HSPA4 Is a Biomarker of Placenta Accreta and Enhances the Angiogenesis Ability of Vessel Endothelial Cells
Abstract
1. Introduction
2. Results
2.1. The Characteristics of the Patients with Placenta Accreta and the Subjects with Normal Pregnancy
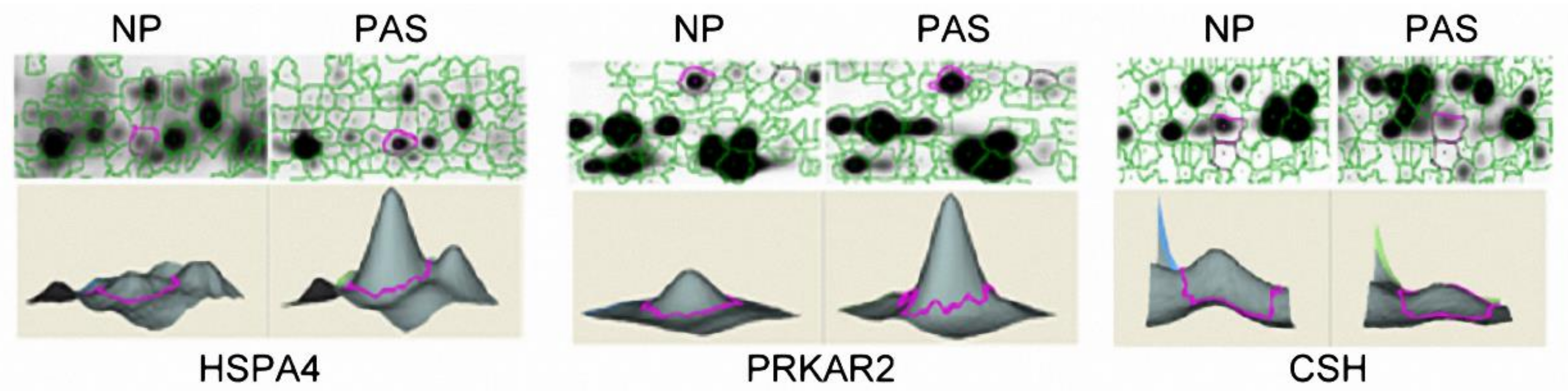
2.2. Protein Abundance Variations between PAS and NP Placenta Tissues
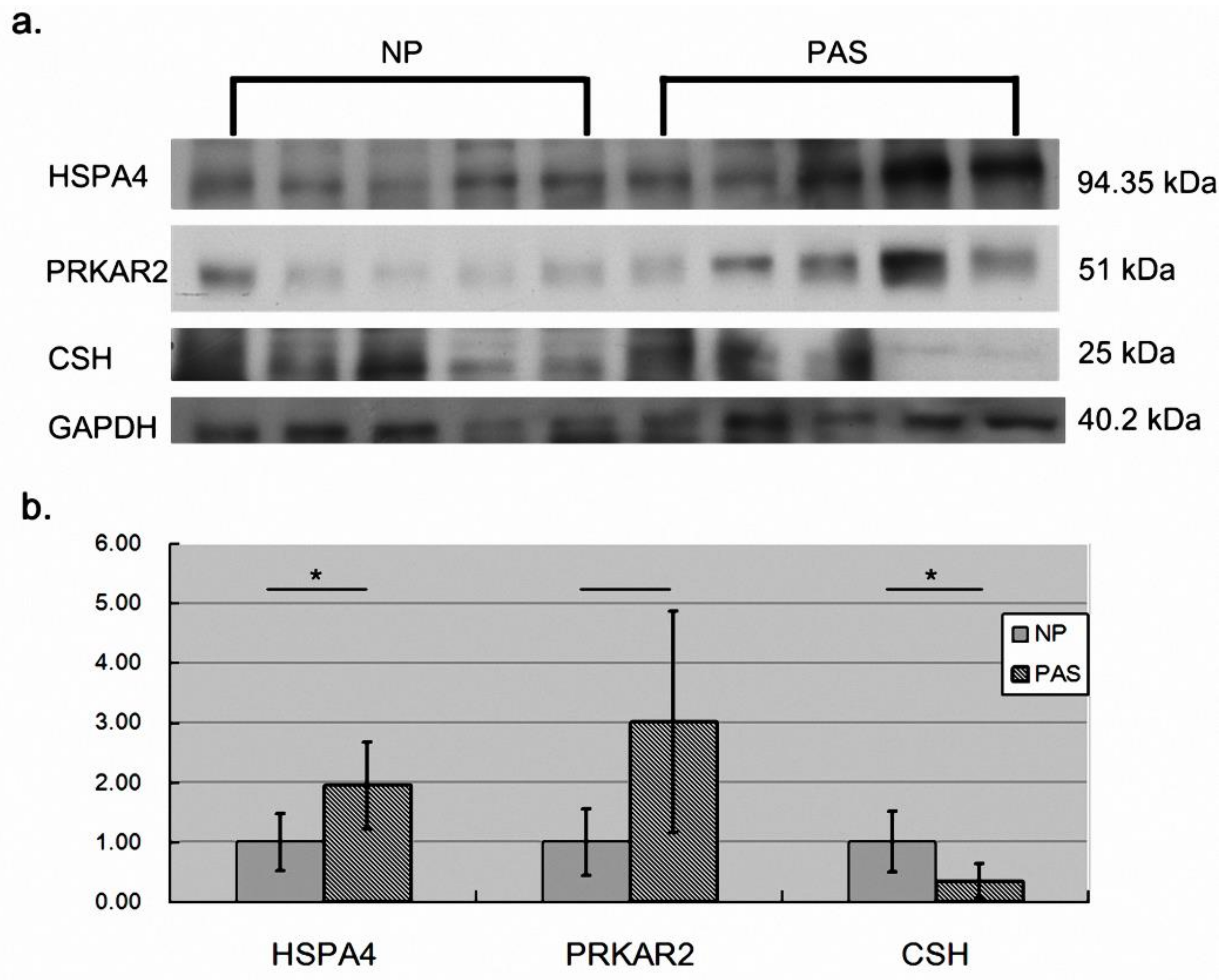
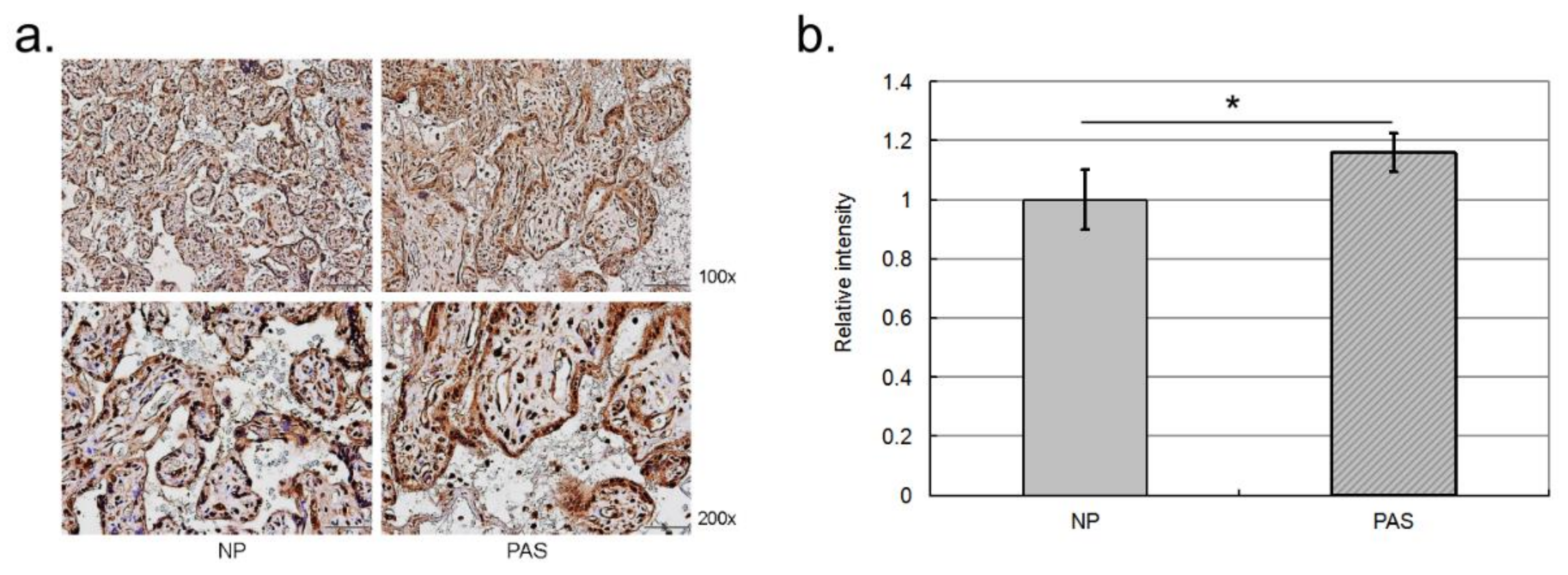
2.3. Validation of the Differentially Abundant Proteins
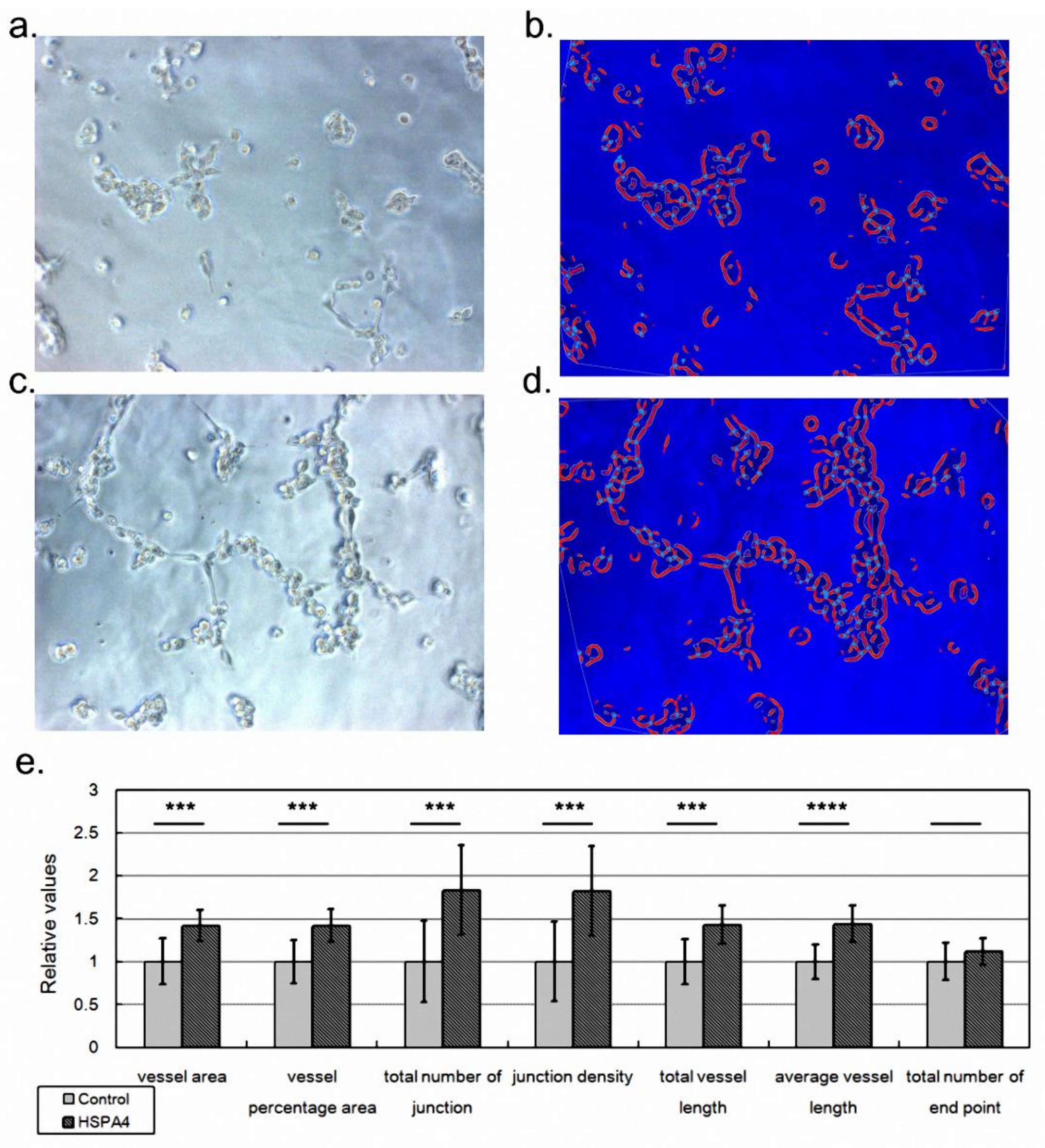
2.4. HSPA4 Overexpression Enhanced the Angiogenesis Ability of Vessel Endothelial Cells
3. Discussion
4. Materials and Methods
4.1. Subject Enrollment and Sample Collection
4.2. Protein Enrichment from Cell Lysate
4.3. Protein Quantification with 2D-DIGE and Identification with MALDI TOF
4.4. Western Blot Assay
4.5. Immunohistochemistry Assay
4.6. Enhancing HSPA4 Expression with Construct
4.7. Evaluating the Angiogenesis Ability of HUVECs with Tube Formation Assay
4.8. Analyzing Cell Growth Morphology with AngioTool
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, S.; Kocherginsky, M.; Hibbard, J.U. Abnormal placentation: Twenty-year analysis. Am. J. Obstet. Gynecol. 2005, 192, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.C.; Ou, H.Y.; Tsang, L.L.; Kung, F.T.; Hsu, T.Y.; Cheng, Y.F. Prophylactic intraoperative uterine artery embolization to control hemorrhage in abnormal placentation during late gestation. Fertil. Steril. 2009, 91, 1951–1955. [Google Scholar] [CrossRef] [PubMed]
