Abstract
C2H2 zinc finger protein (C2H2-ZFP) is one of the most important transcription factor families in higher plants. In this study, a total of 145 C2H2-ZFPs was identified in Sorghum bicolor and randomly distributed on 10 chromosomes. Based on the phylogenetic tree, these zinc finger gene family members were divided into 11 clades, and the gene structure and motif composition of SbC2H2-ZFPs in the same clade were similar. SbC2H2-ZFP members located in the same clade contained similar intron/exon and motif patterns. Thirty-three tandem duplicated SbC2H2-ZFPs and 24 pairs of segmental duplicated genes were identified. Moreover, synteny analysis showed that sorghum had more collinear regions with monocotyledonous plants such as maize and rice than did dicotyledons such as soybean and Arabidopsis. Furthermore, we used quantitative RT-PCR (qRT-PCR) to analyze the expression of C2H2-ZFPs in different organs and demonstrated that the genes responded to cold and drought. For example, Sobic.008G088842 might be activated by cold but is inhibited in drought in the stems and leaves. This work not only revealed an important expanded C2H2-ZFP gene family in Sorghum bicolor but also provides a research basis for determining the role of C2H2-ZFPs in sorghum development and abiotic stress resistance.
1. Introduction
The zinc finger protein family has evolved into a massive transcription factor family in higher plants [1]. Each zinc finger is approximately 23–30 amino acids in length, which is composed of cysteine and histidine combined with zinc ions through hydrogen bonds. It is well known that ZFP types are usually classified based on the number and position of cysteine and histidine residues, and ZFP can be divided into 10 subclasses, namely C2H2, C2HC, C2HC5, C2C2, CCCH, C3HC4, C4, C4HC3, C6, and C8 [2]. Among them, C2H2 zinc finger is one of the most general motifs in ZFP, which was initially reported in the African clawed frog (Xenopus laevis) transcription factor IIIA (TFIIIA) protein, so it is also called TFIIIA zinc finger protein [3]. Furthermore, its sequence feature is CX2-4CX12HX3-5H, where X stands for amino acid, C stands for cysteine, and H stands for histidine, while the number stands for the number of residues [4]. The number of C2H2 zinc fingers has great variability, ranging from one to dozens, which implies that the function of C2H2-ZFPs is very wide [5].
So far, more than 10 species of C2H2-ZFPs have been identified, including Arabidopsis thaliana (176), Glycine max (321), Camellia sinensis (134), Medicago truncatula (218), tomato (112), durum wheat (122), and rice (189) [6,7,8,9,10,11,12]. Because zinc fingers can bind to the main grooves of the DNA sequence, C2H2-ZFPs can affect DNA transcription, but the functions of C2H2-ZFPs are far more than activating or inhibiting transcription. In higher plants, they can also package RNA, assemble proteins, bind lipids, and participate in the regulation of apoptosis [13].
Studies show that C2H2-ZFPs have a wide range of effects on plant growth and development and resistance to abiotic stresses. In tomato, C2H2 zinc finger proteins H and Sh were found to be important transcription factors that controlled the initiation and elongation of type I and III multicellular trichomes [14]. At the same time, SlZFP6 was upregulated by protein H, which increased the density and length of tomato trichomes [15]. Apart from that, when the SUMOylation of the zinc finger transcription factor STOP1 was up-regulated, plants could enhance the resistance to aluminum stress in Arabidopsis [16]. Moreover, in Medicago truncatula, MtSUPERMAN could regulate compound inflorescence and flower development [17]. In addition, POPOVICH played a core part in the development of flower nectar stimulation in Aquilegia [18]. Furthermore, ectopic expression of MdZAT10 in Arabidopsis reduced the tolerance to drought stress [19].
Sweet sorghum (Sorghum bicolor) can be grown in most semi-arid regions of the world and has high sugar production and high photosynthetic conversion rate, so it is often used as a high energy crop and is planted all over the world [20]. The growth and development of sorghum is affected by various abiotic stresses, including drought stress and low temperature stress, which directly restrict the yield and quality of sorghum [21,22]. Although sorghum is drought-tolerant, saline–alkali-tolerant, and sensitive to low temperature, the mechanism is still unclear, and only a few C2H2 zinc finger genes have been identified in sorghum [23,24,25,26]. For example, overexpression of the SbSTOP1 gene can increase sorghum resistance to aluminum stress by activating the transcription of a β-1,3-glucanase gene [25]. Moreover, four STOP1-like proteins (SbSTOP1a, SbSTOP1b, SbSTOP1c, and SbSTOP1d) have been identified to be associated with aluminum tolerance, and SbSTOP1 might function as a homodimer and/or heterodimer [24]. However, most C2H2-ZFPs have not been identified in sorghum. Therefore, the whole genome sequence and expression of SbC2H2-ZFPs were analyzed in this study.
A total of 145 C2H2-type ZFPs was identified in this research. These zinc finger gene family members were divided into 11 clades by the phylogenetic tree. The SbC2H2-ZFP members located in the same clade contained similar intron/exon and motif patterns. Tandem repeats contributed more to the increase in C2H2-ZFP membership in sorghum than segmental repeats. Almost all C2H2-ZFPs of clades were expressed in roots, stems, and leaves, and we used qRT-PCR to determine the responses of C2H2-ZFPs to cold and drought stress. Our results enriched the knowledge of structural information, evolutionary relationship, and the expression patterns of sorghum C2H2-ZFPs and provided a basis for further studies on C2H2-ZFPs in the growth and stress resistance of sorghum.
