Immune Modulation Using Extracellular Vesicles Encapsulated with MicroRNAs as Novel Drug Delivery Systems
Abstract
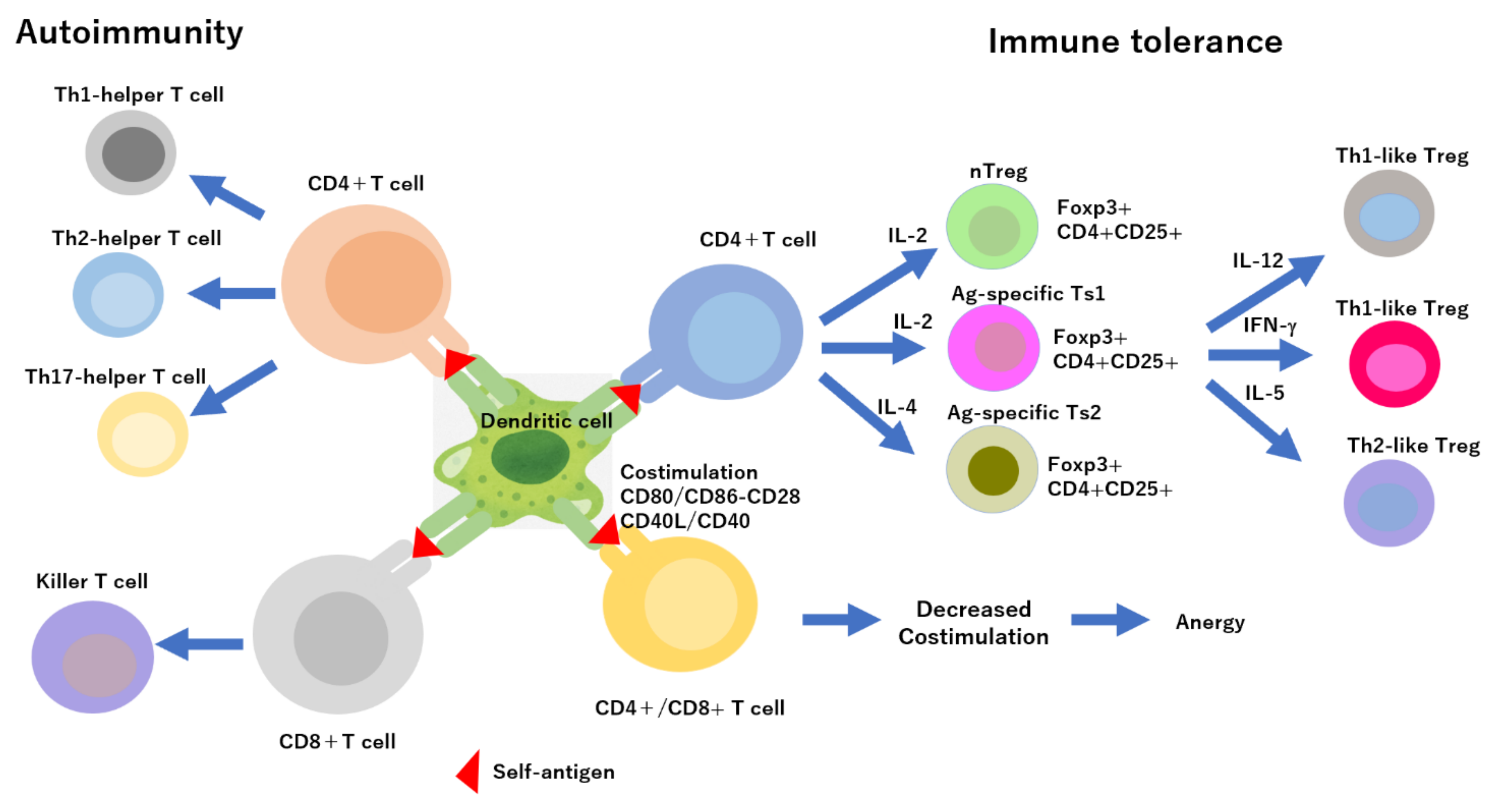
1. Introduction
2. Immune-Cell-Derived Exosome as an Immune Modulator
3. Regulation of Immune Response by Exosomes
4. Exosomal miRNA-Mediated Molecular Pathways in Immune Response
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Tsubata, T. Role of inhibitory B cell co-receptors in B cell self-tolerance to non-protein antigens. Immunol. Rev. 2022, 30, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Alhazmi, A.; Nekoua, M.P.; Michaux, H.; Sane, F.; Halouani, A.; Engelmann, I.; Alidjinou, E.K.; Martens, H.; Jaidane, H.; Geenen, V.; et al. Effect of Coxsackievirus B4 Infection on the Thymus: Elucidating Its Role in the Pathogenesis of Type 1 Diabetes. Microorganisms 2021, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Song, R.H.; Gao, C.Q.; Zhao, J.; Zhang, J.A. An Update Evolving View of Copy Number Variations in Autoimmune Diseases. Front. Genet. 2022, 12, 794348. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, J.; Hidalgo, Y.; Sauma, D.; Rosemblatt, M.; Bono, M.R.; Núñez, S. The Multifaceted Roles of B Cells in the Thymus: From Immune Tolerance to Autoimmunity. Front. Immunol. 2021, 12, 766698. [Google Scholar] [CrossRef] [PubMed]
- Greaves, R.B.; Chen, D.; Green, E.A. Thymic B Cells as a New Player in the Type 1 Diabetes Response. Front. Immunol. 2021, 12, 772017. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Tanaka, M. Programmed cell death and the immune system. Nat. Rev. Immunol. 2017, 17, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Khan, U.; Ghazanfar, H. T Lymphocytes and Autoimmunity. Int. Rev. Cell Mol. Biol. 2018, 341, 125–168. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.; John, S.; Cullen, J.; Luo, W.; Shlomchik, M.J.; Garrett-Sinha, L.A. Requirement for Transcription Factor Ets1 in B Cell Tolerance to Self-Antigens. J. Immunol. 2015, 195, 3574–3583. [Google Scholar] [CrossRef] [PubMed]
- Iriki, H.; Takahashi, H.; Wada, N.; Nomura, H.; Mukai, M.; Kamata, A.; Ito, H.; Yamagami, J.; Matsui, T.; Kurebayashi, Y.; et al. Peripheral tolerance by Treg via constraining OX40 signal in autoreactive T cells against desmoglein 3, a target antigen in pemphigus. Proc. Natl. Acad. Sci. USA 2021, 118, e2026763118. [Google Scholar] [CrossRef] [PubMed]
- Arcangeletti, M.C.; Caselli, E. Recent Advances in Unveiling the Role of Beta-Herpesviruses in Autoimmune Diseases. Microorganisms 2021, 9, 2572. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, N.; Akhtari, M.; Farhadi, E.; Mansouri, R.; Faezi, S.T.; Jamshidi, A.; Mahmoudi, M. Role of the innate and adaptive immune responses in the pathogenesis of systemic lupus erythematosus. Inflamm. Res. 2022; in press. [Google Scholar] [CrossRef] [PubMed]
