Prunetinoside Inhibits Lipopolysaccharide-Provoked Inflammatory Response via Suppressing NF-κB and Activating the JNK-Mediated Signaling Pathway in RAW264.7 Macrophage Cells
Abstract
:1. Introduction
2. Results
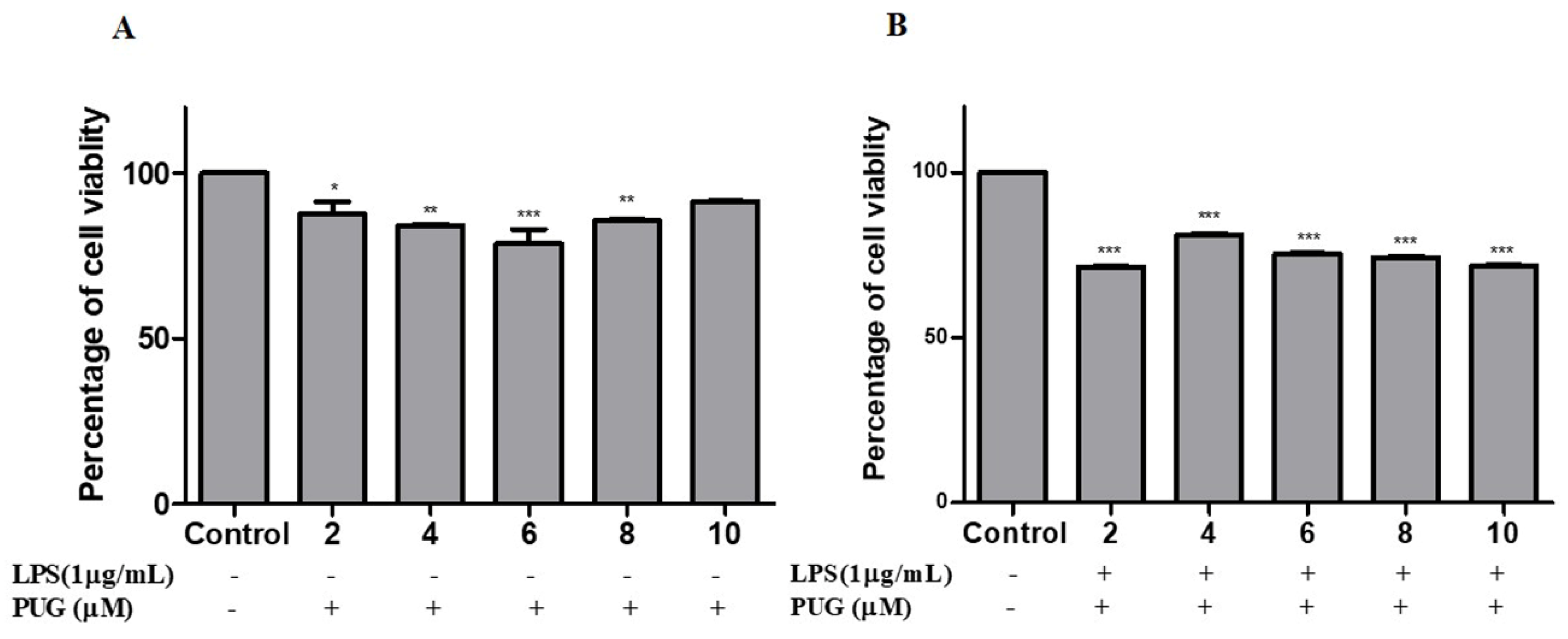
2.1. Cell Cytotoxicity Effect of PUG on Activated Macrophage
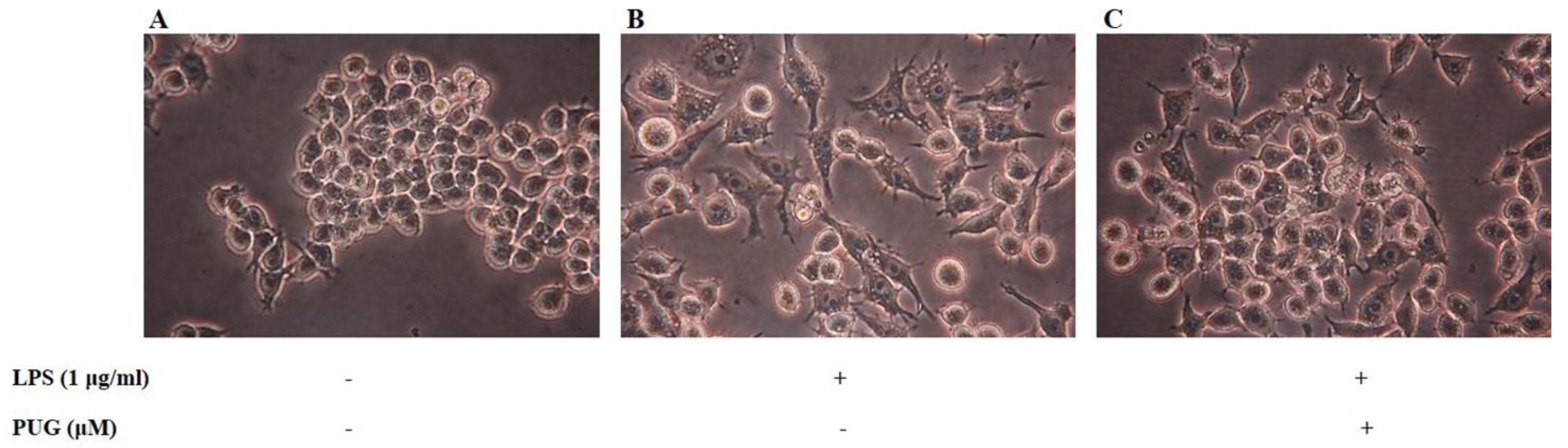
2.2. Morphological Hallmarks Associated with PUG in Macrophage Cells
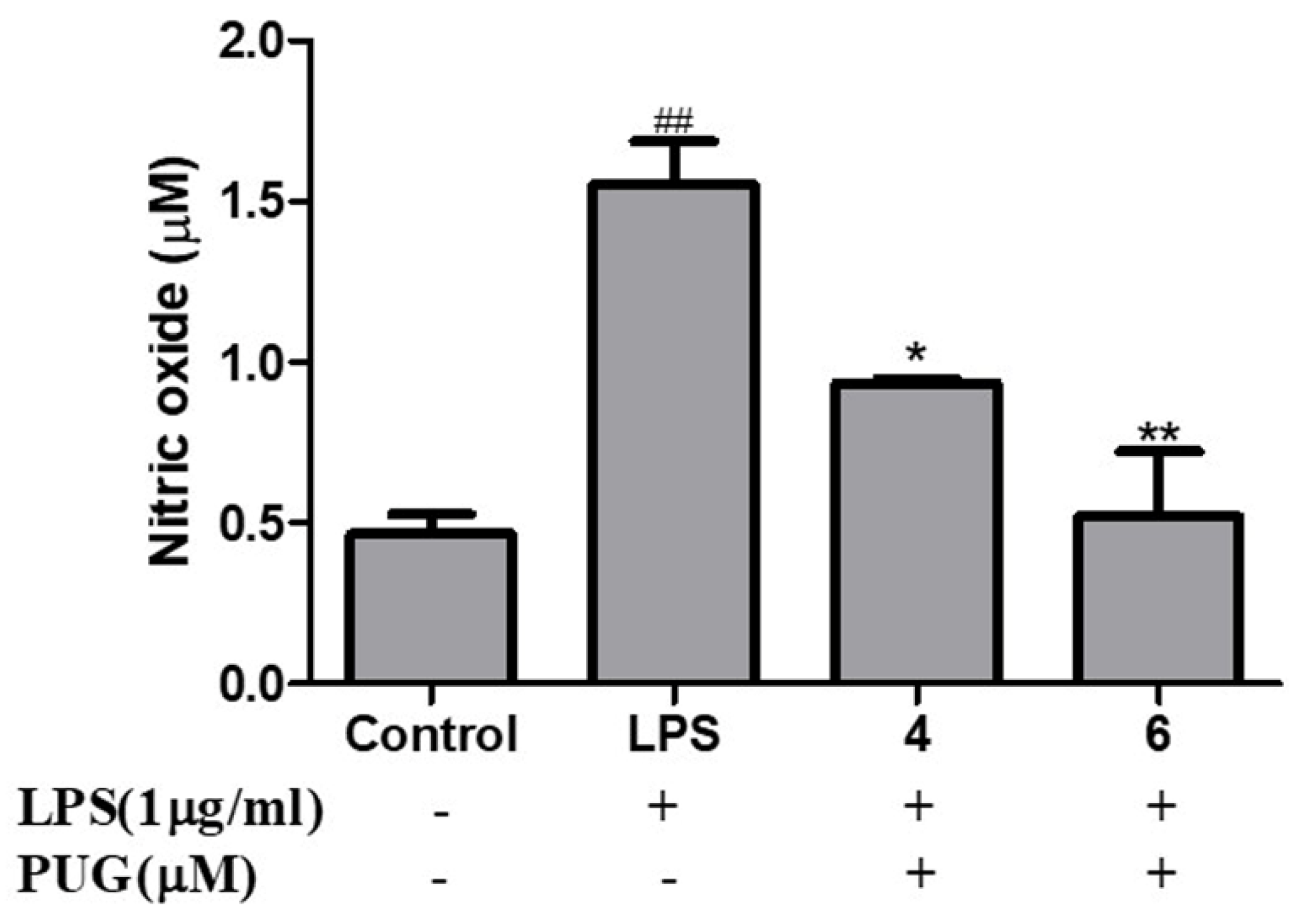
2.3. Effect of PUG on Nitric Oxide (NO) Release on RAW264.7 Macrophage Cells
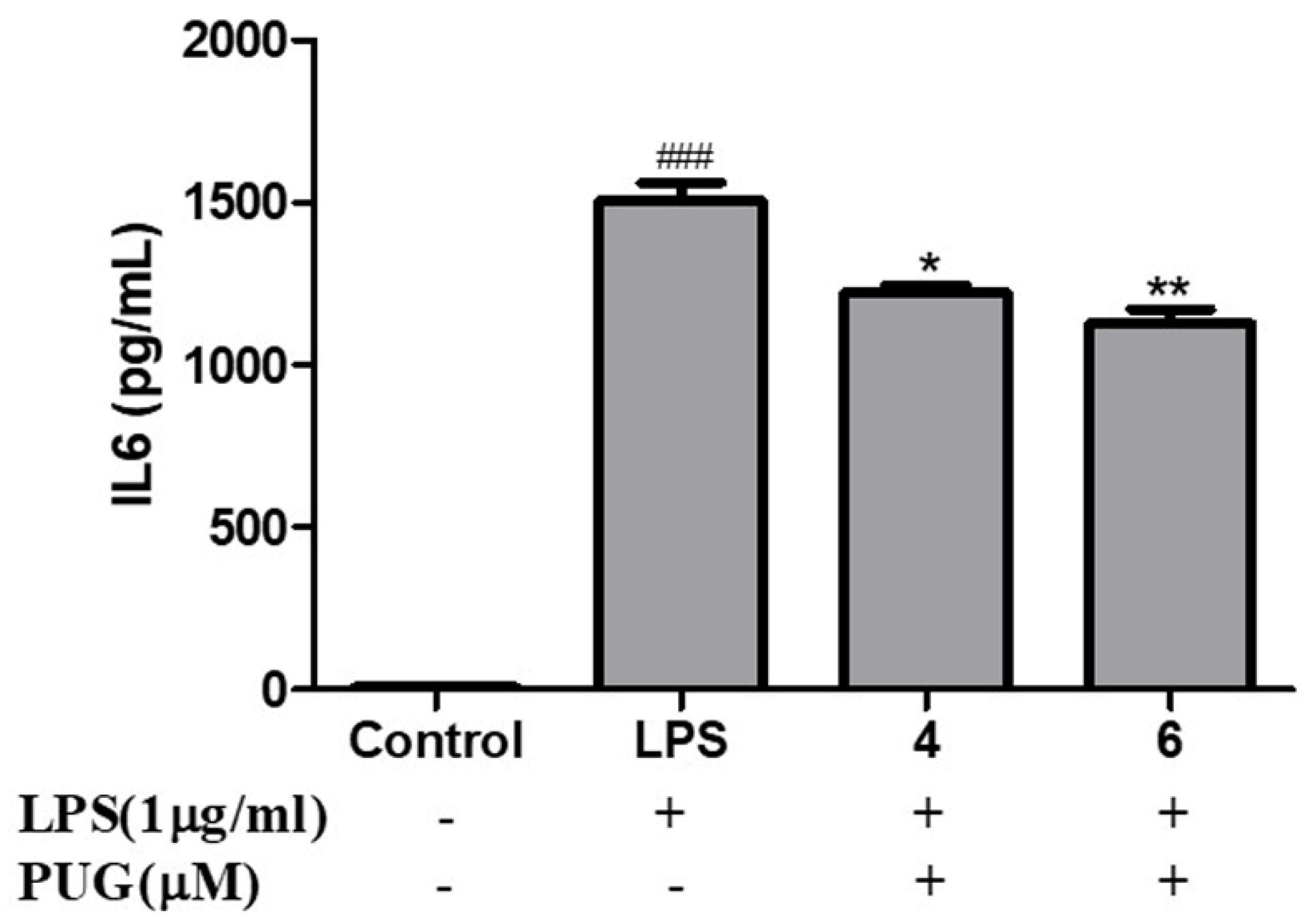
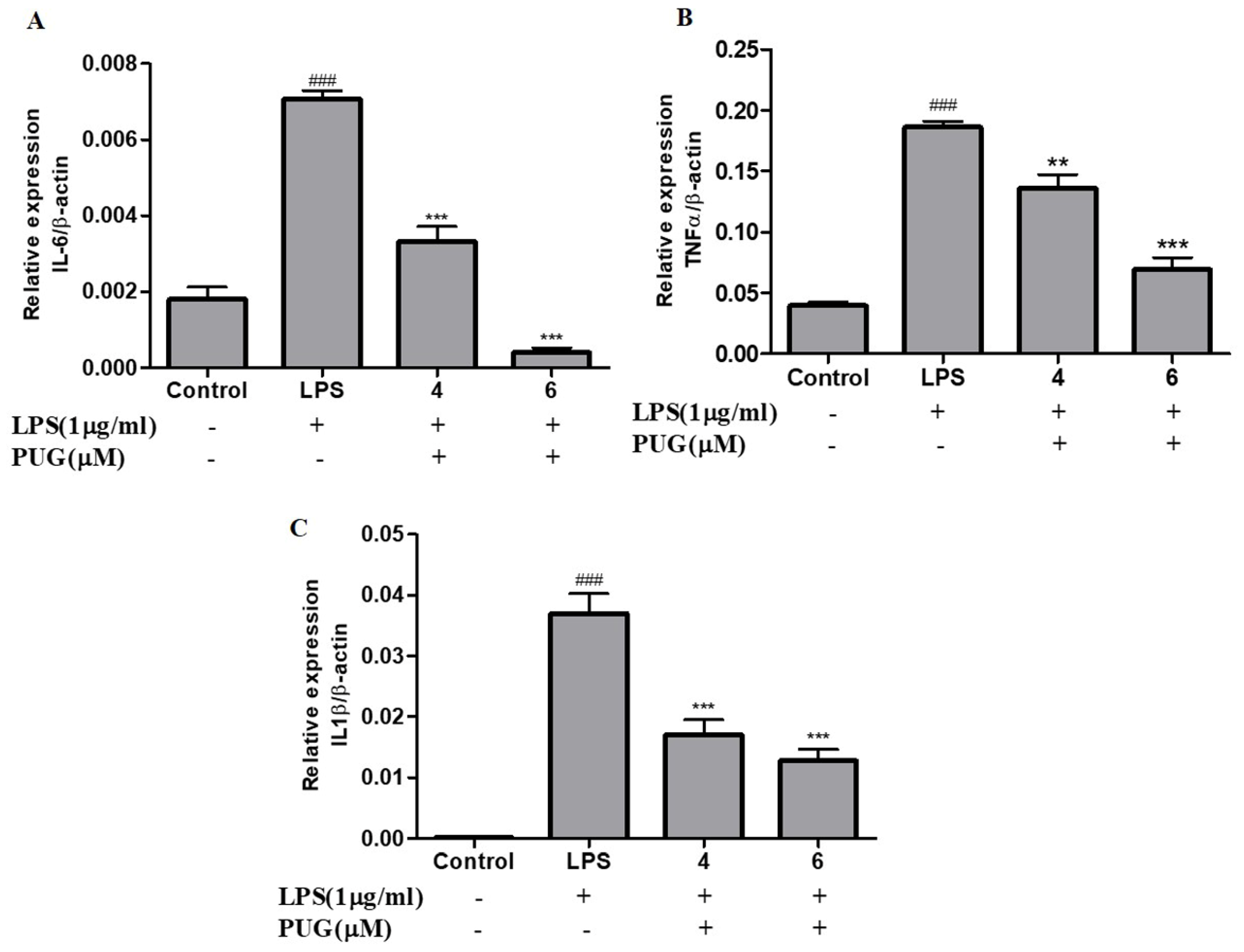
2.4. Effect of PUG on the Pro-Inflammatory Cytokine Interleukin-6 (IL-6) on Activated RAW264.7 Macrophage Cells
2.5. PUG Decreases the mRNA Expression of Pro-Inflammatory Cytokines in Activated RAW264.7 Macrophage Cells
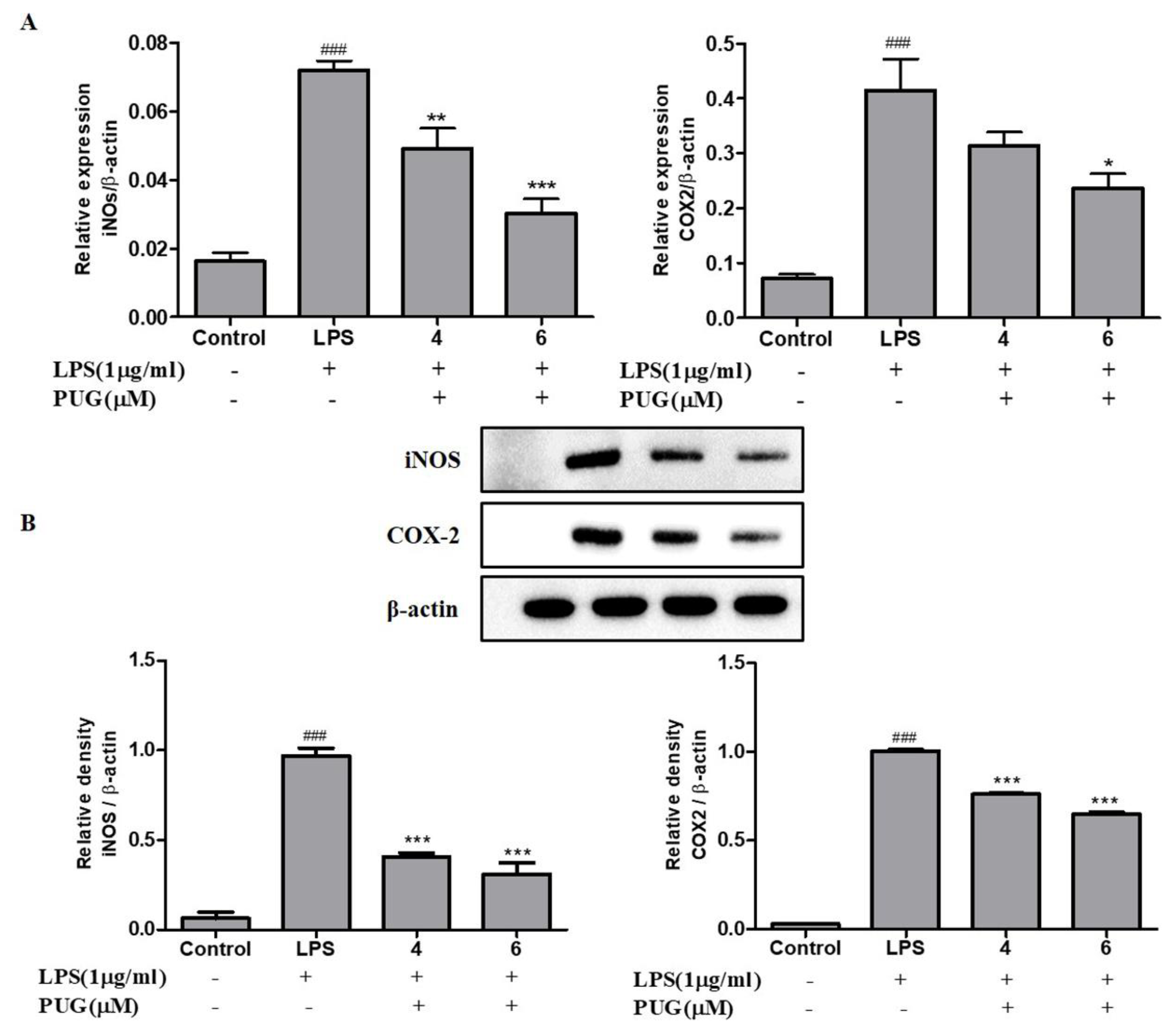
2.6. PUG Decreases the Expression of Pro-Inflammatory Cytokines iNOS and COX2 in Activated RAW264.7 Cells
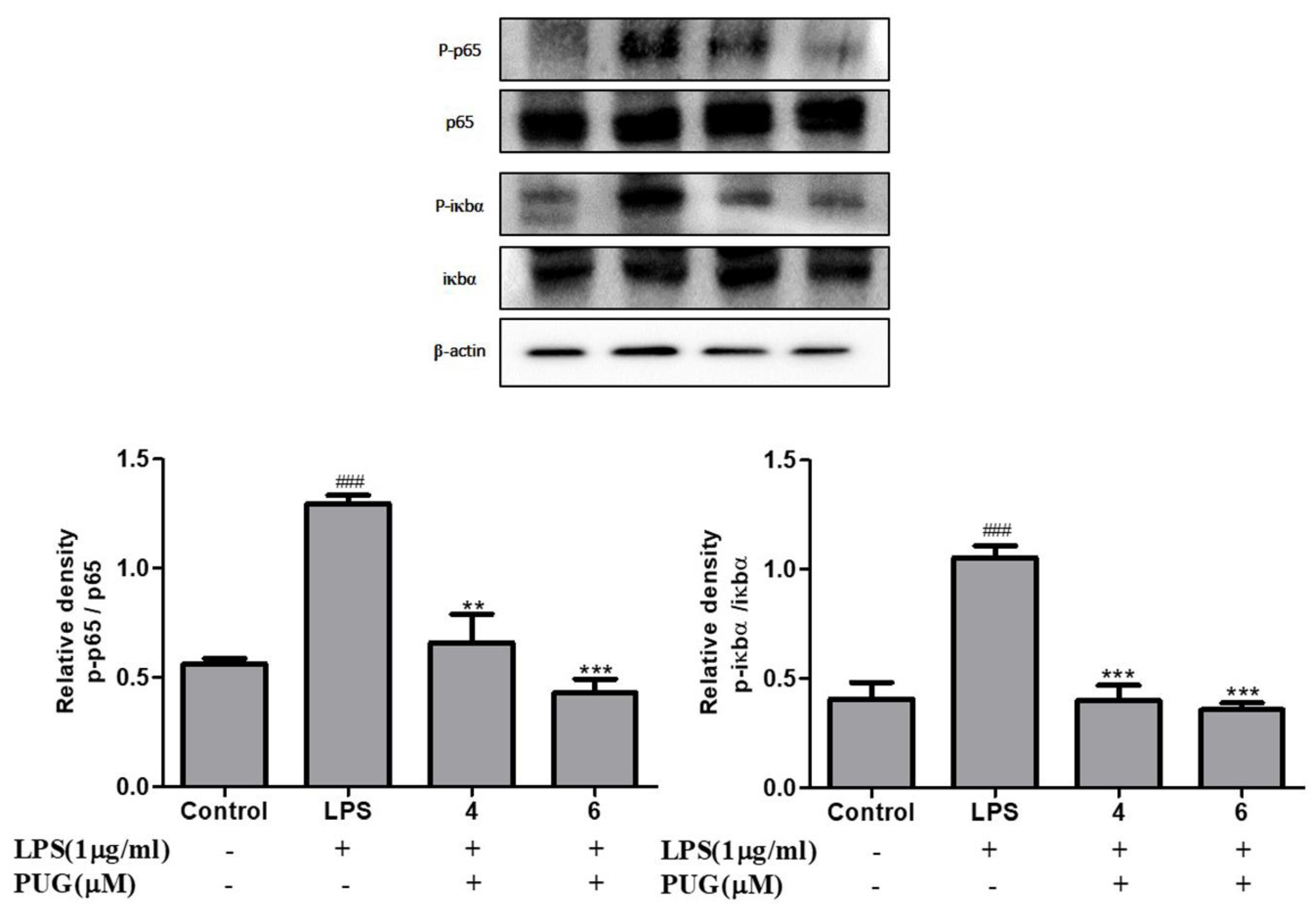
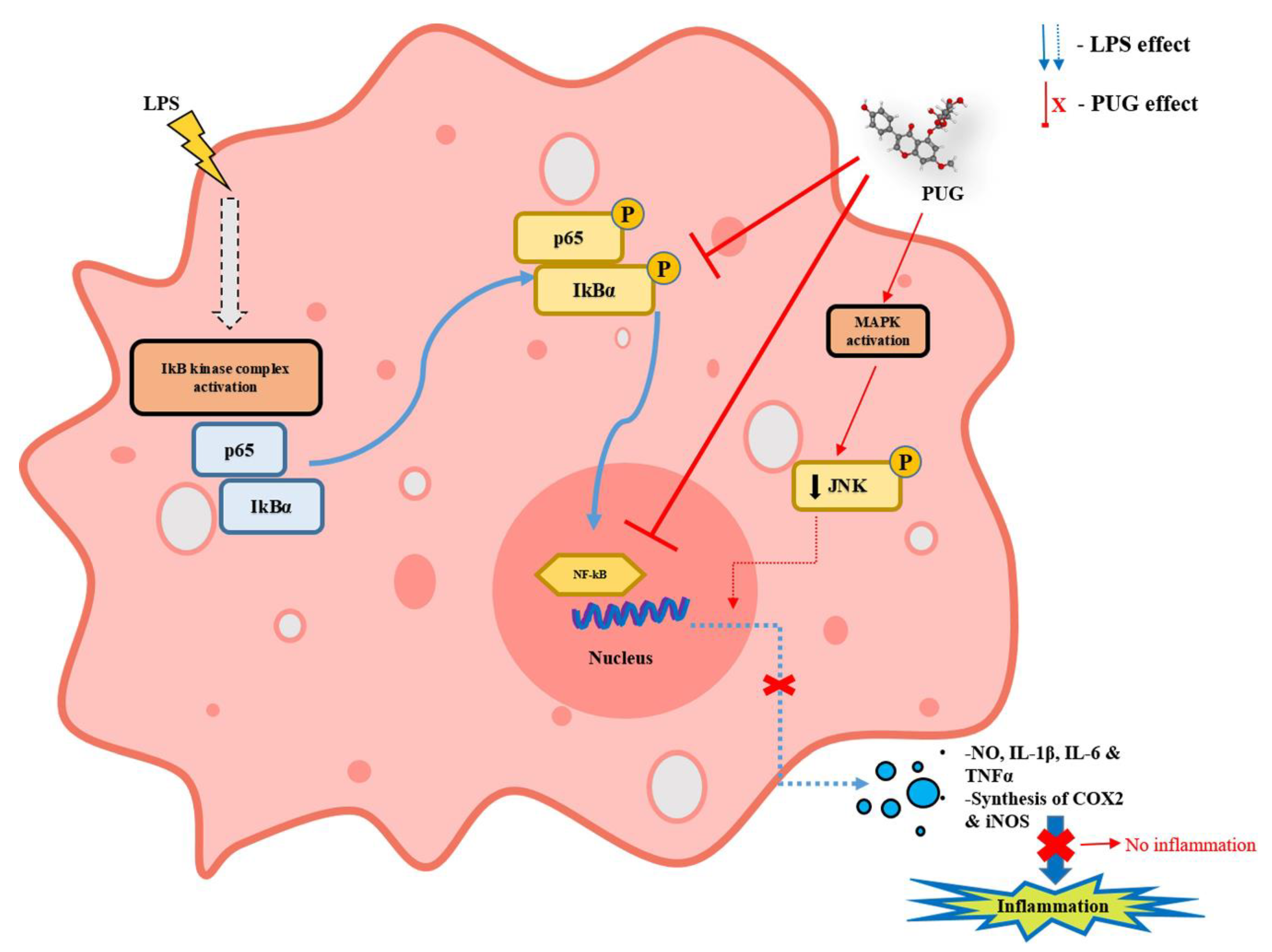
2.7. The Effect of PUG on the NF-κB Pathway
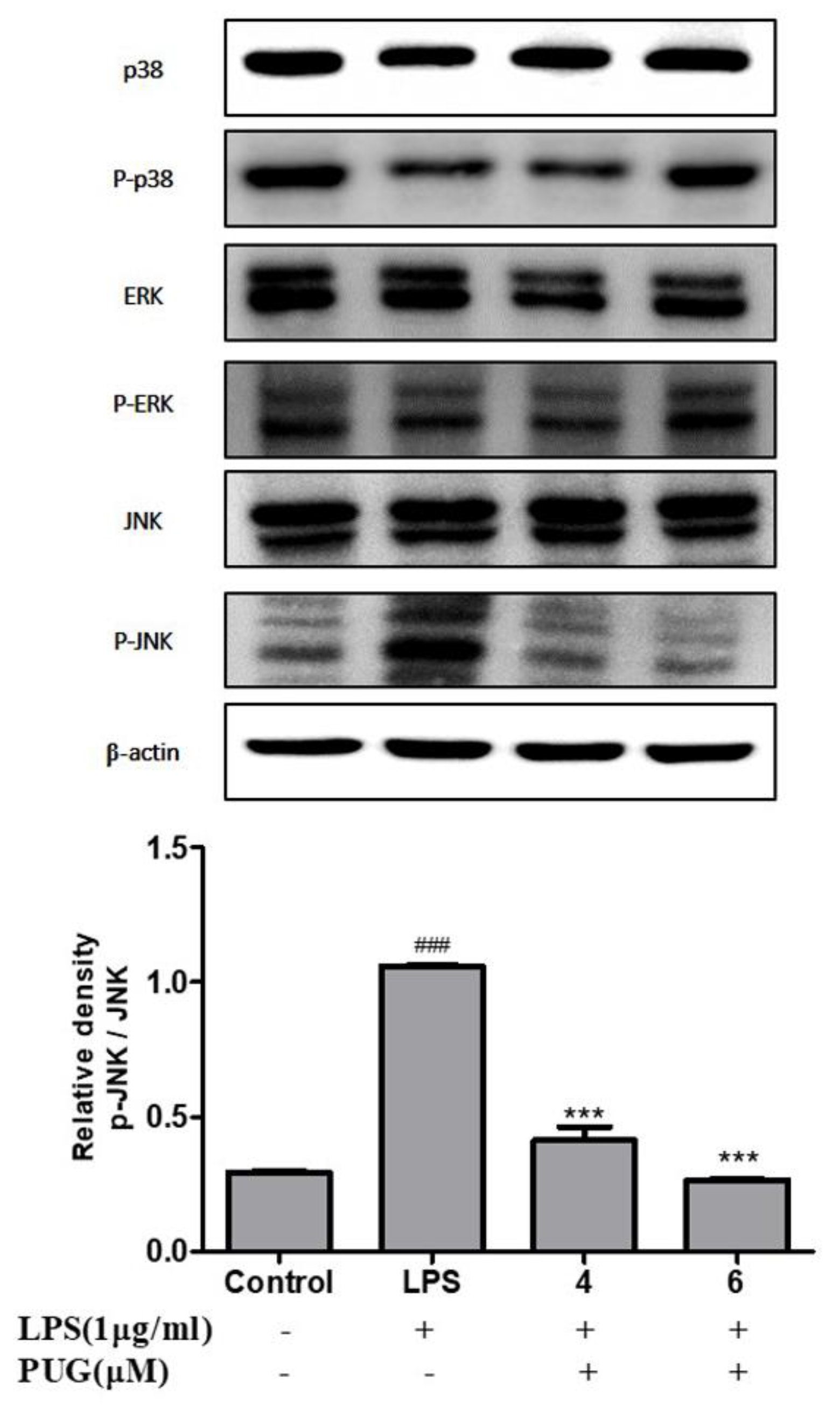
2.8. Effect of PUG on Activating MAPKs in LPS Stimulated RAW264.7 Cells
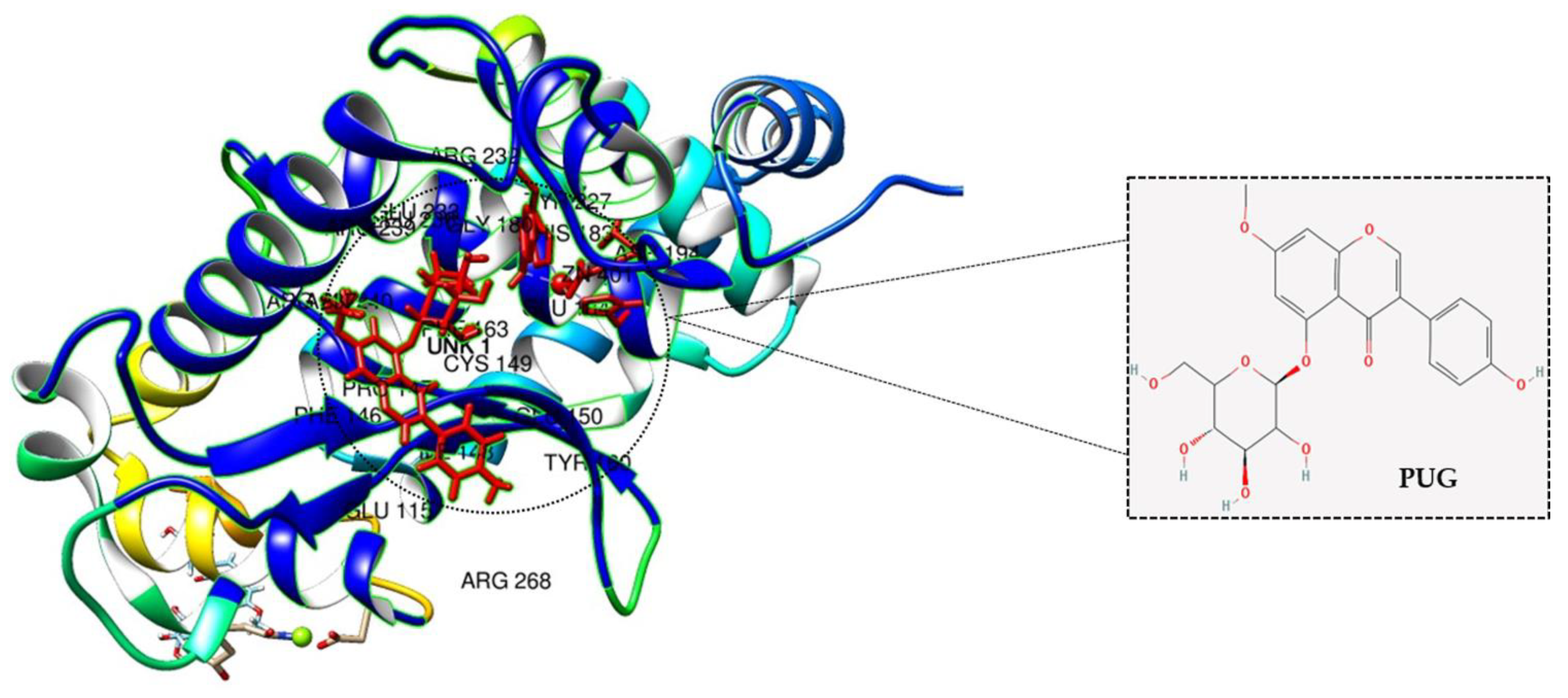
2.9. Molecular Docking Analysis of PUG with NF-κB
3. Discussion
4. Materials and Methods
4.1. Cell Line Maintenance and Compound
4.2. Cytotoxicity Test
4.3. Nitric Oxide (NO) Assay
4.4. Enzyme-Linked Immunoassay (ELISA)