2. Results
2.1. Identification of C2H2-ZFP Genes in S. bicolor
After removing genes containing multiple transcripts, there were found to be only 109 C2H2-ZFPs listed in the sorghum transcription factor database (http://planttfdb.gao-lab.org/family.php?sp=Sbi&fam=C2H2, accessed on 5 September 2021). Given there were 167 C2H2-ZFPs reported in Arabidopsis, we thought that there might be more C2H2-ZFPs in the S. bicolor genome. To identify all C2H2-ZFP members in S. bicolor, the hidden Markov model (HMM) profiles (PF00096, PF13894, PF13912, PF18414, PF16622, and PF18658) from the Pfam database were used for searching SbC2H2-ZFP genes (http://pfam.xfam.org/, accessed on 5 September 2021). The SbC2H2-ZFP genes were identified through alignment against Arabidopsis C2H2-ZFP sequences (e-value < 0.01). Furthermore, the sequences lacking the C2H2-zinc finger motif were removed based on MEME (https://meme-suite.org/meme/tools/meme, accessed on 25 September 2021) search (e-value < 0.05). Eventually, a total of 145 C2H2-ZFP genes was identified in S. bicolor (Table S1). Sobic.002G116600 has a minimum molecular weight (Mw) of 10.82 kDa, while Sobic.004G265500 has a maximum molecular weight of 178.48 kDa. Moreover, isoelectric point (pI) values of the SbC2H2-ZFPs were between 4.54 and 10.25, 47.65% of which were over 7.0.
2.2. Phylogenetic Analysis and Classification of SbC2H2-ZFPs
To predict functions of SbC2H2-ZFPs, a phylogenetic tree containing both A. thaliana and S. bicolor C2H2-ZFPs was built by the maximum likelihood (ML) method. Based on the sequence similarity and topology, SbC2H2-ZFPs were divided into five clades containing clade A, clade B, clade C, clade D, and clade E (Figure 1). Twenty-two SbZFPs and 37 AtZFPs belonged to clade A, 16 SbZFPs and 27 AtZFPs were part of clade B, 37 SbZFPs and 41 AtZFPs were assigned to clade C, 37 SbZFPs and 26 AtZFPs were grouped into clade D, and 33 SbZFPs and 42 AtZFPs were sections of clade E.
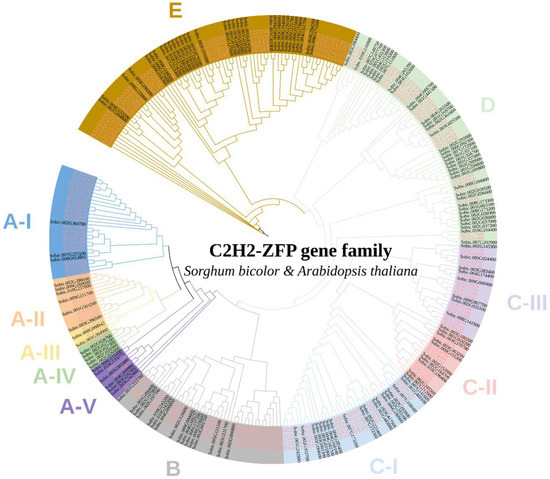
Figure 1.
Phylogenetic tree of C2H2-ZFP members between Arabidopsis thaliana and Sorghum bicolor. The tree was constructed with the maximum likelihood (ML) method. The ranges and branches of the circular tree in 11 clades were marked using different colors. C2H2-ZFP proteins from Arabidopsis and Sorghum bicolor have the prefix “AT” and “Sobic”, respectively.
Clade A was divided into five subclades—clade A-I, A-II, A-III, A-IV, and A-V. Clade A-I contained many ubiquitin carboxyl-terminal hydrolase-related genes such as AT5G61940, AT1G52430, and AT5G02660, as well as the known MAZ1 (AT5G15480), which regulates intine formation and the exine pattern in Arabidopsis [27]. In addition, ELF6 (AT5G04240) in clade A-II and FIS2 (AT2G35670) in clade A-III play roles in pollination and flowering [28,29]. Clade B contained REIL2 (AT2G24500) responding to cold stress and URO (AT3G23140) relevant to IAA homeostasis [30].
In addition, clade C was divided into three subclades—clade C-I, C-II, and C-III. Clade C-I consisted of indeterminate-domain (IDD)-type C2H2-ZFPs, such as BIB (AT3G45260), IDD7 (AT1G55110), IDD11 (AT3G13810), IDD12 (AT4G02670), and IDD1 (AT5G66730). IDD-type C2H2-ZFPs play roles in a variety of plant growth and development processes, including root development, flowering, seed maturation, leaf growth, hormone regulation, and defense against pathogens [31]. Additionally, clade C-II contained STOP and WIP-type C2H2-ZFPs. STOP1 (At1g34370) protects plants against acidic soils [32]. WIP-type AtC2H2-ZFPs, including WIP1 (AT1G34790) and WIP2 (AT3G57670), regulate endothelium differentiation, pollen tube development, and leaf vasculature growth in plants [33,34]. Apart from that, DAZ1 (AT2G17180) and DAZ2 (AT4G35280), which were in clade C-III, are involved in pollen fertility.
What is more, ZFP6 (AT1G67030), ZFP5 (AT1G10480), GIS (AT3G58070), GIS2 (AT5G06650), and ZFP8 (AT2G41940) contribute to trichome branching in clade D. Moreover, clade E included ZAT6 (AT5G04340), relevant to cold stress, and ZAT10 (AT1G27730), responding to salt tolerance [35,36]. Moreover, ZAT7 (At3g46090), the EAR motif of the AtC2H2-type zinc finger protein, inhibits WRKY70 expression under salt stress [37].
2.3. Gene Features and Conserved Motifs of SbC2H2-ZFPs
According to phylogenetic analysis in Figure 2A, 145 SbC2H2-ZFPs were divided into 11 subclasses (Figure 2A). A total of 145 SbC2H2-ZFPs protein sequences was analyzed by MEME (https://meme-suite.org/meme/tools/meme, accessed on 24 December 2021), and 10 conserved motifs were identified (Figure 2B). The details of these motifs are shown in Table S2. Among the 10 motifs, motifs 1, 2, and 6 conformed to the sequence characteristics of C2H2-Zinc finger. As a result, only 145 members had any of motifs 1, 2, and 6 left, although we identified 165 members based on the sequence alignment. In addition, motif 1 was distributed in nearly all of the SbCsC2H2-ZFPs, which implied that motif 1 could be a conserved and important motif among C2H2-ZFPs in S. bicolor. Motif 1 and motif 2 have the sequence “QALGGH”, the symbol of Q-type C2H2-ZFPs, which are specific to plants [38]. Motif 6 was mainly identified in clade A-I, A-V, C-II, and C-III, and motif 7 existed in clade C-II. Moreover, motif 9 and motif 10 were commonly identified in clade A-IV. In addition, there were the most motifs in clade C-I, including motifs 5, 1, 3, 2, 4, and 8, which implied that specific motifs might enable SbC2H2-ZFPs-specific functions. Overall, sequences with similar motif structures were clustered together, indicating the reliability of phylogenetic tree classification.

