Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root
Abstract
1. Introduction
2. Results
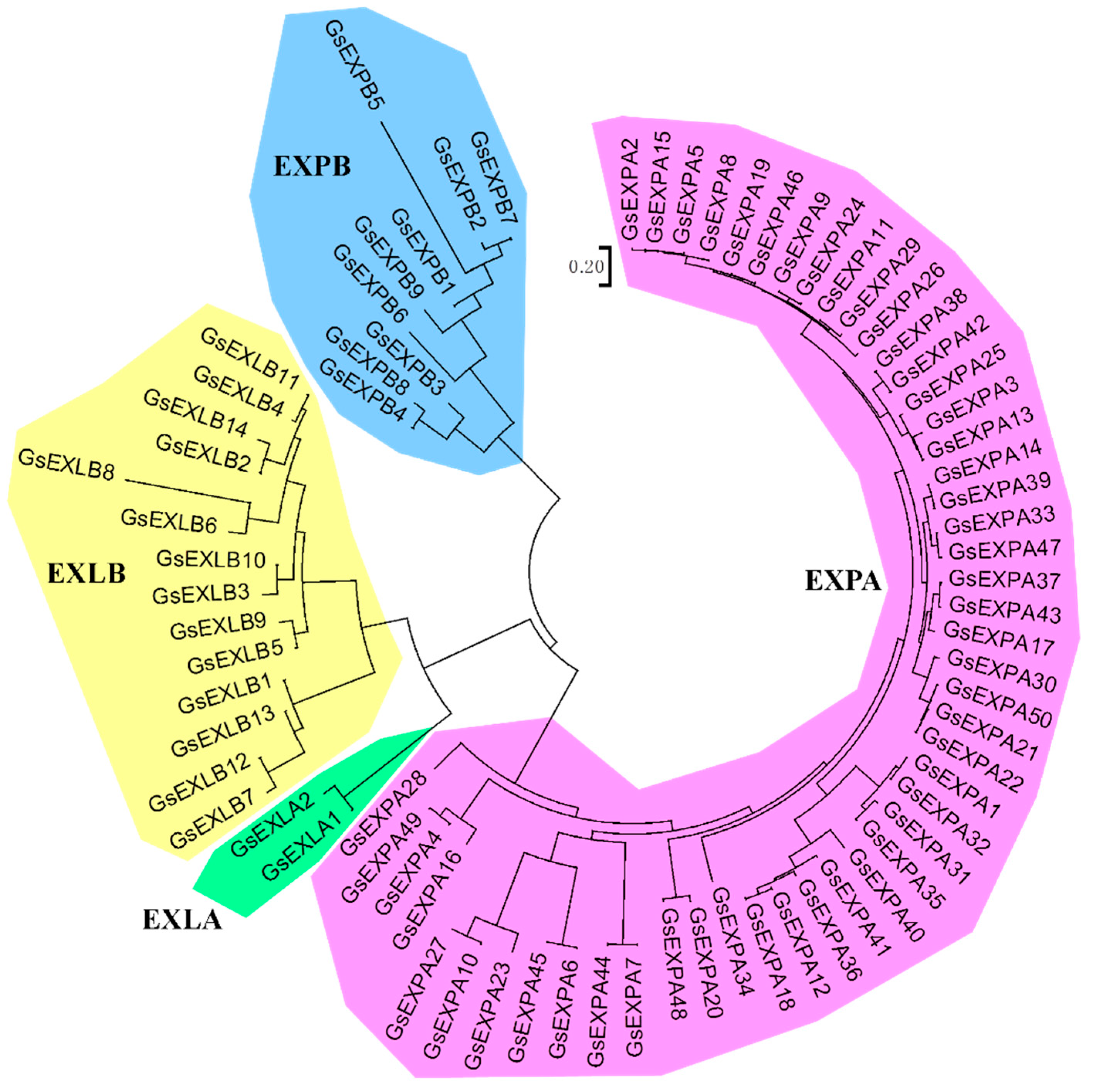
2.1. Identification of Expansin Family Members in Wild Soybean