- Tantbirojn, P.; Crum, C.P.; Parast, M.M. Pathophysiology of placenta creta: The role of decidua and extravillous trophoblast. Placenta 2008, 29, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Silver, R.M.; Landon, M.B.; Rouse, D.J.; Leveno, K.J.; Spong, C.Y.; Thom, E.A.; Moawad, A.H.; Caritis, S.N.; Harper, M.; Wapner, R.J.; et al. Maternal morbidity associated with multiple repeat cesarean deliveries. Obstet. Gynecol. 2006, 107, 1226–1232. [Google Scholar] [CrossRef]
- Miller, D.A.; Chollet, J.A.; Goodwin, T.M. Clinical risk factors for placenta previa-placenta accreta. Am. J. Obstet. Gynecol. 1997, 177, 210–214. [Google Scholar] [CrossRef]
- Chou, M.M.; Ho, E.S.; Lee, Y.H. Prenatal diagnosis of placenta previa accreta by transabdominal color Doppler ultrasound. Ultrasound Obstet. Gynecol. 2000, 15, 28–35. [Google Scholar] [CrossRef]
- Thorp, J.M., Jr.; Councell, R.B.; Sandridge, D.A.; Wiest, H.H. Antepartum diagnosis of placenta previa percreta by magnetic resonance imaging. Obstet. Gynecol. 1992, 80 Pt 2, 506–508. [Google Scholar]
- Du, M.R.; Zhou, W.H.; Yan, F.T.; Zhu, X.Y.; He, Y.Y.; Yang, J.Y.; Li, D.J. Cyclosporine A induces titin expression via MAPK/ERK signalling and improves proliferative and invasive potential of human trophoblast cells. Hum. Reprod. 2007, 22, 2528–2537. [Google Scholar] [CrossRef][Green Version]
- Staun-Ram, E.; Goldman, S.; Shalev, E. Ets-2 and p53 mediate cAMP-induced MMP-2 expression, activity and trophoblast invasion. Reprod. Biol. Endocrinol. 2009, 7, 135. [Google Scholar] [CrossRef]
- Tseng, J.J.; Hsu, S.L.; Wen, M.C.; Ho, E.S.; Chou, M.M. Expression of epidermal growth factor receptor and c-erbB-2 oncoprotein in trophoblast populations of placenta accreta. Am. J. Obstet. Gynecol. 2004, 191, 2106–2113. [Google Scholar] [CrossRef]