- Glauzy, S.; Sng, J.; Bannock, J.M.; Gottenberg, J.E.; Korganow, A.S.; Cacoub, P.; Saadoun, D.; Meffre, E. Defective Early B Cell Tolerance Checkpoints in Sjögren’s Syndrome Patients. Arthritis Rheumatol. 2017, 69, 2203–2208. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, X.; Luo, X.; Wang, H.; Zhu, Y.; Du, G.; Chen, W.; Chen, Z.; Hao, X.; Zhang, Z.; et al. Nanoemulsions Target to Ectopic Lymphoids in Inflamed Joints to Restore Immune Tolerance in Rheumatoid Arthritis. Nano Lett. 2021, 21, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Kunkl, M.; Sambucci, M.; Ruggieri, S.; Amormino, C.; Tortorella, C.; Gasperini, C.; Battistini, L.; Tuosto, L. CD28 Autonomous Signaling Up-Regulates C-Myc Expression and Promotes Glycolysis Enabling Inflammatory T Cell Responses in Multiple Sclerosis. Cells 2019, 8, 575. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.I.; Lee, A.H.; Shin, H.Y.; Song, H.R.; Park, J.H.; Kang, T.B.; Lee, S.R.; Yang, S.H. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Viallard, J.F.; Pellegrin, J.L.; Moreau, J.F. Cytotoxic T lymphocytes and autoimmunity. Curr. Opin. Rheumatol. 2005, 17, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Shlomchik, M.J. Sites and stages of autoreactive B cell activation and regulation. Immunity 2008, 28, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Prüss, H. Autoantibodies in neurological disease. Nat. Rev. Immunol. 2021, 21, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Gronski, M.A.; Weinem, M. Death pathways in T cell homeostasis and their role in autoimmune diabetes. Rev. Diabet. Stud. 2006, 3, 88–95. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iriki, H.; Mukai, M.; Ito, H.; Kurebayashi, Y.; Amagai, M.; Takahashi, H. Imiquimod-induced dermatitis impairs thymic tolerance of autoreactive CD4+ T cells to desmoglein 3. J. Dermatol. Sci. 2020, 100, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Marunouchi, T.; Nishiumi, C.; Iinuma, S.; Yano, E.; Tanonaka, K. Effects of Hsp90 inhibitor on the RIP1-RIP3-MLKL pathway during the development of heart failure in mice. Eur. J. Pharmacol. 2021, 898, 173987. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Tong, J.; Yang, L.; Wei, L.; Stolz, D.B.; Yu, J.; Zhang, J.; Zhang, L. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc. Natl. Acad. Sci. USA 2018, 115, 3930–3935. [Google Scholar] [CrossRef] [PubMed]
- Mirshahidi, S.; Huang, C.T.; Sadegh-Nasseri, S. Anergy in peripheral memory CD4(+) T cells induced by low avidity engagement of T cell receptor. J. Exp. Med. 2001, 194, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Wildner, G.; Diedrichs-Möhring, M. Molecular Mimicry and Uveitis. Front. Immunol. 2020, 11, 580636. [Google Scholar] [CrossRef] [PubMed]
- Martín-Villa, J.M.; Vaquero-Yuste, C.; Molina-Alejandre, M.; Juarez, I.; Suárez-Trujillo, F.; López-Nares, A.; Palacio-Gruber, J.; Barrera-Gutiérrez, L.; Fernández-Cruz, E.; Rodríguez-Sainz, C.; et al. HLA-G: Too Much or Too Little? Role in Cancer and Autoimmune Disease. Front. Immunol. 2022, 13, 796054. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. T-cell cross-reactivity may explain the large variation in how cancer patients respond to checkpoint inhibitors. Scand. J. Immunol. 2018, 87. [Google Scholar] [CrossRef] [PubMed]