4.5. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
4.6. Western Blotting
4.7. Molecular Docking
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, C.; Zhao, W.; Zhang, X.; Chen, X. Neocryptotanshinone inhibits lipopolysaccharide-induced inflammation in RAW264. 7 macrophages by suppression of NF-κB and iNOS signaling pathways. Acta Pharm. Sin. B 2015, 5, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allavena, P.; Garlanda, C.; Borrello, M.G.; Sica, A.; Mantovani, A. Pathways connecting inflammation and cancer. Curr. Opin. Genet. Dev. 2008, 18, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gu, J.; Zong, M.; Zhang, Q.; Li, H.; Li, D.; Mou, X.; Liu, P.; Liu, Y.; Qiu, F. Anti-inflammatory action of physalin A by blocking the activation of NF-κB signaling pathway. J. Ethnopharmacol. 2021, 267, 113490. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Park, S.J.; Yun, K.-J.; Cho, Y.-W.; Park, H.-J.; Lee, K.-T. Isoliquiritigenin isolated from the roots of Glycyrrhiza uralensis inhibits LPS-induced iNOS and COX-2 expression via the attenuation of NF-κB in RAW 264.7 macrophages. Eur. J. Pharmacol. 2008, 584, 175–184. [Google Scholar] [CrossRef]
- Kang, S.R.; Park, K.I.; Park, H.S.; Lee, D.H.; Kim, J.A.; Nagappan, A.; Kim, E.H.; Lee, W.S.; Shin, S.C.; Park, M.K. Anti-inflammatory effect of flavonoids isolated from Korea Citrus aurantium L. on lipopolysaccharide-induced mouse macrophage RAW 264.7 cells by blocking of nuclear factor-kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) signalling pathways. Food Chem. 2011, 129, 1721–1728. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef] [Green Version]