- Goh, W.; Yamamoto, S.Y.; Thompson, K.S.; Bryant-Greenwood, G.D. Relaxin, Its Receptor (RXFP1), and Insulin-Like Peptide 4 Expression Through Gestation and in Placenta Accreta. Reprod. Sci. 2013, 20, 968–980. [Google Scholar] [CrossRef]
- Duncan, M.W.; Hunsucker, S.W. Proteomics as a tool for clinically relevant biomarker discovery and validation. Exp. Biol. Med. 2005, 230, 808–817. [Google Scholar] [CrossRef]
- Marko-Varga, G.; Fehniger, T.E. Proteomics and disease—The challenges for technology and discovery. J. Proteome Res. 2004, 3, 167–178. [Google Scholar] [CrossRef]
- Bartels, H.C.; Postle, J.D.; Downey, P.; Brennan, D.J. Placenta Accreta Spectrum: A Review of Pathology, Molecular Biology, and Biomarkers. Dis. Markers 2018, 2018, 1507674. [Google Scholar] [CrossRef]
- Wu, C.Y.; Lin, C.T.; Wu, M.Z.; Wu, K.J. Induction of HSPA4 and HSPA14 by NBS1 overexpression contributes to NBS1-induced in vitro metastatic and transformation activity. J. Biomed. Sci. 2011, 18, 1. [Google Scholar] [CrossRef]
- Zhong, M.A.; Zhang, H.; Qi, X.Y.; Lu, A.G.; You, T.G.; Gao, W.; Guo, X.L.; Zhou, Z.Q.; Yang, Y.; Wang, C.J. ShRNA-mediated gene silencing of heat shock protein 70 inhibits human colon cancer growth. Mol. Med. Rep. 2011, 4, 805–810. [Google Scholar]
- Lee, F.Y.; Sun, C.K.; Sung, P.H.; Chen, K.H.; Chua, S.; Sheu, J.J.; Chung, S.Y.; Chai, H.T.; Chen, Y.L.; Huang, T.H.; et al. Daily melatonin protects the endothelial lineage and functional integrity against the aging process, oxidative stress, and toxic environment and restores blood flow in critical limb ischemia area in mice. J. Pineal Res. 2018, 65, e12489. [Google Scholar] [CrossRef]
