoto, R.I. A cellular perspective on conformational disease: The role of genetic background and proteostasis networks. Curr. Opin. Struct. Biol. 2010, 20, 23–32. [Google Scholar] [CrossRef]
- Gregersen, N.; Bolund, L.; Bross, P. Protein misfolding, aggregation, and degradation in disease. Mol. Biotechnol. 2005, 31, 141–150. [Google Scholar] [CrossRef]
- Dirix, C.; Meersman, F.; MacPhee, C.E.; Dobson, C.M.; Heremans, K. High hydrostatic pressure dissociates early aggregates of TTR105-115, but not the mature amyloid fibrils. J. Mol. Biol. 2005, 347, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Smeller, L.; Meersman, F.; Fidy, J.; Heremans, K. High-pressure FTIR study of the stability of horseradish peroxidase. Effect of heme substitution, ligand binding, Ca++ removal, and reduction of the disulfide bonds. Biochemistry 2003, 42, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Gekko, K.; Araga, M.; Kamiyama, T.; Ohmae, E.; Akasaka, K. Pressure dependence of the apparent specific volume of bovine serum albumin: Insight into the difference between isothermal and adiabatic compressibilities. Biophys. Chem. 2009, 144, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Gekko, K. Compressibility gives new insight into protein dynamics and enzyme function. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 2002, 1595, 382–386. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Totrov, M.; Abagyan, R.; Breslauer, K.J. The hydration of globular proteins as derived from volume and compressibility measurements: Cross correlating thermodynamic and structural data. J. Mol. Biol. 1996, 260, 588–603. [Google Scholar] [CrossRef]
- Schay, G.; Kaposi, A.D.; Smeller, L.; Szigeti, K.; Fidy, J.; Herenyi, L. Dissimilar flexibility of alpha and beta subunits of human adult hemoglobin influences the protein dynamics and its alteration induced by allosteric effectors. PLoS ONE 2018, 13, e0194994. [Google Scholar] [CrossRef]
- Balog, E.; Galantai, R.; Kohler, M.; Laberge, M.; Fidy, J. Metal coordination influences substrate binding in horseradish peroxidase. Eur. Biophys. J. Biophys. Lett. 2000, 29, 429–438. [Google Scholar] [CrossRef]
- Simonato, F.; Campanaro, S.; Lauro, F.M.; Vezzi, A.; D’Angelo, M.; Vitulo, N.; Valle, G.; Bartlett, D.H. Piezophilic adaptation: A genomic point of view. J. Biotechnol. 2006, 126, 11–25. [Google Scholar] [CrossRef]
- Jin, M.; Gai, Y.B.; Guo, X.; Hou, Y.P.; Zeng, R.Y. Properties and applications of extremozymes from deep-sea extremophilic microorganisms: A mini review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef]
- Abe, F.; Minegishi, H. Global screening of genes essential for growth in high-pressure and cold environments: Searching for basic adaptive strategies using a yeast deletion library. Genetics 2008, 178, 851–872. [Google Scholar] [CrossRef]
- Morris, J.P.; Thatje, S.; Ravaux, J.; Shillito, B.; Fernando, D.; Hauton, C. Acute combined pressure and temperature exposures on a shallow-water crustacean: Novel insights into the stress response and high pressure neurological syndrome. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2015, 181, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Sieg, J.; Sandmeier, C.C.; Lieske, J.; Meents, A.; Lemmen, C.; Streit, W.R.; Rarey, M. Analyzing structural features of proteins from deep-sea organisms. Proteins 2022. [Google Scholar] [CrossRef] [PubMed]
- Schummel, P.H.; Haag, A.; Kremer, W.; Kalbitzer, H.R.; Winter, R. Cosolvent and crowding effects on the temperature and pressure dependent conformational dynamics and stability of globular actin. J. Phys. Chem. B 2016, 120, 6575–6586. [Google Scholar] [CrossRef] [PubMed]