- Venkatesan, T.; Park, E.-J.; Choi, Y.-W.; Lee, J.; Kim, Y.-K. Anti-inflammatory activity of Ternstroemia gymnanthera stem bark extracts in bacterial lipopolysaccharide-stimulated RAW264. 7 murine macrophage cells. Pharm. Biol. 2017, 55, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.A.; DuBois, R.N. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat. Rev. Cancer 2001, 1, 11–21. [Google Scholar] [CrossRef]
- Kim, S.-C.; Kang, S.-H.; Jeong, S.-J.; Kim, S.-H.; Ko, H.S.; Kim, S.-H. Inhibition of c-Jun N-terminal kinase and nuclear factor κ B pathways mediates fisetin-exerted anti-inflammatory activity in lipopolysccharide-treated RAW264. 7 cells. Immunopharmacol. Immunotoxicol. 2012, 34, 645–650. [Google Scholar] [CrossRef]
- Kothari, P.; Pestana, R.; Mesraoua, R.; Elchaki, R.; Khan, K.F.; Dannenberg, A.J.; Falcone, D.J. IL-6–mediated induction of matrix metalloproteinase-9 is modulated by JAK-dependent IL-10 expression in macrophages. J. Immunol. 2014, 192, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Madonna, R.; de Caterina, R. Relevance of new drug discovery to reduce NF-κB activation in cardiovascular disease. Vasc. Pharmacol. 2012, 57, 41–47. [Google Scholar] [CrossRef]