- Einerson, B.D.; Branch, D.W. Surgical Management of Placenta Accreta Spectrum. Clin. Obstet. Gynecol. 2018, 61, 774–782. [Google Scholar] [CrossRef]
- Silver, R.M.; Branch, D.W. Placenta Accreta Spectrum. N. Engl. J. Med. 2018, 378, 1529–1536. [Google Scholar] [CrossRef]
- Morisaki, T.; Yashiro, M.; Kakehashi, A.; Inagaki, A.; Kinoshita, H.; Fukuoka, T.; Kasashima, H.; Masuda, G.; Sakurai, K.; Kubo, N.; et al. Comparative proteomics analysis of gastric cancer stem cells. PLoS ONE 2014, 9, e110736. [Google Scholar] [CrossRef]
- Josimovich, J.B.; Atwood, B.L.; Goss, D.A. Luteotrophic, Immunologic and Electrophoretic Properties of Human Placental Lactogen. Endocrinology 1963, 73, 410–420. [Google Scholar] [CrossRef]
- Sibiak, R.; Jankowski, M.; Gutaj, P.; Mozdziak, P.; Kempisty, B.; Wender-Ożegowska, E. Placental Lactogen as a Marker of Maternal Obesity, Diabetes, and Fetal Growth Abnormalities: Current Knowledge and Clinical Perspectives. J. Clin. Med. 2020, 9, 1142. [Google Scholar] [CrossRef]
- Corbacho, A.M.; Martinez De La Escalera, G.; Clapp, C. Roles of prolactin and related members of the prolactin/growth hormone/placental lactogen family in angiogenesis. J. Endocrinol. 2002, 173, 219–238. [Google Scholar] [CrossRef]
- Lo, T.K.; Yung, W.K.; Lau, W.L.; Law, B.; Lau, S.; Leung, W.C. Planned conservative management of placenta accrete—Experience of a regional general hospital. J. Matern.-Fetal Neonatal Med. 2014, 27, 291–296. [Google Scholar] [CrossRef]