Figure 2.
Motif distributions and gene-structure analysis of SbC2H2-ZFP genes. (A) The phylogenetic tree was built by the ML method with a bootstrap value of 1000. (B) The conserved motifs in SbC2H2-ZFP proteins (1–10) are in different colors. The black lines represent relative protein lengths. (C) Exons, introns, and untranslated region (UTR) are represented by yellow rectangles, gray lines, and green rectangles, respectively. The length of SbC2H2-ZFPs is shown below.
Furthermore, in order to understand the characteristics of SbC2H2-ZFPs, we analyzed their gene structures, including the number of introns and exons. In general, there were more exons than introns (Figure 2C). Among the 145 SbC2H2-ZFPs, a total of 79 members did not contain introns, accounting for 54.48%; a total of 38 members (26.21%) had one or two introns; a total of 28 members (19.31%) gained more than two introns. In all subclasses, there were members that contained introns and those that did not contain introns. Overall, the number of introns varied greatly in different SbC2H2-ZFPs, reflecting their diversity in structure and function. The details are listed in Table S1.
2.4. Chromosomal Distribution and Gene Duplication in SbC2H2-ZFP Genes
Chromosomal distribution of SbC2H2-ZFP genes was constructed according to S. bicolor genome information (Figure 3, Tables S1 and S3). There were C2H2-ZFP genes on each chromosome. Chr2 harbored the most SbC2H2-ZFP genes (32 genes, ~22.07%), followed by Chr1 (26, ~17.93%), while Chr5 contained the least (6 genes, ~4.14%). Chr3, Chr4, and Chr8 contained 19 (~13.10%), 15 (~10.34%), and 7 (~4.83%) SbC2H2-ZFP genes, respectively. Chr7 and Chr10 contained the same number of SbC2H2-ZFP genes (11 each, ~7.59%); furthermore, Chr6 and Chr9 each contained 9 (~6.21%) SbC2H2-ZFP genes. Moreover, if two genes were situated in the same chromosome within 100 kb of distance and separated by five or fewer genes, they would be regarded as tandemly duplicated genes [38]. As a result, we identified 19 tandem duplication events containing 33 SbC2H2-ZFP genes on Chr1, 2, 3, 8, and 10 (Figure 3). Interestingly, some SbC2H2-ZFPs participated in more than one tandem repeat event, such as Sobic.001G356800, Sobic.001G357000, Sobic.002G026800, Sobic.002G026900, and Sobic.002G036600. Moreover, all genes in tandem repeat events came from the same subfamily, suggesting the accuracy in the subfamily classification of the evolutionary tree (Figure 3, Table S3).
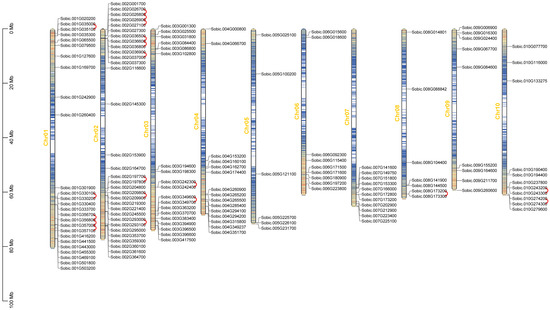
Figure 3.
Chromosomal location of SbC2H2-ZFP genes in the S. bicolor genome. The length of chromosomes is measured in Mb. Genes of SbC2H2-ZFP are marked in black. Tandemly duplicated genes are represented with red wavy lines.
Apart from that, we found 24 segmental duplication events involving 34 SbC2H2-ZFP genes (Figure 4, Table S4). The SbC2H2-ZFP genes were distributed in 10 linkage groups (LGs). LG01 had the largest number of SbC2H2-ZFP genes (8, ~23.53%), followed by LG02 (6, ~17.65%), whereas LG05 did not contain any SbC2H2-ZFP genes. LG04, LG06, LG07, and LG09 contained 5 (~14.71%), 4 (~11.76%), 5 (~14.71%), and 3 (~8.82%) SbC2H2-ZFP genes, respectively. Moreover, LG03, LG08, and LG10 contained the least SbC2H2-ZFP genes (2 each, ~5.88%). In addition, out of all identified SbC2H2-ZFP genes, clade E had the most linked genes (25/34, ~73.53%).
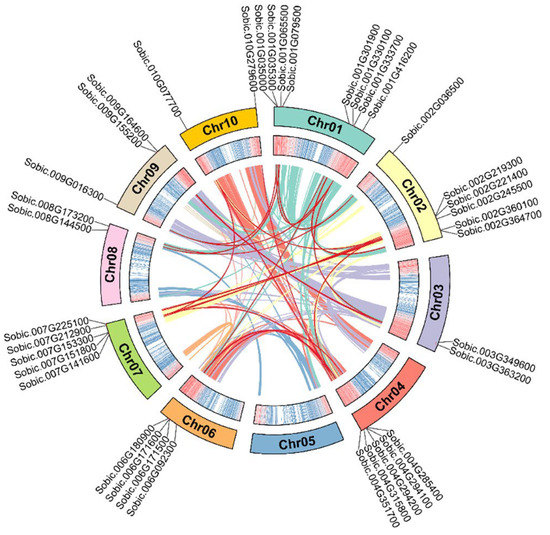
Figure 4.