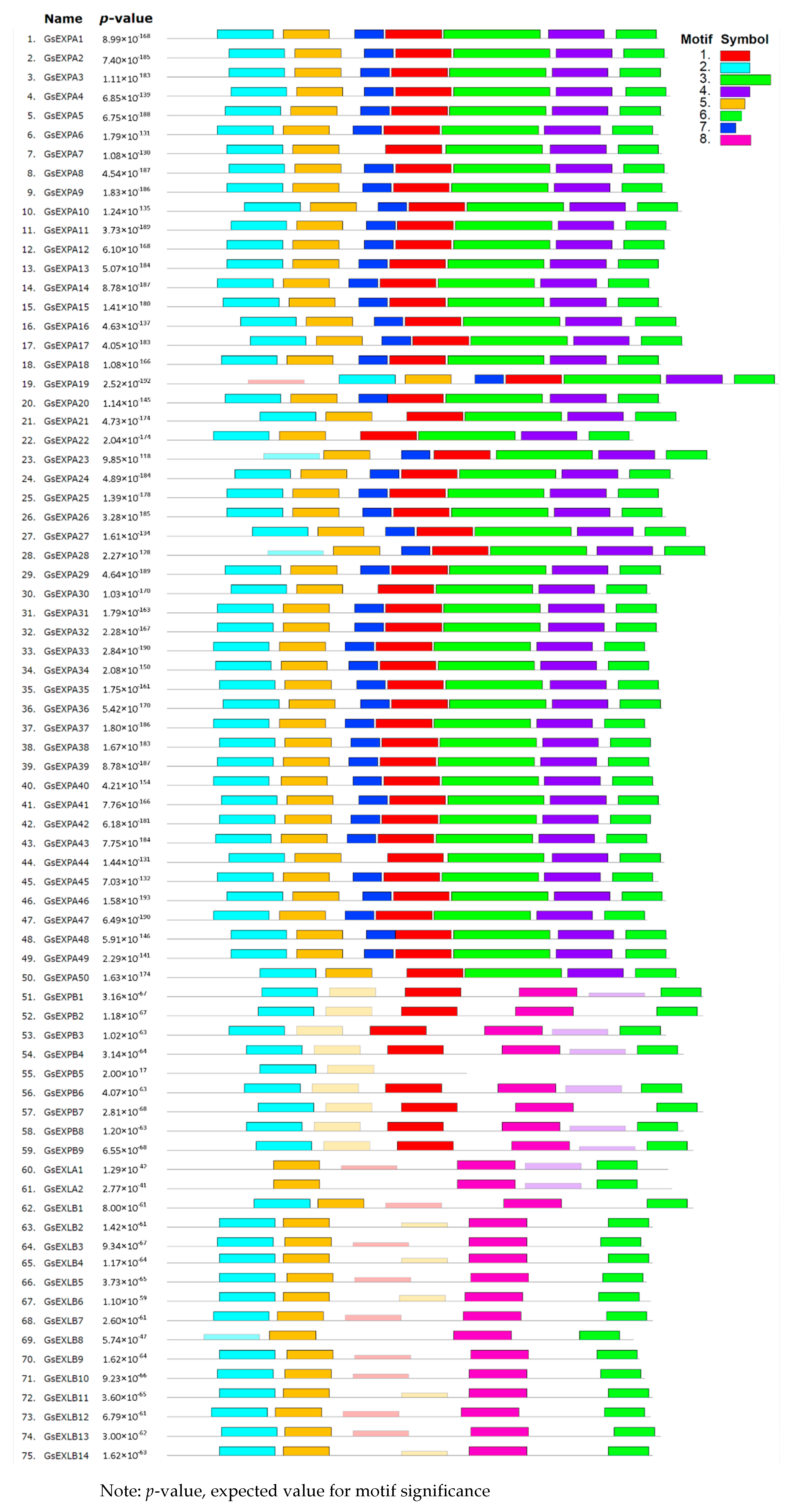
2.2. Gene Structure and Conserved Motifs