- Huang, K.L.; Leung-Chit Tsang, L.; Cheng, Y.F.; Huang, F.J.; Fu, H.C.; Kung, F.T.; Tsai, C.C.; Cheng, H.H.; Lai, Y.J.; Ou, C.Y.; et al. Planned conservative management of placenta increta and percreta with prophylactic transcatheter arterial embolization and leaving placenta in situ for women who desire fertility preservation. Placenta 2020, 97, 51–57. [Google Scholar] [CrossRef]
- Hsu, T.Y.; Lin, H.; Hung, H.N.; Yang, K.D.; Ou, C.Y.; Tsai, C.C.; Cheng, H.H.; Chung, S.H.; Cheng, B.H.; Wong, Y.H.; et al. Two-Dimensional Differential Gel Electrophoresis to Identify Protein Biomarkers in Amniotic Fluid of Edwards Syndrome (Trisomy 18) Pregnancies. PLoS ONE 2016, 11, e0145908. [Google Scholar] [CrossRef]
- Hsu, T.Y.; Tsai, K.W.; Lan, K.C.; Hung, H.N.; Lai, Y.J.; Cheng, H.H.; Tsai, C.C.; Li, S.C. Identifying the potential protein biomarkers of preterm birth in amniotic fluid. Taiwan J. Obstet. Gynecol. 2020, 59, 366–371. [Google Scholar] [CrossRef]
- Zudaire, E.; Gambardella, L.; Kurcz, C.; Vermeren, S. A computational tool for quantitative analysis of vascular networks. PLoS ONE 2011, 6, e27385. [Google Scholar] [CrossRef]

| Normal Pregnancy (NP, n = 5) | Placenta Accreta Spectrum (PAS, n = 5) | p-Value | |
|---|---|---|---|
| Age (years old) | 31 ± 3.9 | 33.6 ± 1.2 | NS |
| GA (week) | 36 ± 1.1 | 35.8 ± 2.3 | NS |
| NW (g) | 2940 ± 224.9 | 2684 ± 514.6 | NS |
| Case | Age | Gestational Age | Type of Placenta Accreta | Blood Loss (mL) |
|---|---|---|---|---|
| 1 | 32 | 37 | Percreta | 6000 |
| 2 | 33 | 38 | Increta | 14,110 |
| 3 | 33 | 33 | Percreta | 4500 |