A schematic diagram of the synteny relationship of the SbC2H2-ZFP genes. Colors represent all collinear segments in the S. bicolor genome, and red lines represent duplicated C2H2-ZFP gene pairs. The outermost circle shows the chromosome number, and the second outer circle shows the density of each chromosome.
2.5. Synteny Analysis of SbC2H2-ZFP Genes
To further investigate evolution mechanisms of SbC2H2-ZFP genes, we analyzed syntenic relationships of S. bicolor with four representative species: two dicotyledon plant species (A. thaliana and G. max), and two monocotyledon plant species (O. sativa and Z. mays) (Figure 5, Table S5). A total of 110 SbC2H2-ZFP genes were syntenic with those in A. thaliana (14), followed by G. max (60), O. sativa (88), and Z. mays (148) (Table S5). The numbers of orthologous gene pairs between sorghum and the other four species (A. thaliana, G. max, O. sativa, and Z. mays) were 23, 85, 123, and 202, respectively.
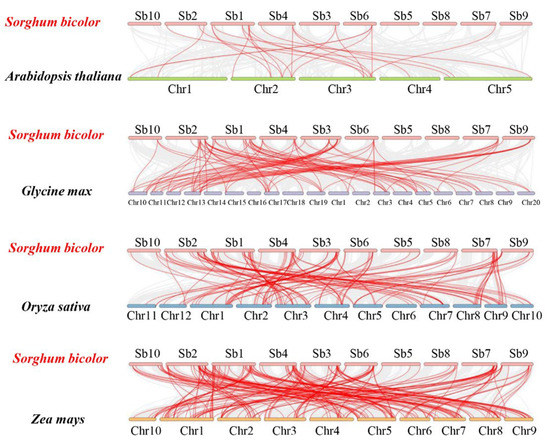
Figure 5.
Synteny analyses of C2H2-ZFP genes between S. bicolor and four representative species. Gray lines represent the collinear regions within S. bicolor and other genomes, and red lines indicate the syntenic SbC2H2-ZFP gene pairs.
Some SbC2H2-ZFP genes were associated with more than four syntenic gene pairs between S. bicolor and Z. mays, such as Sobic.001G416200, Sobic.002G036500, Sobic.002G219300, Sobic.004G315800, and Sobic.007G151800. This may indicate that before ancestors diverged, these orthologous gene pairs had already existed, suggesting that these genes are vital to the evolution of the C2H2-ZFP gene family. Apart from that, there were 55 gene pairs identified between S. bicolor and the other two monocotyledonous plants not existing between S. bicolor and two dicotyledonous plants, such as Sobic.002G360100, Sobic.006G115400, and Sobic.001G503200. As a result, these gene pairs may have formed after the divergence of monocots and dicots (Table S5).
2.6. Expression Patterns of SbC2H2-ZFPs in Several Tissues
To study the potential functions of SbC2H2-ZFP genes, we randomly selected one gene in each clade and analyzed their expression in three vegetative organs (roots, stems, leaves) by qRT-PCR (Figure 6A). Different SbC2H2-ZFP genes had different expression patterns in different organs, whereas almost all genes were expressed in all tissues. Several genes had the highest expression in the roots, such as Sobic.004G153200, Sobic.005G121100, Sobic.007G202900, Sobic.007G225100, and Sobic.009G024400. However, Sobic.001G501800 and Sobic.008G088842 had the highest expression in the stems. This indicated that the transcriptional abundance of different SbC2H2-ZFP genes varied in different organs, implying that SbC2H2-ZFPs play various roles in the growth and development of sorghum. Interestingly, the expression of some genes in the organs was quite correlated to that of others, implying that they might have synergism. For example, the expression of Sobic.005G121100, Sobic.007G202900, and Sobic.007G225100 was significantly positively correlated in the roots, and they were all highly expressed. However, they were significantly negatively correlated with Sobic.001G501800.
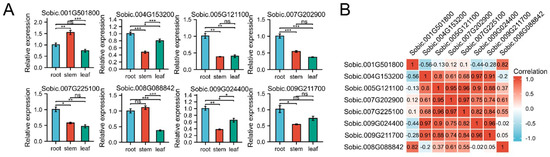
Figure 6.
Tissue-specific expression of 8 SbC2H2-ZFP genes and the relevance of their expression. (A) Expression patterns of 8 SbC2H2-ZFP genes in root, stem, and leaf were analyzed by qRT-PCR. Error bars are obtained from three biological replicates, and standard error is selected as the value of the bar. Asterisks display significant expression differences of genes in different organs (* p < 0.05, ** p < 0.01, *** p < 0.001; one-way ANOVA). (B) Positive numbers: positive correlations; negative numbers: negative correlations.
2.7. Expression Patterns of SbC2H2-ZFPs in Response to Cold and Drought Stress
To analyze the potential roles of SbC2H2-ZFPs in sorghum responding to cold and drought stress, we performed qRT-PCR experiments under two abiotic stresses (Figure 7). As shown in Figure 7, some SbC2H2-ZFPs were significantly induced but others were extremely repressed. It was obvious that some SbC2H2-ZFPs showed various changes in different tissues and under different stresses. For example, under cold, the expression level of Sobic.008G088842 increased in the stems and leaves, whereas it was down-regulated under drought stress. This indicated that Sobic.008G088842 might be activated by cold but inhibited in drought. Expression of most genes in the roots was not affected by cold and drought stress, except Sobic.005G121100, which was significantly up-regulated under cold stress. Interestingly, a majority of genes was activated in the leaves under cold, such as Sobic.001G501800, Sobic.007G202900, Sobic.007G225100, Sobic.009G211700, Sobic.008G088842, and Sobic.004G153200, but more were up-regulated in the stems under drought.