- Schummel, P.H.; Anders, C.; Jaworek, M.W.; Winter, R. Cosolvent and crowding effects on the temperature- and pressure-dependent dissociation process of the/-tubulin heterodimer. ChemPhysChem 2019, 20, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Morita, T. Structure-based analysis of high pressure adaptation of alpha-actin. J. Biol. Chem. 2003, 278, 28060–28066. [Google Scholar] [CrossRef] [PubMed]
- Roobab, U.; Abida, A.; Afzal, R.; Madni, G.M.; Zeng, X.A.; Rahaman, A.; Aadil, R.M. Impact of high-pressure treatments on enzyme activity of fruit-based beverages: An overview. Int. J. Food Sci. Technol. 2022, 57, 801–815. [Google Scholar] [CrossRef]
- Houben, K.; Kermani, Z.J.; van Buggenhout, S.; Jolie, R.P.; van Loey, A.M.; Hendrickx, M.E. Thermal and high-pressure stability of pectinmethylesterase, polygalacturonase, beta-galactosidase and alpha-arabinofuranosidase in a tomato matrix: Towards the creation of specific endogenous enzyme populations through processing. Food Bioprocess Technol. 2013, 6, 3368–3380. [Google Scholar] [CrossRef]
- Sila, D.N.; Smout, C.; Satara, Y.; Truong, V.; van Loey, A.; Hendrickx, M. Combined thermal and high pressure effect on carrot pectinmethylesterase stability and catalytic activity. J. Food Eng. 2007, 78, 755–764. [Google Scholar] [CrossRef]
- Timpmann, K.; Linnanto, J.M.; Yadav, D.; Kangur, L.; Freiberg, A. Hydrostatic high-pressure-induced denaturation of LH2 membrane proteins. J. Phys. Chem. B 2021, 125, 9979–9989. [Google Scholar] [CrossRef]
- Winter, R.; Jeworrek, C. Effect of pressure on membranes. Soft Matter 2009, 5, 3157–3173. [Google Scholar] [CrossRef]
- Singh, H.; Emberley, J.; Morrow, M.R. Pressure induces interdigitation differently in DPPC and DPPG. Eur. Biophys. J. Biophys. Lett. 2008, 37, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Skanes, I.D.; Stewart, J.; Keough, K.M.W.; Morrow, M.R. Effect of chain unsaturation on bilayer response to pressure. Phys. Rev. E 2006, 74, 051913. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.L.G.; Sulc, M.; Winter, R. Compressibilities and volume fluctuations of archaeal tetraether liposomes. Biophys. J. 2010, 99, 3319–3326. [Google Scholar] [CrossRef] [PubMed]
- Winter, R. Pressure effects on artificial and cellular membranes. In High Pressure Bioscience; Akasaka, K., Matsuki, H., Eds.; Subcellular Biochemistry; Springer: New York, NY, USA, 2015; Volume 72, pp. 345–370. [Google Scholar]
- Eisenblatter, J.; Winter, R. Pressure effects on the structure and phase behavior of DMPC-gramicidin lipid bilayers: A synchrotron SAXS and H-2-NMR spectroscopy study. Biophys. J. 2006, 90, 956–966. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sugar, I.P.; Bonanno, A.P.; Chong, P.L.G. Gramicidin lateral distribution in phospholipid membranes: Fluorescence phasor plots and statistical mechanical model. Int. J. Mol. Sci. 2018, 19, 3690. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, H.; Klose, G.; Heremans, K. FTIR spectroscopy study of the pressure-dependent behaviour of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1-palmitoyl-2-oleolyl-sn-glycero-3-phosphocholine (POPC) at low degrees of hydration. Chem. Phys. Lipids 2013, 170, 33–40. [Google Scholar] [CrossRef]
- Brooks, N.J. Pressure effects on lipids and bio-membrane assemblies. IUCrJ 2014, 1, 470–477. [Google Scholar] [CrossRef]
- Bonanno, A.; Chong, P.L.G. Certain, but not all, tetraether lipids from the thermoacidophilic archaeon sulfolobus acidocaldarius can form black lipid membranes with remarkable stability and exhibiting mthk channel activity with unusually high Ca2+ sensitivity. Int. J. Mol. Sci. 2021, 22, 12941. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Macgregor, R.B.; Wu, J.Q.; Najaf-Zadeh, R. Sequence, salt, charge, and the stability of DNA at high pressure. In High Pressure Effects in Molecular Biophysics and Enzymology; Markley, J.L., Northrop, D.B., Royer, C.A., Eds.; Oxford University Press: Oxford, UK, 1996; pp. 149–170. [Google Scholar]