| 4 | 35 | 33 | Increta | 1900 |
| 5 | 35 | 38 | Increta | 1500 |
| Spot No. | SwissPort Entry Name | Protein Name | Theoretical pI/Mr | Sequence Coverage, MS% | Score | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|
| 1 | A2KLM6 | Immunoglobulin heavy chain variable region | 5.25/11 | 77 | 68 | −1.69 | 0.052 |
| 2 | HSPA4 | Heat shock 70 kDa protein 4 | 5.11/94.2 | 20 | 57 | 4.92 | 0.045 |
| 3 | PRKAR2 | cAMP-dependent protein kinase type II-alpha regulatory subunit | 4.96/45.4 | 33 | 68 | 7.42 | 0.023 |
| 4 | GSTP1 | Glutathione S-transferase P | 5.43/23.3 | 47 | 66 | −1.50 | 0.054 |
| 5 | MYNN | Myoneurin | 5.72/10.6 | 79 | 69 | −1.66 | 0.038 |
| 6 | ZN211 | Zinc finger protein 211 | 8.83/64.4 | 27 | 56 | −1.65 | 0.041 |
| 7 | UTS2 | Urotensin-2 isoform a preproprotein | 7/16 | 41 | 66 | 2.00 | 0.0052 |
| 8 | ZN157 | Zinc finger protein 157 | 8.83/58.2 | 29 | 62 | 1.98 | 0.025 |
| 9 | ACTB | Actin, cytoplasmic 1 | 5.29/41.7 | 25 | 58 | 5.36 | 0.021 |
| 10 | CSH | Chorionic somatomammotropin hormone | 5.34/25 | 57 | 147 | −1.63 | 0.028 |
| 11 | IQEC1 | IQ motif and SEC7 domain-containing protein 1 | 6.49/108 | 17 | 58 | 2.54 | 0.053 |
| 12 | CLIC1 | Chloride intracellular channel protein 1 | 5.09/26.9 | 51 | 140 | 1.54 | 0.037 |
| 13 | PMFBP | Polyamine-modulated factor 1-binding protein 1 | 5.94/118 | 22 | 59 | 1.91 | 0.010 |
| 14 | DDX4 | Probable ATP-dependent RNA helicase DDX4 | 5.62/79.2 | 20 | 57 | −1.51 | 0.014 |
| 15 | PLXB2 | Plexin-B2 | 5.85/204 | 15 | 58 | −1.63 | 0.044 |
| 16 | D3DT17 | hCG2038441 | 9.28/16.3 | 65 | 67 | 1.59 | 0.035 |