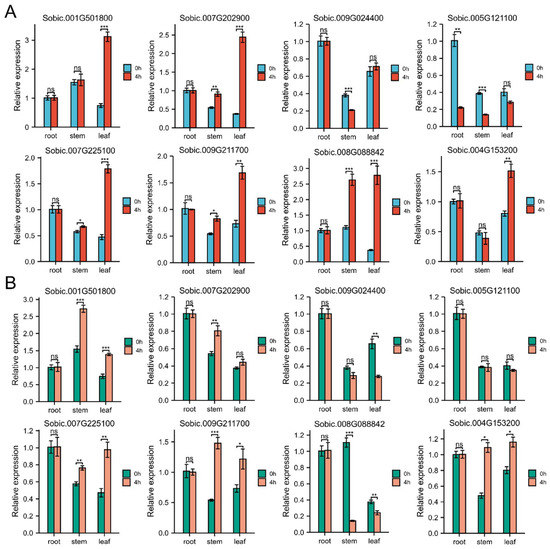
Figure 7.
Expression of 8 SbC2H2-ZFP genes under cold (A) and PEG (B) stresses at the seedling stage. Error bars are obtained from three biological replicates, and standard error is selected as the value of the bar. Asterisks display significant expression differences of genes after treatment (* p < 0.05, ** p < 0.01, *** p < 0.001; t-test).
3. Discussion
C2H2-type zinc finger proteins are one of the most abundant transcription factor families in higher plants. Previous reports indicate that they play an important role in cucumber, Arabidopsis, wheat, and tomato [39,40,41,42]. Therefore, many researchers have tried to perform genome-wide analysis on C2H2-ZFPs in various species, such as tomato, wheat, grape, and oyster mushroom [43,44,45,46], but little is known about sorghum C2H2-ZFP proteins. In this study, we carried out a genome-wide study of the S. bicolor C2H2-ZFP family and identified a total of 145 SbC2H2-ZFP members. Then we analyzed the evolutionary relationship of C2H2-ZFP between sorghum and Arabidopsis to infer the possible function of SbC2H2-ZFPs using a phylogenetic tree. What is more, motif composition, gene structure, chromosomal location, gene duplication events, and the expression of SbC2H2-ZFPs in different vegetative organs were analyzed, and their responses to cold and drought stress were investigated.
In this study, motifs 1, 2, and 6 were the characteristics of C2H2-zinc fingers. In addition to C2H2-type motifs, SbC2H2-ZFPs also contained many other motifs, suggesting that SbC2H2-ZFPs play an extensive role in higher plants. Motif 1 and Motif 2 had the sequence “QALGGH”, which is the symbol for the plant-specific Q-type C2H2-ZFPs [38]. Q-type C2H2-ZFPs have been reported to be involved in the growth, development, and organogenesis of a variety of plants, as well as in response to stresses and defense [47,48,49,50,51]. Motif 6 was mainly present in the clades A-I, A-V, C-II, and C-III, while motif 7 was present in clade C-II. Furthermore, motif 9 and motif 10 were common in the clade A-IV. Interestingly, most motifs were present in clade C-I, including motifs 5, 1, 3, 2, 4, and 8, implying that specific motifs may enable specific functions of SbC2H2-ZFPs.
Thirty-three SbC2H2-ZFPs (22.76%) were identified as tandem repeat genes, and 34 SbC2H2-ZFPs (23.45%) were identified as segmental repeat genes. Among them, several SbC2H2-ZFPs were involved in more than one tandem repeat event. These results suggested that gene duplication contributed to the expansion of a new gene family in the evolution of plant genome [52] and played a significant role in the evolution of the SbC2H2-ZFP genes.
The expression of a gene is often used to predict its function. Previous findings have shown that the expression of C2H2-ZFP genes was affected by tissue differences and various abiotic stresses [46,53]. Our results showed that among the selected SbC2H2-ZFPs, most genes were expressed in roots higher than those in leaves or stems, but almost all SbC2H2-ZFP members were expressed in roots, stems, and leaves. This indicated that the transcript abundances of different SbC2H2-ZFP genes were different in different organs, suggesting that SbC2H2-ZFP plays different roles in the growth and development of sorghum. It has been reported that plant growth and development are affected by the transcript abundance of C2H2-ZFP genes [26,54,55]. In addition, it was found that some SbC2H2-ZFPs showed different changes in different tissues or stresses by analyzing the expression of SbC2H2-ZFP under cold and drought stress. For example, under cold stress, the expression level of Sobic.008G088842 was increased in stems and leaves, while it was down-regulated under drought stress. This indicated that the response of Sobic.008G088842 to different stresses might be worth further study. In addition, most genes were activated in leaves under cold stress, but more genes were up-regulated in stems under drought treatment. As a result, sorghum leaves may be suitable materials to study cold stress in the future, but stems might be better for drought stress research.
4. Materials and Methods
4.1. Identification of C2H2 ZFPs in Sorghum bicolor
The S. bicolor genome sequence was downloaded from the Phytozome v13 database (https://phytozome-next.jgi.doe.gov/info/Sbicolor_v3_1_1, accessed on 5 September 2021) [56]. The protein sequences of C2H2 ZFPs in Arabidopsis were downloaded from TAIR (https://www.arabidopsis.org/servlets/Search?search_action=sendToSequenceDLAll&type=gene&pageNum=0&size=4&query_id=41631445, accessed on 5 September 2021). All possible C2H2 ZFPs in S. bicolor were identified according to HMM profiles including PF00096, PF13894, PF13912, PF18414, PF16622, and PF18658 by using TBtools [57]. After this step, we obtained 184 S. bicolor C2H2-ZFPs. Then, all potential protein sequences were submitted to the NCBI-BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 25 September 2021) and were validated through UniprotKB/Swiss-Prot(swissprot) and MEME. Eventually, 145 S. bicolor C2H2-ZFPs were reserved.
4.2. C2H2-ZFP Gene Structure and Conserved Motifs
To calculate molecular weights (Mws) and theoretical isoelectric points (pIs) of SbC2H2-ZFPs, all gene sequences were submitted to ExPASy (http://web.expasy.org/, accessed on 30 September 2021). Moreover, the gene structure of SbC2H2-ZFPs was obtained using the “Gene Location Visualize (Advanced)” function in TBtools. Apart from that, we used MEME 5.4.1 (https://meme-suite.org/meme/tools/meme, accessed on 24 December 2021) to analyze the motifs of SbC2H2-ZFP proteins. The parameters we used were as follows: motif sites distribution, zero or one occurrence per sequence; the maximum number of motifs, 10; minimum sites of each motif, 21; maximum sites of each motif, 30 [58].