- Nordmeier, E. Effects of pressure on the helix coil transition of calf thymus DNA. J. Phys. Chem. 1992, 96, 1494–1501. [Google Scholar] [CrossRef]
- Sugimoto, N. Noncanonical structures and their thermodynamics of DNA and RNA under molecular crowding: Beyond the watson-crick double helix. Int. Rev. Cell Mol. Biol. 2014, 307, 205–273. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-quadruplexes and their ligands: Biophysical methods to unravel G-quadruplex/ligand interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Marchand, A.; Amrane, S.; Gabelica, V.; Mergny, J.L. Quadruplex turncoats: Cation-dependent folding and stability of quadruplex-DNA double switches. J. Am. Chem. Soc. 2016, 138, 2780–2792. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Fujii, T.; Podbevsek, P.; Plavec, J.; Sugimoto, N. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 2017, 166, 190–198. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef]
- Zhang, D.H.; Fujimoto, T.; Saxena, S.; Yu, H.Q.; Miyoshi, D.; Sugimoto, N. Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry 2010, 49, 4554–4563. [Google Scholar] [CrossRef]
- Fry, M. Tetraplex DNA and its interacting proteins. Front. Biosci.-Landmark 2007, 12, 4336–4351. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sugimoto, N. Effect of pressure on thermal stability of G-quadruplex DNA and double-stranded DNA structures. Molecules 2013, 18, 13297–13319. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sugimoto, N. Effect of pressure on the stability of G-quadruplex DNA: Thermodynamics under crowding conditions. Angew. Chem. Int. Edit. 2013, 52, 13774–13778. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sugimoto, N. Volumetric contributions of loop regions of G-quadruplex DNA to the formation of the tertiary structure. Biophys. Chem. 2017, 231, 146–154. [Google Scholar] [CrossRef]
- Takahashi, S.; Bhowmik, S.; Sugimoto, N. Volumetric analysis of formation of the complex of G-quadruplex DNA with hemin using high pressure. J. Inorg. Biochem. 2017, 166, 199–207. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Ruggiero, E.; Zanin, I.; Terreri, M.; Richter, S.N. G-quadruplex targeting in the fight against viruses: An update. Int. J. Mol. Sci. 2021, 22, 984. [Google Scholar] [CrossRef]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-quadruplex/helicase world as a potential antiviral approach against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef]
- Tluckova, K.; Marusic, M.; Tothova, P.; Bauer, L.; Sket, P.; Plavec, J.; Viglasky, V. Human papillomavirus G-quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef]
- Somkuti, J.; Molnar, O.R.; Grad, A.; Smeller, L. Pressure perturbation studies of noncanonical viral nucleic acid structures. Biology 2021, 10, 1173. [Google Scholar] [CrossRef]
- Biswas, B.; Kandpal, M.; Vivekanandan, P. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017, 45, 11268–11280. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palu, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Dubins, D.N.; Le, D.M.N.T.; Leung, K.; Macgregor, R.B. The role of loops and cation on the volume of unfolding of G-quadruplexes related to HTel. Biophys. Chem. 2017, 231, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Chalikian, T.V.; Macgregor, R.B. Volumetric properties of four-stranded DNA structures. Biology 2021, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Abu-Ghazalah, R.M.; Rutledge, S.; Lau, L.W.Y.; Dubins, D.N.; Macgregor, R.B.; Helmy, A.S. Concentration-dependent structural transitions of human telomeric DNA sequences. Biochemistry 2012, 51, 7357–7366. [Google Scholar] [CrossRef] [PubMed]