- Lawrence, T.; Gilroy, D.W.; Colville-Nash, P.R.; Willoughby, D.A. Possible new role for NF-κB in the resolution of inflammation. Nat. Med. 2001, 7, 1291–1297. [Google Scholar] [CrossRef]
- Tak, P.P.; Firestein, G.S. NF-κB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.-E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- He, J.; Lu, X.; Wei, T.; Dong, Y.; Cai, Z.; Tang, L.; Liu, M. Asperuloside and asperulosidic acid exert an anti-inflammatory effect via suppression of the NF-κB and MAPK signaling pathways in LPS-induced RAW 264.7 macrophages. Int. J. Mol. Sci. 2018, 19, 2027. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Huang, Y.; Heasley, L.E.; Berl, T.; Schnermann, J.B.; Briggs, J.P. MAPK mediation of hypertonicity-stimulated cyclooxygenase-2 expression in renal medullary collecting duct cells. J. Biol. Chem. 2000, 275, 23281–23286. [Google Scholar] [CrossRef] [Green Version]
- Spagnuolo, C.; Moccia, S.; Russo, G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 2018, 153, 105–115. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Y.; Patel, G.; Xue, Q.; Njateng, G.S.S.; Cai, S.; Cheng, G.; Kai, G. In vitro and in vivo anti-inflammatory effects of different extracts from Epigynum auritum through down-regulation of NF-κB and MAPK signaling pathways. J. Ethnopharmacol. 2020, 261, 113105. [Google Scholar] [CrossRef]
- Zhan, R.; Xia, L.; Shao, J.; Wang, C.; Chen, D. Polysaccharide isolated from Chinese jujube fruit (Zizyphus jujuba cv. Junzao) exerts anti-inflammatory effects through MAPK signaling. J. Funct. Foods 2018, 40, 461–470. [Google Scholar] [CrossRef]