4.3. Chromosomal Location and Gene Duplication of C2H2-ZFPs in S. bicolor
The chromosomal locations of SbC2H2-ZFPs were visualized by MCScanX and TBtools [57,59]. The default e-value cutoff of MCScanX is 1 × e−10. The origin of C2H2-ZFP members was analyzed by MCScanX with default parameters. Furthermore, if two genes were located in the same chromosome within 100 kb of distance, and separated by five or fewer genes, they would be identified as tandemly duplicated genes [38]. Apart from that, we studied C2H2-ZFP homology between S. bicolor and four other plants (A. thaliana, O. sativa subsp. indica, Z. mays, and G. max) by Dual Synteny Plotter [57]. The genome sequences of four species were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/, accessed on 30 December 2021) [60,61,62,63,64].
4.4. Phylogenetic Analysis of C2H2-ZFPs in S. bicolor
The full-length protein sequences of S. bicolor and A. thaliana C2H2-ZFPs were used for phylogenetic analysis. Multiple sequence alignments were performed with MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/, accessed on 25 October 2021), and the resulting sequences were trimmed using trimAl. Then, a phylogenetic tree was inferred using IQ-TREE with a bootstrap value of 1000. Apart from that, the phylogenetic tree was visualized and annotated by iTOL (https://itol.embl.de/, accessed on 30 October 2021).
4.5. Plant Materials and Abiotic Stress in S. bicolor
Sorghum bicolor BTx623 was used in this study. S. bicolor was grown in uniformly mixed Pindstrup substrate (www.pindstrup.com, accessed on 30 October 2021) [65] in a light incubator with a 16 h/30 °C day and 8 h/25 °C night regime. The roots, stems, and leaves from five plants were collected, quickly placed in liquid nitrogen, and stored at −80 °C until further use. Apart from that, sorghum plants at 40 days were selected for drought by treating with 10% PEG6000 for 4 h and cold stress by being placed in 4 °C for 4 h. Each treatment had five replicates, and all samples collected were stored at −80 °C.
4.6. Total RNA Extraction and qRT-PCR Analysis
Total RNA of S. bicolor samples was extracted using a Plant RNA Kit (Omega Bio-tek Inc., Norcross, GA, USA) and reverse transcribed by a PerfectStart Uni RT&qPCR Kit (Transgen Biotech, Beijing, China). Specific primers were designed by Oligo 7.0 (Table S6). The qRT-PCR was conducted, and each selected gene was assayed at least three times. We used the EIF4α (eukaryotic initiation factor 4-α) gene as the control, the expression of which was stable in almost all growth stages and tissues [66]. The expression data were calculated according to the 2−(ΔΔCT) method and visualized using R 3.6.3 with the ggplot2 package (Version 3.3.5, Wickham, 2016, https://cran.r-project.org/web/packages/ggplot2/index.html, accessed on 5 September 2021).
5. Conclusions
We identified 145 C2H2-ZFP members that were randomly distributed on 10 chromosomes in S. bicolor. These members were divided into 11 clades based on the phylogenetic tree, and the genes in the same clade contained similar intron/exon and motif patterns. Furthermore, thirty-three tandem duplicated SbC2H2-ZFPs and 24 pairs of segmental duplicated genes were identified. Moreover, synteny analysis showed that sorghum had more collinear regions with monocotyledonous plants such as maize and rice than with dicotyledonous plants such as soybean and Arabidopsis. In addition, qRT-PCR analysis showed that several genes had the highest expression in the roots, such as Sobic.004G153200 and Sobic.005G121100, while Sobic.001G501800 and Sobic.008G088842 had the highest expression in the stems. The experiment was also helpful for understanding the mechanisms of how C2H2-ZFPs regulated sorghum resistance to cold and drought stresses. For example, Sobic.008G088842 may play an important role in sorghum resistance to cold stress, while Sobic.004G153200 may improve drought tolerance. In conclusion, it provided important information for further study of the C2H2-ZFP family and a framework for stress-resistance research in sorghum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23105571/s1.
Author Contributions
Data curation, H.C., J.C., M.L., H.Z., S.Z. and D.L.; funding acquisition, S.C.; investigation, H.C.; writing—original draft, H.C.; writing—review and editing, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Intergovernmental International Cooperation on Science and Technology Innovation under the Ministry of Science and Technology of PRC (approval no. 2018YFE0112400) (to S.C.) and the National Natural Science Foundation of China (approval no. 31770207) (to S.C.).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The Sorghum bicolor whole genome sequence information is from the Phytozome v13 database (https://phytozome-next.jgi.doe.gov/info/Sbicolor_v3_1_1, accessed on 5 September 2021). The Sorghum bicolor materials (DALISHI) used in this study were purchased from ChangJingZhongYe company (https://www.cmeii.com/, accessed on 20 September 2021). The datasets supporting the conclusions of this article are included in the article and its Supplementary Materials.
Acknowledgments
We would like to acknowledge all researchers in our laboratory for their help.
Conflicts of Interest
The authors have no conflict of interest to declare in relation to this article.