- Shearer, A.E.H.; Chen, K.; Hoover, D.G. High-pressure effects on viruses. In High Pressure Processing of Food: Principles, Technology and Applications; Balasubramaniam, V.M., BarbosaCanovas, G.V., Lelieveld, H.L.M., Eds.; Food Engineering Series; Springer: Berlin/Heidelberg, Germany, 2016; pp. 295–315. [Google Scholar]
- Lou, F.F.; Neetoo, H.; Chen, H.Q.; Li, J.R. High hydrostatic pressure processing: A promising nonthermal technology to inactivate viruses in high-risk foods. Ann. Rev. Food Sci. Technol. 2015, 6, 389–409. [Google Scholar] [CrossRef]
- Ishimaru, D.; Sa-Carvalho, D.; Silva, J.L. Pressure-inactivated FMDV: A potential vaccine. Vaccine 2004, 22, 2334–2339. [Google Scholar] [CrossRef]
- Bonafe, C.F.S.; Vital, C.M.R.; Telles, R.C.B.; Goncalves, M.C.; Matsuura, M.S.A.; Pessine, F.B.T.; Freitas, D.R.C.; Vega, J. Tobacco mosaic virus disassembly by high hydrostatic pressure in combination with urea and low temperature. Biochemistry 1998, 37, 11097–11105. [Google Scholar] [CrossRef]
- Pontes, L.; Cordeiro, Y.; Giongo, V.; Villas-Boas, M.; Barreto, A.; Araujo, J.R.; Silva, J.L. Pressure-induced formation of inactive triple-shelled rotavirus particles is associated with changes in the spike protein VP4. J. Mol. Biol. 2001, 307, 1171–1179. [Google Scholar] [CrossRef]
- Heinz, V.; Buckow, R. Food preservation by high pressure. J. Consum. Prot. Food Saf. 2010, 5, 73–81. [Google Scholar] [CrossRef]
- Ludwig, H. Effects of high pressure on bacteria and fungi. In Proceedings of the 2nd International Conference on High Pressure Bioscience and Biotechnology, Dortmund, Germany, 16–19 September 2002; pp. 259–265. [Google Scholar]
- Kilimann, K.V.; Hartmann, C.; Delgado, A.; Vogel, R.F.; Ganzle, M.G. Combined high pressure and temperature induced lethal and sublethal injury of Lactococcus lactis—Application of multivariate statistical analysis. Int. J. Food Microbiol. 2006, 109, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Reineke, K.; Schlumbach, K.; Baier, D.; Mathys, A.; Knorr, D. The release of dipicolinic acid—The rate-limiting step of Bacillus endospore inactivation during the high pressure thermal sterilization process. Int. J. Food Microbiol. 2013, 162, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, K.; Maeno, A.; Yamazaki, A. Direct high-pressure NMR observation of dipicolinic acid leaking from bacterial spore: A crucial step for thermal inactivation. Biophys. Chem. 2017, 231, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Modugno, C.; Peltier, C.; Simonin, H.; Dujourdy, L.; Capitani, F.; Sandt, C.; Perrier-Cornet, J.M. Understanding the effects of high pressure on bacterial spores using synchrotron infrared spectroscopy. Front. Microbiol. 2020, 10, 3122. [Google Scholar] [CrossRef]
- Barba, F.J.; Terefe, N.S.; Buckow, R.; Knorr, D.; Orlien, V. New opportunities and perspectives of high pressure treatment to improve health and safety attributes of foods. A review. Food Res. Int. 2015, 77, 725–742. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smeller, L. Biomolecules under Pressure: Phase Diagrams, Volume Changes, and High Pressure Spectroscopic Techniques. Int. J. Mol. Sci. 2022, 23, 5761. https://doi.org/10.3390/ijms23105761
Smeller L. Biomolecules under Pressure: Phase Diagrams, Volume Changes, and High Pressure Spectroscopic Techniques. International Journal of Molecular Sciences. 2022; 23(10):5761. https://doi.org/10.3390/ijms23105761
Chicago/Turabian StyleSmeller, László. 2022. "Biomolecules under Pressure: Phase Diagrams, Volume Changes, and High Pressure Spectroscopic Techniques" International Journal of Molecular Sciences 23, no. 10: 5761. https://doi.org/10.3390/ijms23105761
APA StyleSmeller, L. (2022). Biomolecules under Pressure: Phase Diagrams, Volume Changes, and High Pressure Spectroscopic Techniques. International Journal of Molecular Sciences, 23(10), 5761. https://doi.org/10.3390/ijms23105761