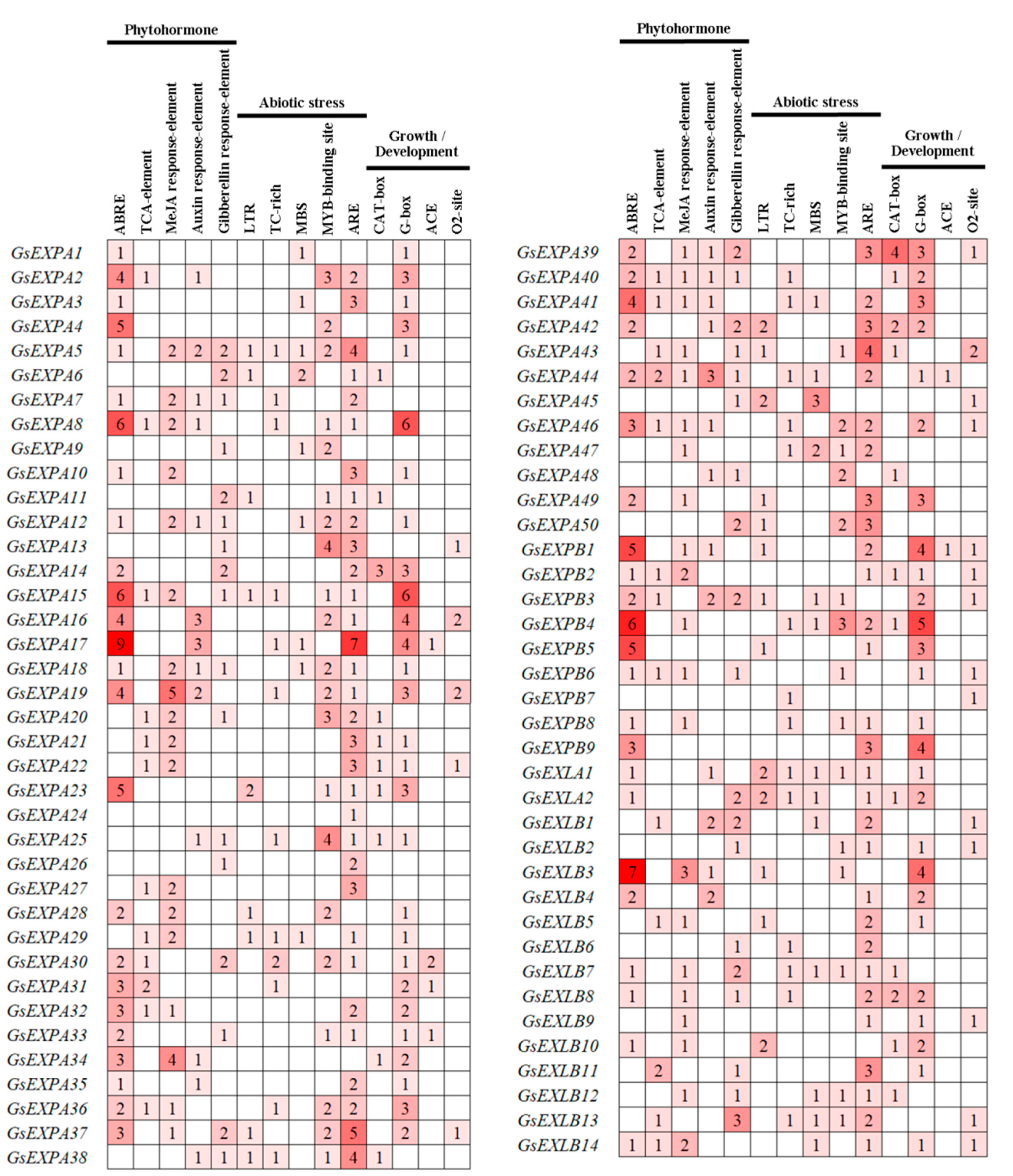
2.3. Cis-Elements
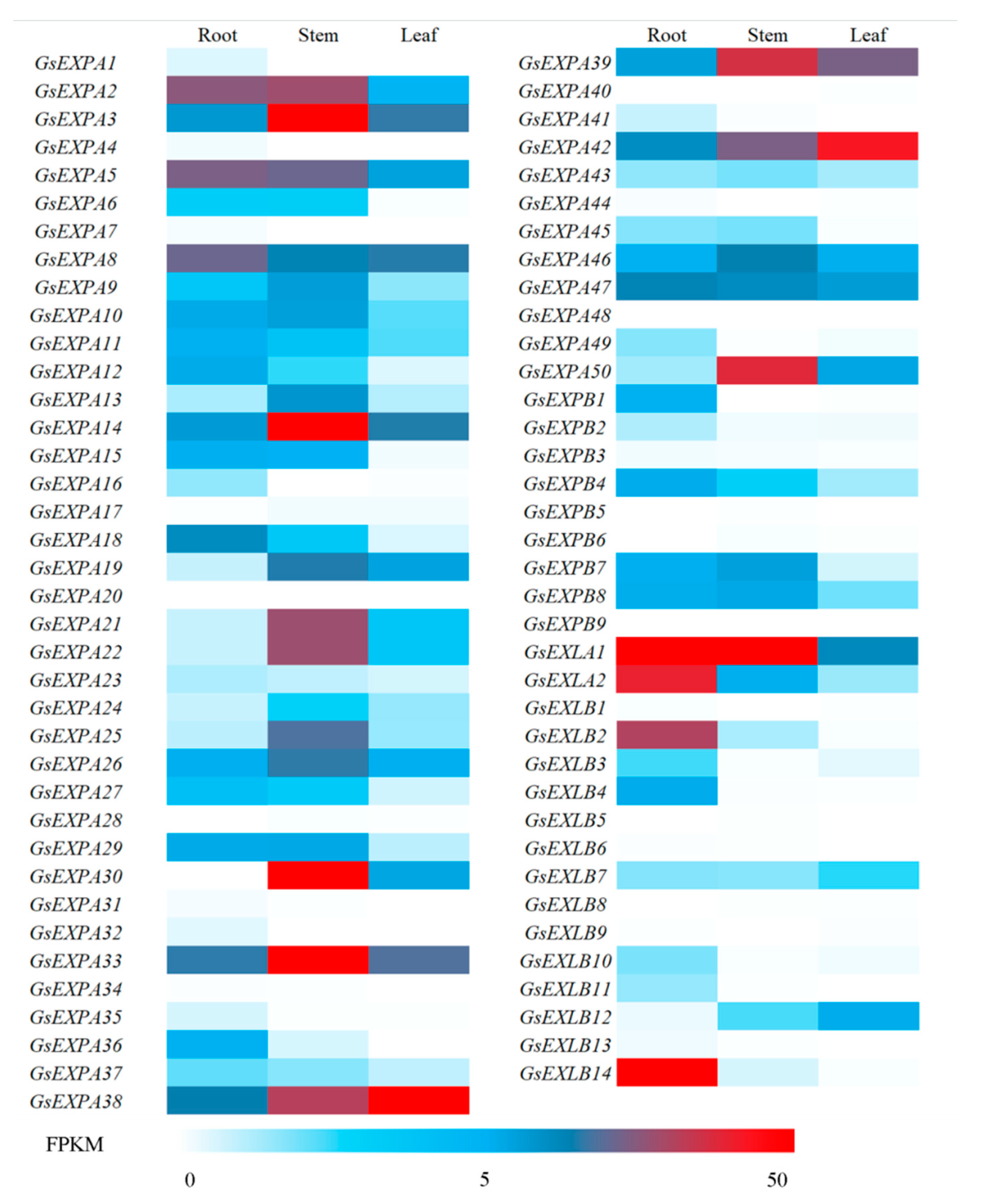