- Koenig, A.; Buskiewicz-Koenig, I.A. Redox Activation of Mitochondrial DAMPs and the Metabolic Consequences for Development of Autoimmunity. Antioxid. Redox Signal. 2022, 36, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Fillatreau, S.; Manfroi, B.; Dörner, T. Toll-like receptor signalling in B cells during systemic lupus erythematosus. Nat. Rev. Rheumatol. 2021, 17, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xie, Z.; Lu, Q.; Chang, C.; Zhou, Z. Beyond Genetics: What Causes Type 1 Diabetes. Clin. Rev. Allergy Immunol. 2017, 52, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Effraimidis, G.; Wiersinga, W.M. Mechanisms in endocrinology: Autoimmune thyroid disease: Old and new players. Eur. J. Endocrinol. 2014, 170, R241–R252. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ni, Y.Q.; Liu, Y.S. Mechanisms of Action of MiRNAs and LncRNAs in Extracellular Vesicle in Atherosclerosis. Front. Cardiovasc. Med. 2021, 8, 733985. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Tandon, S.; Tandon, C. Emerging role of exosomes in arterial and renal calcification. Hum. Exp. Toxicol. 2021, 40, 1385–1402. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Peng, J.; Wang, S.; Liu, X.; Zhang, K.; Zhang, Z.; Wang, C.; Jing, X.; Zhou, C.; Wang, Y. Applications of stem cell-derived exosomes in tissue engineering and neurological diseases. Rev. Neurosci. 2018, 29, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, Y.; Li, H.J. Advances in mesenchymal stem cell exosomes: A review. Stem Cell Res. Ther. 2021, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, H.; Peng, H.; Huyan, T.; Cacalano, N.A. Exosomes: Versatile Nano Mediators of Immune Regulation. Cancers 2019, 11, 1557. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gil, Z. The Role of Extracellular Vesicles in Cancer-Nerve Crosstalk of the Peripheral Nervous System. Cells 2022, 11, 1294. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, Q. Potential clinical applications of exosomes in the diagnosis, treatment, and prognosis of cardiovascular diseases: A narrative review. Ann. Transl. Med. 2022, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Yaghoubi, Y.; Movassaghpour, A.; Zamani, M.; Talebi, M.; Mehdizadeh, A.; Yousefi, M. Human umbilical cord mesenchymal stem cells derived-exosomes in diseases treatment. Life Sci. 2019, 233, 116733. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.D.; Peiris, H.N.; Kobayashi, M.; Koh, Y.Q.; Duncombe, G.; Illanes, S.E.; Rice, G.E.; Salomon, C. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015, 213, S173–S181. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wiener Medizinische Wochenschrift 2016, 166, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.B.; Dammer, E.B.; Ren, R.J.; Wang, G. The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Gruenberg, J. Life in the lumen: The multivesicular endosome. Traffic 2020, 21, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Rentero, C.; Blanco-Muñoz, P.; Meneses-Salas, E.; Grewal, T.; Enrich, C. Annexins-Coordinators of Cholesterol Homeostasis in Endocytic Pathways. Int. J. Mol. Sci. 2018, 19, 1444. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Cai, S.; Shen, J.; Peng, H. Tetraspanins: Novel Molecular Regulators of Gastric Cancer. Front. Oncol. 2021, 11, 702510. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef] [PubMed]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [PubMed]