- Cai, B.; Seong, K.-J.; Bae, S.-W.; Chun, C.; Kim, W.-J.; Jung, J.-Y. A synthetic diosgenin primary amine derivative attenuates LPS-stimulated inflammation via inhibition of NF-κB and JNK MAPK signaling in microglial BV2 cells. Int. Immunopharmacol. 2018, 61, 204–214. [Google Scholar] [CrossRef]
- Kang, H.; Kwak, T.-K.; Kim, B.-G.; Lee, K.-J. The anti-inflammatory effect of Prunus yedoensis bark extract on adipose tissue in diet-induced obese mice. Evid. Based Complement. Altern. Med. 2015, 2015, 937904. [Google Scholar] [CrossRef] [Green Version]
- Vetrivel, P.; Murugesan, R.; Bhosale, P.B.; Ha, S.E.; Kim, H.H.; Heo, J.D.; Kim, G.S. A Network pharmacological approach to reveal the pharmacological targets and its associated biological mechanisms of prunetin-5-O-glucoside against gastric cancer. Cancers 2021, 13, 1918. [Google Scholar] [CrossRef]
- Coghlan, M.J.; Jacobson, P.B.; Lane, B.; Nakane, M.; Lin, C.W.; Elmore, S.W.; Kym, P.R.; Luly, J.R.; Carter, G.W.; Turner, R. A novel antiinflammatory maintains glucocorticoid efficacy with reduced side effects. Mol. Endocrinol. 2003, 17, 860–869. [Google Scholar] [CrossRef] [Green Version]
- Kandasamy, V.; Balasundaram, U. Caesalpinia bonduc (L.) Roxb. As a promising source of pharmacological compounds to treat Poly Cystic Ovary Syndrome (PCOS): A review. J. Ethnopharmacol. 2021, 279, 114375. [Google Scholar] [CrossRef]