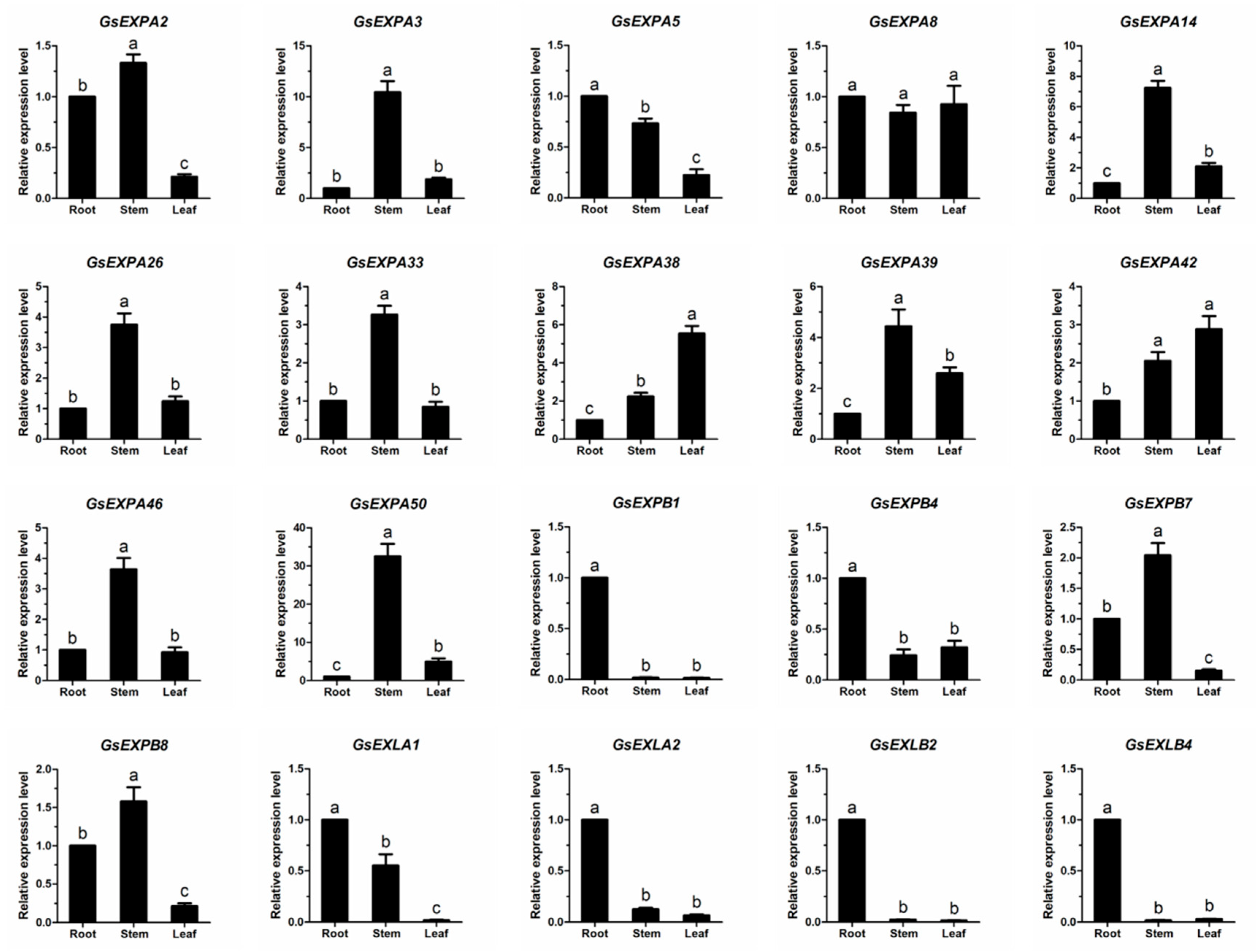
2.4. Wild Soybean Vegetative Organ Basal Transcriptome