References
- Takatsuji, H. Zinc-finger proteins, the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 1999, 39, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Kielbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012, 185, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes (Reprinted from EMBO Journal, vol 4, pg 1609–1614, 1985). J. Trace Elem. Exp. Med. 2001, 14, 157–169. [Google Scholar] [CrossRef]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef]
- Tsai, R.Y.L.; Reed, R.R. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 1998, 18, 6447–6456. [Google Scholar] [CrossRef]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef]
- Yuan, S.; Li, X.; Li, R.; Wang, L.; Zhang, C.; Chen, L.; Hao, Q.; Zhang, X.; Chen, H.; Shan, Z.; et al. Genome-Wide Identification and Classification of Soybean C2H2 Zinc Finger Proteins and Their Expression Analysis in Legume-Rhizobium Symbiosis. Front. Microbiol. 2018, 9, 126. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, J.; Zhong, G.; Wang, B. Genome-Wide Identification and Expression Patterns of the C2H2-Zinc Finger Gene Family Related to Stress Responses and Catechins Accumulation in Camellia sinensis [L.] O. Kuntze. Int. J. Mol. Sci. 2021, 22, 4197. [Google Scholar] [CrossRef]
- Jiao, Z.; Wang, L.; Du, H.; Wang, Y.; Wang, W.; Liu, J.; Huang, J.; Huang, W.; Ge, L. Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula. BMC Plant Biol. 2020, 20, 401. [Google Scholar] [CrossRef]
- Ming, N.; Ma, N.; Jiao, B.; Lv, W.; Meng, Q. Genome Wide Identification of C2H2-Type Zinc Finger Proteins of Tomato and Expression Analysis Under Different Abiotic Stresses. Plant Mol. Biol. Rep. 2019, 38, 75–94. [Google Scholar] [CrossRef]
- Faraji, S.; Rasouli, S.H.; Kazemitabar, S.K. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp Durum), insights into the roles in biological processes especially stress response. BioMetals 2018, 31, 1019–1042. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins, new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Li, R.; Wang, X.; Zhang, S.; Liu, X.; Zhou, Z.; Liu, Z.; Wang, K.; Tian, Y.; Wang, H.; Zhang, Y.; et al. Two zinc-finger roteins control the initiation and elongation of long stalk trichomes in tomato. J. Genet. Genom. 2021, 48, 1057–1069. [Google Scholar] [CrossRef]
- Zheng, F.; Cui, L.; Li, C.; Xie, Q.; Ai, G.; Wang, J.; Yu, H.; Wang, T.; Zhang, J.; Ye, Z.; et al. Hair (H) interacts with SlZFP8-like to regulate the initiation and elongation of trichomes by modulating SlZFP6 expression in tomato. J. Exp. Bot. 2021, 73, 228–244. [Google Scholar] [CrossRef]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; Burg, H.A.V.D.; Huang, C.-F. Regulation of Aluminum Resistance in Arabidopsis Involves the SUMOylation of the Zinc Finger Transcription Factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef]
- Rodas, A.L.; Roque, E.; Hamza, R.; Gómez-Mena, C.; Minguet, E.G.; Wen, J.; Mysore, K.S.; Beltrán, J.P.; Cañas, L.A. MtSUPERMAN plays a key role in compound inflorescence and flower development in Medicago truncatula. Plant J. 2020, 105, 816–830. [Google Scholar] [CrossRef]
- Ballerini, E.S.; Min, Y.; Edwards, M.B.; Kramer, E.M.; Hodges, S.A. POPOVICH, encoding a C2H2 zinc-finger transcription factor, plays a central role in the development of a key innovation, floral nectar spurs, in Aquilegia. Proc. Natl. Acad. Sci. USA 2020, 117, 22552–22560. [Google Scholar] [CrossRef]
- Yang, K.; Li, C.-Y.; An, J.-P.; Wang, D.-R.; Wang, X.; Wang, C.-K.; You, C.-X. The C2H2-type zinc finger transcription factor MdZAT10 negatively regulates drought tolerance in apple. Plant Physiol. Biochem. 2021, 167, 390–399. [Google Scholar] [CrossRef]
- Naoura, G.; Emendack, Y.; Baloua, N.; Brocke, K.V.; Hassan, M.A.; Sawadogo, N.; Nodjasse, A.D.; Djinodji, R.; Trouche, G.; Laza, H.E. Characterization of semi-arid Chadian sweet sorghum accessions as potential sources for sugar and ethanol production. Sci. Rep. 2020, 10, 14947. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Ullah, F.; Ben-Hur, A.; Reddy, A.S.N. Transcriptome Analysis of Drought-Resistant and Drought-Sensitive Sorghum (Sorghum bicolor) Genotypes in Response to PEG-Induced Drought Stress. Int. J. Mol. Sci. 2020, 21, 772. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.; Hu, J.; Salas Fernandez, M.G. Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J. Exp. Bot. 2017, 68, 4545–4557. [Google Scholar] [CrossRef] [PubMed]
- Kusvuran, A.; Bilgici, M.; Kusvuran, S.; Nazli, R.I. The effect of different organic matters on plant growth regulation and nutritional components under salt stress in sweet sorghum [Sorghum bicolor (L.) Moench.]. Maydica 2021, 66, 9. [Google Scholar]
- Huang, S.; Gao, J.; You, J.; Liang, Y.; Guan, K.; Yan, S.; Zhan, M.; Yang, Z. Identification of STOP1-Like Proteins Associated with Aluminum Tolerance in Sweet Sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 258. [Google Scholar] [CrossRef]
- Gao, J.; Yan, S.; Yu, H.; Zhan, M.; Guan, K.; Wang, Y.; Yang, Z. Sweet sorghum (Sorghum bicolor L.) SbSTOP1 activates the transcription of a beta-1,3-glucanase gene to reduce callose deposition under Al toxicity, A novel pathway for Al tolerance in plants. Biosci. Biotechnol. Biochem. 2018, 83, 446–455. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins, Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Lyu, T.; Hu, Z.; Liu, W.; Cao, J. Arabidopsis Cys(2)/His(2) zinc-finger protein MAZ1 is essential for intine formation and exine pattern. Biochem. Biophys. Res. Commun. 2019, 518, 299–305. [Google Scholar] [CrossRef]
- Keyzor, C.; Mermaz, B.; Trigazis, E.; Jo, S.; Song, J. Histone Demethylases ELF6 and JMJ13 Antagonistically Regulate Self-Fertility in Arabidopsis. Front. Plant Sci. 2021, 12, 640135. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, D.; Zhang, H.; Skaggs, M.I.; Lloyd, A.; Ran, D.; An, L.; Schumaker, K.S.; Drews, G.N.; Yadegari, R. FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 Plays a Dual Role in Regulating Type I MADS-Box Genes in Early Endosperm Development. Plant Physiol. 2018, 177, 285–299. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, Y.; Yuan, Z.; Müller, J.L.; Yu, C.; Xu, Y.; Shao, X.; Li, X.; Decker, E.L.; Reski, R.; et al. Overexpression of the Arabidopsis Gene UPRIGHT ROSETTE Reveals a Homeostatic Control for Indole-3-Acetic Acid. Plant Physiol. 2010, 153, 1311–1320. [Google Scholar] [CrossRef][Green Version]