| 17 | B7ZC06 | Golgi autoantigen, golgin subfamily a, 2 | 6.05/53.3 | 20 | 67 | 1.62 | 0.0032 |
| 18 | AMBP | Protein AMBP | 5.95/38.9 | 38 | 56 | −1.67 | 0.025 |
| 19 | ANXA2 | Annexin A2 | 7.57/38.5 | 28 | 86 | −1.56 | 0.0060 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.-C.; Lan, K.-C.; Hung, H.-N.; Huang, W.-T.; Lai, Y.-J.; Cheng, H.-H.; Tsai, C.-C.; Huang, K.-L.; You, H.-L.; Hsu, T.-Y. HSPA4 Is a Biomarker of Placenta Accreta and Enhances the Angiogenesis Ability of Vessel Endothelial Cells. Int. J. Mol. Sci. 2022, 23, 5682. https://doi.org/10.3390/ijms23105682
Li S-C, Lan K-C, Hung H-N, Huang W-T, Lai Y-J, Cheng H-H, Tsai C-C, Huang K-L, You H-L, Hsu T-Y. HSPA4 Is a Biomarker of Placenta Accreta and Enhances the Angiogenesis Ability of Vessel Endothelial Cells. International Journal of Molecular Sciences. 2022; 23(10):5682. https://doi.org/10.3390/ijms23105682
Chicago/Turabian StyleLi, Sung-Chou, Kuo-Chung Lan, Hsuan-Ning Hung, Wan-Ting Huang, Yun-Ju Lai, Hsin-Hsin Cheng, Chih-Chang Tsai, Kun-Long Huang, Huey-Ling You, and Te-Yao Hsu. 2022. "HSPA4 Is a Biomarker of Placenta Accreta and Enhances the Angiogenesis Ability of Vessel Endothelial Cells" International Journal of Molecular Sciences 23, no. 10: 5682. https://doi.org/10.3390/ijms23105682
APA StyleLi, S.-C., Lan, K.-C., Hung, H.-N., Huang, W.-T., Lai, Y.-J., Cheng, H.-H., Tsai, C.-C., Huang, K.-L., You, H.-L., & Hsu, T.-Y. (2022). HSPA4 Is a Biomarker of Placenta Accreta and Enhances the Angiogenesis Ability of Vessel Endothelial Cells. International Journal of Molecular Sciences, 23(10), 5682. https://doi.org/10.3390/ijms23105682