- Abusaliya, A.; Ha, S.E.; Bhosale, P.B.; Kim, H.H.; Park, M.Y.; Vetrivel, P.; Kim, G.S. Glycosidic flavonoids and their potential applications in cancer research: A review. Mol. Cell. Toxicol. 2021, 18, 9–16. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, H.; Li, Y.; Chen, W.; Li, H.; Peng, K.; Zhang, Z.; Sun, X. Targeting NF-kB signaling with polymeric hybrid micelles that co-deliver siRNA and dexamethasone for arthritis therapy. Biomaterials 2017, 122, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Vetrivel, P.; Kim, H.H.; Ha, S.E.; Venkatarame Gowda Saralamma, V.; Kim, G.S. Artemisia iwayomogi (Dowijigi) inhibits lipopolysaccharide-induced inflammation in RAW264. 7 macrophages by suppressing the NF-κB signaling pathway. Exp. Ther. Med. 2020, 19, 2161–2170. [Google Scholar] [PubMed] [Green Version]
- Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2002, 2, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Dijsselbloem, N.; Berghe, W.V.; de Naeyer, A.; Haegeman, G. Soy isoflavone phyto-pharmaceuticals in interleukin-6 affections: Multi-purpose nutraceuticals at the crossroad of hormone replacement, anti-cancer and anti-inflammatory therapy. Biochem. Pharmacol. 2004, 68, 1171–1185. [Google Scholar] [CrossRef]
- Ma, C.; Wang, Y.; Dong, L.; Li, M.; Cai, W. Anti-inflammatory effect of resveratrol through the suppression of NF-κB and JAK/STAT signaling pathways. Acta Biochim. Biophys. Sin. 2015, 47, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Han, J.; Hui, L. MAPK signaling in inflammation-associated cancer development. Protein Cell 2010, 1, 218–226. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Ham, I.; Choi, H.-Y. Anti-inflammatory effect of prunetin via the suppression of NF-κB pathway. Food Chem. Toxicol. 2013, 58, 124–132. [Google Scholar] [CrossRef]
- Qawoogha, S.S.; Shahiwala, A. Identification of potential anticancer phytochemicals against colorectal cancer by structure-based docking studies. J. Recept. Signal Transduct. 2020, 40, 67–76. [Google Scholar] [CrossRef]
- Rampogu, S.; Parameswaran, S.; Lemuel, M.R.; Lee, K.W. Exploring the therapeutic ability of fenugreek against type 2 diabetes and breast cancer employing molecular docking and molecular dynamics simulations. Evid. Based Complement. Altern. Med. 2018, 2018, 1943203. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| iNOS | F | TCCTACACCACACCAAAC |
| R | CTCCAATCTCTGCCTATC | |
| COX2 | F | CCTCTGCGATGCTCTTCC |
| R | TCACACTTATACTGGTCAAATCC | |
| IL-6 | F | GAGGATACCACTCCCAACAGACC |
| R | AAGTGCATCATCGTTGTTCATACA | |
| IL-1β | F | TGCAGAGTTCCCCAACTGGTACATC |
| R | GTGCTGCCTAATGTCCCCTTGAATC | |
| TNFα | F | TGGAGTCATTGCTCTGTGAAGGGA |
| R | AGTCCTTGATGGTGGTGCATGAGA | |
| β-Actin | F | TACTGCCCTGGCTCCTAGCA |
| R | TGGACAGTGAGGCCAGGATAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abusaliya, A.; Bhosale, P.B.; Kim, H.H.; Ha, S.E.; Park, M.Y.; Jeong, S.H.; Vetrivel, P.; Park, J.-S.; Kim, G.S. Prunetinoside Inhibits Lipopolysaccharide-Provoked Inflammatory Response via Suppressing NF-κB and Activating the JNK-Mediated Signaling Pathway in RAW264.7 Macrophage Cells. Int. J. Mol. Sci. 2022, 23, 5442. https://doi.org/10.3390/ijms23105442
Abusaliya A, Bhosale PB, Kim HH, Ha SE, Park MY, Jeong SH, Vetrivel P, Park J-S, Kim GS. Prunetinoside Inhibits Lipopolysaccharide-Provoked Inflammatory Response via Suppressing NF-κB and Activating the JNK-Mediated Signaling Pathway in RAW264.7 Macrophage Cells. International Journal of Molecular Sciences. 2022; 23(10):5442. https://doi.org/10.3390/ijms23105442
Chicago/Turabian StyleAbusaliya, Abuyaseer, Pritam Bhagwan Bhosale, Hun Hwan Kim, Sang Eun Ha, Min Yeong Park, Se Hyo Jeong, Preethi Vetrivel, Joon-Suk Park, and Gon Sup Kim. 2022. "Prunetinoside Inhibits Lipopolysaccharide-Provoked Inflammatory Response via Suppressing NF-κB and Activating the JNK-Mediated Signaling Pathway in RAW264.7 Macrophage Cells" International Journal of Molecular Sciences 23, no. 10: 5442. https://doi.org/10.3390/ijms23105442
APA StyleAbusaliya, A., Bhosale, P. B., Kim, H. H., Ha, S. E., Park, M. Y., Jeong, S. H., Vetrivel, P., Park, J.-S., & Kim, G. S. (2022). Prunetinoside Inhibits Lipopolysaccharide-Provoked Inflammatory Response via Suppressing NF-κB and Activating the JNK-Mediated Signaling Pathway in RAW264.7 Macrophage Cells. International Journal of Molecular Sciences, 23(10), 5442. https://doi.org/10.3390/ijms23105442