- Carayon, K.; Chaoui, K.; Ronzier, E.; Lazar, I.; Bertrand-Michel, J.; Roques, V.; Balor, S.; Terce, F.; Lopez, A.; Salomé, L.; et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. J. Biol. Chem. 2011, 286, 34426–34439. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Fatima, A.; Fan, X.X.; Malik, S.I. REVIEW-The Biological importance of cells secreted Exosomes. Pak. J. Pharm. Sci. 2021, 34, 2273–2279. [Google Scholar] [PubMed]
- Zeng, F.; Morelli, A.E. Extracellular vesicle-mediated MHC cross-dressing in immune homeostasis, transplantation, infectious diseases, and cancer. Semin. Immunopathol. 2018, 40, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef] [PubMed]
- Shefler, I.; Salamon, P.; Hershko, A.Y.; Mekori, Y.A. Mast cells as sources and targets of membrane vesicles. Curr. Pharm. Des. 2011, 17, 3797–3804. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, U.; Näslund, T.I.; Hiltbrunner, S.; Larssen, P.; Gabrielsson, S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin. Cancer Biol. 2014, 28, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Näslund, T.I.; Gehrmann, U.; Qazi, K.Q.; Karlsson, M.C.I.; Gabrielsson, S. Dendritic cell-derived exosomes need to activate both T and B cells to induce antitumor immunity. J. Immunol. 2013, 190, 2712–2719. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yang, F.; Yu, L.; Yu, Z.; Jiang, L.; Wang, Q.; Yang, Y.; Wang, L.; Cao, X.; Wang, J. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J. Immunol. 2012, 188, 5954–5961. [Google Scholar] [CrossRef] [PubMed]
- Ostman, S.; Taube, M.; Telemo, E. Tolerosome-induced oral tolerance is MHC dependent. Immunology 2005, 116, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.H.; Ziegler, S.F. Regulating the immune system: The induction of regulatory T cells in the periphery. Arthritis Res. Ther. 2004, 6, 215–222. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Hogquist, K.A. T-cell tolerance: Central and peripheral. Cold Spring Harb. Perspect. Biol. 2012, 4, a006957. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, M.H.; Negahdari, B. Clinical and diagnostic potential of regulatory T cell markers: From bench to bedside. Transpl. Immunol. 2022, 70, 101518. [Google Scholar] [CrossRef] [PubMed]
- Irla, M. Instructive Cues of Thymic T Cell Selection. Annu. Rev. Immunol. 2022, 40, 95–119. [Google Scholar] [CrossRef] [PubMed]
- Nikfarjam, S.; Rezaie, J.; Kashanchi, F.; Jafari, R. Dexosomes as a cell-free vaccine for cancer immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 258. [Google Scholar] [CrossRef] [PubMed]
- El-Awady, A.R.; Elashiry, M.; Morandini, A.C.; Meghil, M.M.; Cutler, C.W. Dendritic cells a critical link to alveolar bone loss and systemic disease risk in periodontitis: Immunotherapeutic implications. Periodontology 2000 2022, 89, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Ouyang, S.; Li, Y.; Xiao, B.; Yang, H. Immature dendritic cell-derived exosomes: A promise subcellular vaccine for autoimmunity. Inflammation 2013, 36, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Moyana, T.; Xiang, J. Review: Cancer immunotherapy by exosome-based vaccines. Cancer Biother. Radiopharm. 