- Prochetto, S.; Reinheimer, R. Step by step evolution of Indeterminate Domain (IDD) transcriptional regulators, from algae to angiosperms. Ann. Bot. 2020, 126, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed]
- Sagasser, M.; Lu, G.-H.; Hahlbrock, K.; Weisshaar, B. A-thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002, 16, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.V.G.; Izhaq, F.; Verdenaud, M.; Eleblu, J.; Haraghi, A.; Sommard, V.; Chambrier, P.; Latrasse, D.; Jegu, T.; Benhamed, M.; et al. Integrative genome-wide analysis reveals the role of WIP proteins in inhibition of growth and development. Commun. Biol. 2020, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.T.; Chan, Z.L. The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J. Pineal Res. 2014, 57, 185–191. [Google Scholar] [CrossRef]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.-K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006, 580, 6537–6542. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef]
- Liu, Z.; Coulter, J.A.; Li, Y.; Zhang, X.; Meng, J.; Zhang, J.; Liu, Y. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 153, 327–340. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Zhu, H.; Zhang, M.; Wang, D.; Xie, K.; Fan, P.; Dou, J.; Liu, D.; Liu, B.; et al. A novel HD-Zip I/C2H2-ZFP/WD-repeat complex regulates the size of spine base in cucumber. New Phytol. 2022, 233, 2643–2658. [Google Scholar] [CrossRef]
- Ma, X.; Wu, Y.; Zhang, G. Formation pattern and regulatory mechanisms of pollen wall in Arabidopsis. J. Plant Physiol. 2021, 260, 153388. [Google Scholar] [CrossRef]
- Cheuk, A.; Ouellet, F.; Houde, M. The barley stripe mosaic virus expression system reveals the wheat C2H2 zinc finger protein TaZFP1B as a key regulator of drought tolerance. BMC Plant Biol. 2020, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Hua, B.; Chang, J.; Han, X.; Xu, Z.; Hu, S.; Li, S.; Wang, R.; Yang, L.; Yang, M.; Wu, S.; et al. H and HL synergistically regulate jasmonate-triggered trichome formation in tomato. Hortic. Res. 2022, 9, uhab080. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, L.; Zhu, S.; Zheng, F.; Yang, C. Identification, genomic organization, and expression profiles of single C2H2 zinc finger transcription factors in tomato (Solanum lycopersicum). J. Appl. Genet. 2020, 62, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, A.; Wu, Q.; Zou, X.; Chen, F.; Cai, R.; Xie, H.; Zhang, M.; Guo, X. Comprehensive genomic survey, structural classification and expression analysis of C2H2-type zinc finger factor in wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 380. [Google Scholar] [CrossRef]
- Arrey-Salas, O.; Caris-Maldonado, J.C.; Hernández-Rojas, B.; Gonzalez, E. Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis vinifera L.), Insights into the Roles in the Pollen Development Regulation. Genes 2021, 12, 302. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Zhao, H.; Zhu, P.; Jiang, X.; Nie, F.; Li, G. Genome-wide identification and expression analyses of C2H2 zinc finger transcription factors in Pleurotus ostreatus. PeerJ 2022, 10, e12654. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Bonthala, V.S.; Mishra, A.K.; Khandelwal, R.; Khan, Y.; Roy, R.; Prasad, M. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genom. 2014, 14, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Kam, J.; Gresshoff, P.M.; Shorter, R.; Xue, G.-P. The Q-type C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress. Plant Mol. Biol. 2008, 67, 305–322. [Google Scholar] [CrossRef]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.J.; Jeong, J.C.; Lee, H.S.; Kwak, S.S. Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol. Biochem. 2013, 70, 445–454. [Google Scholar] [CrossRef]
- Wang, F.; Tong, W.; Zhu, H.; Kong, W.; Peng, R.; Liu, Q.; Yao, Q. A novel Cys(2)/His(2) zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 2015, 243, 783–797. [Google Scholar] [CrossRef]
- Wang, L.-J.; He, S.-Z.; Zhai, H.; Liu, D.-G.; Wang, Y.-N.; Liu, Q.-C. Molecular Cloning and Functional Characterization of a Salt Tolerance-Associated Gene IbNFU1 from Sweetpotato. J. Integr. Agric. 2013, 12, 27–35. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; Odongo, M.R.; Gong, W.; He, S.; Du, X. Genome-wide analysis of cotton C2H2-zinc finger transcription factor family and their expression analysis during fiber development. BMC Plant Biol. 2019, 19, 400. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-C.; Fu, C.-C. Cold-inducible MaC2H2s are associated with cold stress response of banana fruit via regulating MaICE1. Plant Cell Rep. 2019, 38, 673–680. [Google Scholar] [CrossRef] [PubMed]
- McCormick, R.F.; Truong, S.K.; Sreedasyam, A.; Jenkins, J.; Shu, S.; Sims, D.; Kennedy, M.; Amirebrahimi, M.; Weers, B.D.; Mckinley, B.; et al. The Sorghum bicolor reference genome, improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018, 93, 338–354. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools, An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX, a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome, a comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR), improved gene annotation and new tools. Nucleic Acids Res. 2011, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource, improvements and new features. Nucleic Acids Res. 2006, 35, D883–D887. [Google Scholar] [CrossRef]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.-S.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, X.; Yin, P.; Zhang, M.; Jiang, C. A domestication-associated reduction in K+-preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytol. 2019, 222, 301–317. [Google Scholar] [CrossRef]
- Reddy, P.E.; Reddy, D.E.; Sivasakthi, K.; Ebhatnagar-Mathur, P.; Evadez, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).