2007, 22, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Tomiyama, T.; Yang, G.X.; Zhao, M.; Zhang, W.; Tanaka, H.; Wang, J.; Leung, P.S.; Okazaki, K.; He, X.S.; Lu, Q.; et al. The modulation of co-stimulatory molecules by circulating exosomes in primary biliary cirrhosis. Cell Mol. Immunol. 2017, 14, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Shenoda, B.B.; Ajit, S.K. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin. Med. Insights Pathol. 2016, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Gao, W.; Yao, K.; Ge, J. Roles of Exosomes Derived From Immune Cells in Cardiovascular Diseases. Front. Immunol. 2019, 10, 648. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Pu, S.D.; Li, X.; Yu, Z.W.; Zhang, Y.T.; Tong, X.W.; Shan, Y.Y.; Gao, X.Y. Exosomal ncRNAs: Novel therapeutic target and biomarker for diabetic complications. Pharmacol. Res. 2022, 178, 106135. [Google Scholar] [CrossRef] [PubMed]
- Amiri, A.; Bagherifar, R.; Ansari Dezfouli, E.; Kiaie, S.H.; Jafari, R.; Ramezani, R. Exosomes as bio-inspired nanocarriers for RNA delivery: Preparation and applications. J. Transl. Med. 2022, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, A.; Purvinsh, L.; Brodskaia, A.; Vasin, A. Exosomes as Natural Nanocarriers for RNA-Based Therapy and Prophylaxis. Nanomaterials 2022, 12, 524. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021, 35, e21916. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Palanisamy, V. Horizontal transfer of RNAs: Exosomes as mediators of intercellular communication. Wiley Interdiscip. Rev. RNA 2012, 3, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.M.; Carneiro, F.; Machado, J.C.; Melo, S.A. Exosomes and Immune Response in Cancer: Friends or Foes? Front. Immunol. 2018, 9, 730. [Google Scholar] [CrossRef] [PubMed]
- Tavakkoli, S.; Sotoodehnejadnematalahi, F.; Fathollahi, A.; Bandehpour, M.; Haji Molla Hoseini, M.; Yeganeh, F. EL4-derived Exosomes Carry Functional TNF-related Apoptosis-inducing Ligand that are Able to Induce Apoptosis and Necrosis in the Target Cells. Int. J. Mol. Cell Med. 2020, 9, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Zou, Y.P.; Hu, S.Q.; Zhang, T.W.; Zhou, H.; Liang, B.; Zhuang, C.Y.; Wang, H.R.; Jiang, L.B.; Li, X.L. TNF-α-stimulated nucleus pulposus cells induce cell apoptosis through the release of exosomal miR-16 targeting IGF-1 and IGF-1R in rats. Ann. Transl. Med. 2021, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Song, R.; Du, P.; Gao, C.; Yao, Q.; Zhang, J.A. Systemic Proteomic Analysis Reveals Distinct Exosomal Protein Profiles in Rheumatoid Arthritis. J. Immunol. Res. 2021, 2021, 9421720. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, R.; Sarvnaz, H.; Arabpour, M.; Ramshe, S.M.; Asef-Kabiri, L.; Yousefi, H.; Akbari, M.E.; Eskandari, N. Cancer exosomes and natural killer cells dysfunction: Biological roles, clinical significance and implications for immunotherapy. Mol. Cancer 2022, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Czernek, L.; Düchler, M. Functions of Cancer-Derived Extracellular Vesicles in Immunosuppression. Arch. Immunol. Ther. Exp. 2017, 65, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Mincheva-Nilsson, L.; Baranov, V. Cancer exosomes and NKG2D receptor-ligand interactions: Impairing NKG2D-mediated cytotoxicity and anti-tumour immune surveillance. Semin. Cancer Biol. 2014, 28, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, J.; Karlson Tde, L.; Glader, P.; Telemo, E.; Valadi, H. Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS ONE 2012, 7, e49723. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.; Mu, W.; Barry, J.; Han, A.; Carpenter, R.L.; Jiang, B.H.; Peiper, S.C.; Mahoney, M.G.; Aplin, A.E.; et al. Reactive oxygen species reprogram macrophages to suppress antitumor immune response through the exosomal miR-155-5p/PD-L1 pathway. J. Exp. Clin. Cancer Res. 2022, 41, 41. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The effect of tumor-derived exosomes on immune regulation and cancer immunotherapy. Future Oncol. 2017, 13, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Shen, H.; Li, Z.; Wang, T.; Wang, S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J. Hematol. Oncol. 2019, 12, 84. [Google Scholar] [CrossRef] [PubMed]
- De Kouchkovsky, D.; Esensten, J.H.; Rosenthal, W.L.; Morar, M.M.; Bluestone, J.A.; Jeker, L.T. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J. Immunol. 2013, 191, 1594–1605. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, J.; Cheng, Y.; Zheng, P. Exosomal Transfer of Macrophage-Derived miR-223 Confers Doxorubicin Resistance in Gastric Cancer. Onco Targets Ther. 2020, 13, 12169–12179. [Google Scholar] [CrossRef] [PubMed]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Green, J.; Bodogai, M.; Mode, N.A.; Bæk, R.; Jørgensen, M.M.; Freeman, D.W.; Witwer, K.W.; Zonderman, A.B.; Biragyn, A.; et al. Age-Related Changes in Plasma Extracellular Vesicle Characteristics and Internalization by Leukocytes. Sci. Rep. 2017, 7, 1342. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 5335. [Google Scholar] [CrossRef] [PubMed]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef] [PubMed]
- Mycko, M.P.; Cichalewska, M.; Machlanska, A.; Cwiklinska, H.; Mariasiewicz, M.; Selmaj, K.W. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc. Natl. Acad. Sci. USA 2012, 109, E1248–E1257. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Song, H.; Zhou, Z.; Chen, X.; Li, H.; Zhang, Y.; Wang, J.; Ren, X.; Wang, X. Promotion or inhibition of extracellular vesicle release: Emerging therapeutic opportunities. J. Control. Release 2021, 340, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Alabasi, G.; Sandaltzopoulos, R.; Marcu, K.B.; Kolettas, E. Roles of NF-κB Signaling in the Regulation of miRNAs Impacting on Inflammation in Cancer. Biomedicines 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Jing, Z.; Cheng, G. MicroRNAs: New regulators of Toll-like receptor signalling pathways. Biomed. Res. Int. 2014, 2014, 945169. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Ma, L.; He, C. MicroRNA-23a-5p regulates cell proliferation, migration and inflammation of TNF-α-stimulated human fibroblast-like MH7A synoviocytes by targeting TLR4 in rheumatoid arthritis. Exp. Ther. Med. 2021, 21, 479. [Google Scholar] [CrossRef] [PubMed]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef] [PubMed]
- McGettrick, A.F.; O’Neill, L.A.J. The Role of HIF in Immunity and Inflammation. Cell Metab. 2020, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhao, H.; Zhang, M.; He, Y.; Li, X.; Xu, Y.; Liu, X. Hypoxia Induced Changes of Exosome Cargo and Subsequent Biological Effects. Front. Immunol. 2022, 13, 824188. [Google Scholar] [CrossRef] [PubMed]
- Jafari, R.; Rahbarghazi, R.; Ahmadi, M.; Hassanpour, M.; Rezaie, J. Hypoxic exosomes orchestrate tumorigenesis: Molecular mechanisms and therapeutic implications. J. Transl. Med. 2020, 18, 474. [Google Scholar] [CrossRef] [PubMed]
- Dian, D.; Xie, Y.; Huang, M.; Gu, J. Tumor-derived exosomes in hypoxic microenvironment: Release mechanism, biological function and clinical application. J. Cancer 2022, 13, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tan, J.; Miao, Y.; Zhang, Q. MicroRNA in extracellular vesicles regulates inflammation through macrophages under hypoxia. Cell Death Discov. 2021, 7, 285. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Deep, G. Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities. Cancer Lett. 2020, 479, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; O’Dea, E.L.; Doedens, A.; Kim, J.W.; Weidemann, A.; Stockmann, C.; Asagiri, M.; Simon, M.C.; Hoffmann, A.; Johnson, R.S. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010, 24, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.F.; Thomas, M.F.; Hu, J.K.; Yang, Z.; Babiarz, J.E.; Allen, C.D.; Matloubian, M.; Blelloch, R.; Ansel, K.M. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 2011, 35, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Qian, C.; Qian, L.; Ma, F.; Hou, J.; Chen, Y.; Wang, Q.; Cao, X. Integrin CD11b negatively regulates TLR9-triggered dendritic cell cross-priming by upregulating microRNA-146a. J. Immunol. 2012, 188, 5293–5302. [Google Scholar] [CrossRef] [PubMed]
- Murugina, N.E.; Budikhina, A.S.; Dagil, Y.A.; Maximchik, P.V.; Balyasova, L.S.; Murugin, V.V.; Melnikov, M.V.; Sharova, V.S.; Nikolaeva, A.M.; Chkadua, G.Z.; et al. Glycolytic reprogramming of macrophages activated by NOD1 and TLR4 agonists: No association with proinflammatory cytokine production in normoxia. J. Biol. Chem. 2020, 295, 3099–3114. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.; O’Neill, L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreño, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2020, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 781815. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, G.F.; Kuzmina, D.O.; Nuzhina, J.; Shtil, A.A.; Dukhinova, M.S. How Macrophages Become Transcriptionally Dysregulated: A Hidden Impact of Antitumor Therapy. Int. J. Mol. Sci. 2021, 22, 2662. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Shan, Q.; Gu, P.; Wang, X.M.; Tai, L.W.; Sun, M.; Luo, X.; Sun, L.; Cheung, C.W. miRNA-23a/CXCR4 regulates neuropathic pain via directly targeting TXNIP/NLRP3 inflammasome axis. J. Neuroinflamm. 2018, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Hsu, P.Y.; Su, M.C.; Chin, C.H.; Liou, C.W.; Wang, T.Y.; Lin, Y.Y.; Lee, C.P.; Lin, M.C.; Hsiao, C.C. miR-21-5p Under-Expression in Patients with Obstructive Sleep Apnea Modulates Intermittent Hypoxia with Re-Oxygenation-Induced-Cell Apoptosis and Cytotoxicity by Targeting Pro-Inflammatory TNF-α-TLR4 Signaling. Int. J. Mol. Sci. 2020, 21, 999. [Google Scholar] [CrossRef] [PubMed]
- Krzywinska, E.; Stockmann, C. Hypoxia, Metabolism and Immune Cell Function. Biomedicines 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Xue, Y.; Wang, P.; Lin, L.; Liu, Q.; Li, N.; Xu, J.; Cao, X. IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b. J. Immunol. 2014, 193, 3036–3044. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Guo, R.; Shi, Y.; Qi, F.; Guo, C.; Yang, L. miR-155 Regulated Inflammation Response by the SOCS1-STAT3-PDCD4 Axis in Atherogenesis. Mediat. Inflamm. 2016, 2016, 8060182. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Abak, A.; Tavakkoli Avval, S.; Shoorei, H.; Taheri, M.; Samadian, M. The impact of non-coding RNAs on macrophage polarization. Biomed. Pharmacother. 2021, 142, 112112. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Luo, Y.H.; Li, A.L.; Tsai, J.C.; Wu, K.L.; Chung, P.J.; Ma, N. miRNA as a Modulator of Immunotherapy and Immune Response in Melanoma. Biomolecules 2021, 11, 1648. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Brandt, S.; Medeiros, A.; Wang, S.; Wu, H.; Dent, A.; Serezani, C.H. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS ONE 2015, 10, e0115855. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, T.; Li, M.Q.; Zheng, X.L.; Zhao, G.J. Transcriptional Regulation of Macrophages Polarization by MicroRNAs. Front. Immunol. 2018, 9, 1175. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, L.; Hu, Y.; Huang, Y.; Zhang, Y.; Zheng, X.; Wang, S.; Wang, Y.; Yu, Y.; Zhang, M.; et al. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci. Rep. 2016, 6, 25602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, X.; Hu, J.; Chen, F.; Qiao, S.; Sun, X.; Gao, L.; Xie, J.; Xu, B. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc. Res. 2019, 115, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.; Lee, E.J.; Nam, G.H.; Hong, Y.; Cho, E.; Yang, Y.; Kim, I.S. Exosome-SIRPα, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 2017, 121, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Majer, O.; Liu, B.; Kreuk, L.S.M.; Krogan, N.; Barton, G.M. UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity. Nature 2019, 575, 366–370. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaka, Y.; Yashiro, R. Immune Modulation Using Extracellular Vesicles Encapsulated with MicroRNAs as Novel Drug Delivery Systems. Int. J. Mol. Sci. 2022, 23, 5658. https://doi.org/10.3390/ijms23105658
Matsuzaka Y, Yashiro R. Immune Modulation Using Extracellular Vesicles Encapsulated with MicroRNAs as Novel Drug Delivery Systems. International Journal of Molecular Sciences. 2022; 23(10):5658. https://doi.org/10.3390/ijms23105658
Chicago/Turabian StyleMatsuzaka, Yasunari, and Ryu Yashiro. 2022. "Immune Modulation Using Extracellular Vesicles Encapsulated with MicroRNAs as Novel Drug Delivery Systems" International Journal of Molecular Sciences 23, no. 10: 5658. https://doi.org/10.3390/ijms23105658
APA StyleMatsuzaka, Y., & Yashiro, R. (2022). Immune Modulation Using Extracellular Vesicles Encapsulated with MicroRNAs as Novel Drug Delivery Systems. International Journal of Molecular Sciences, 23(10), 5658. https://doi.org/10.3390/ijms23105658