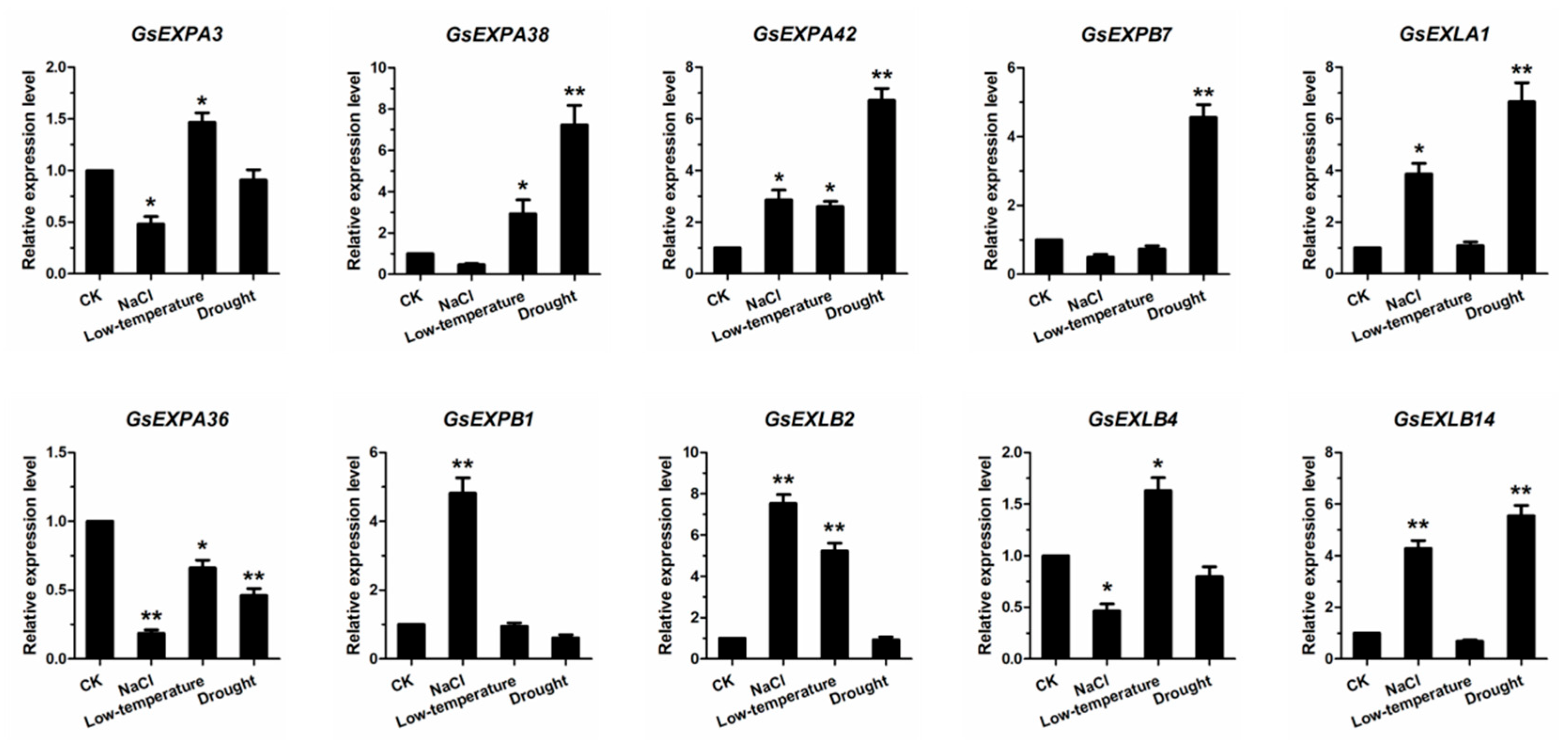
2.5. Expression Profiles
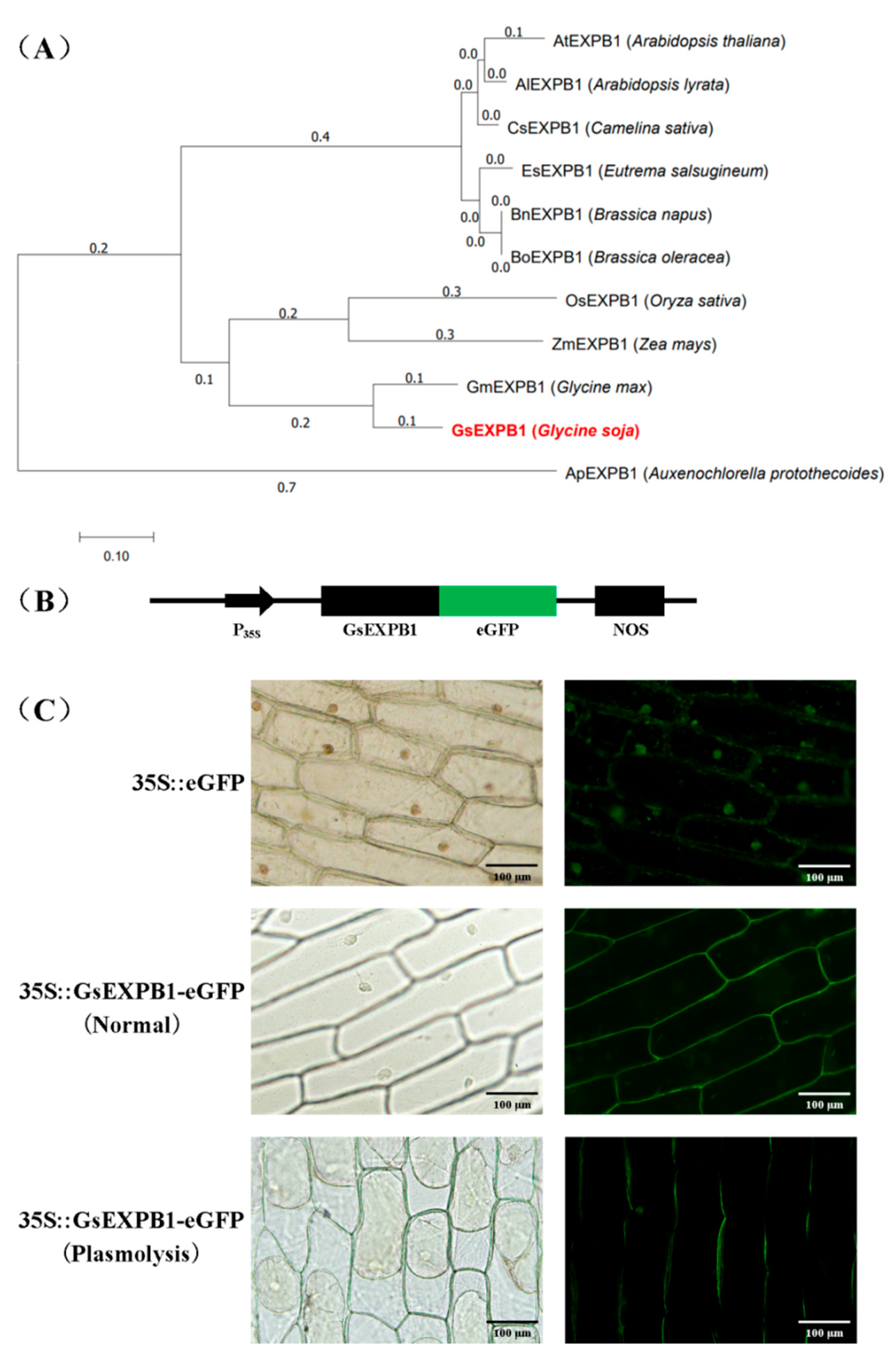
2.6. Amino Acid Evolutionary Tree and Subcellular Localization of GsEXPB1
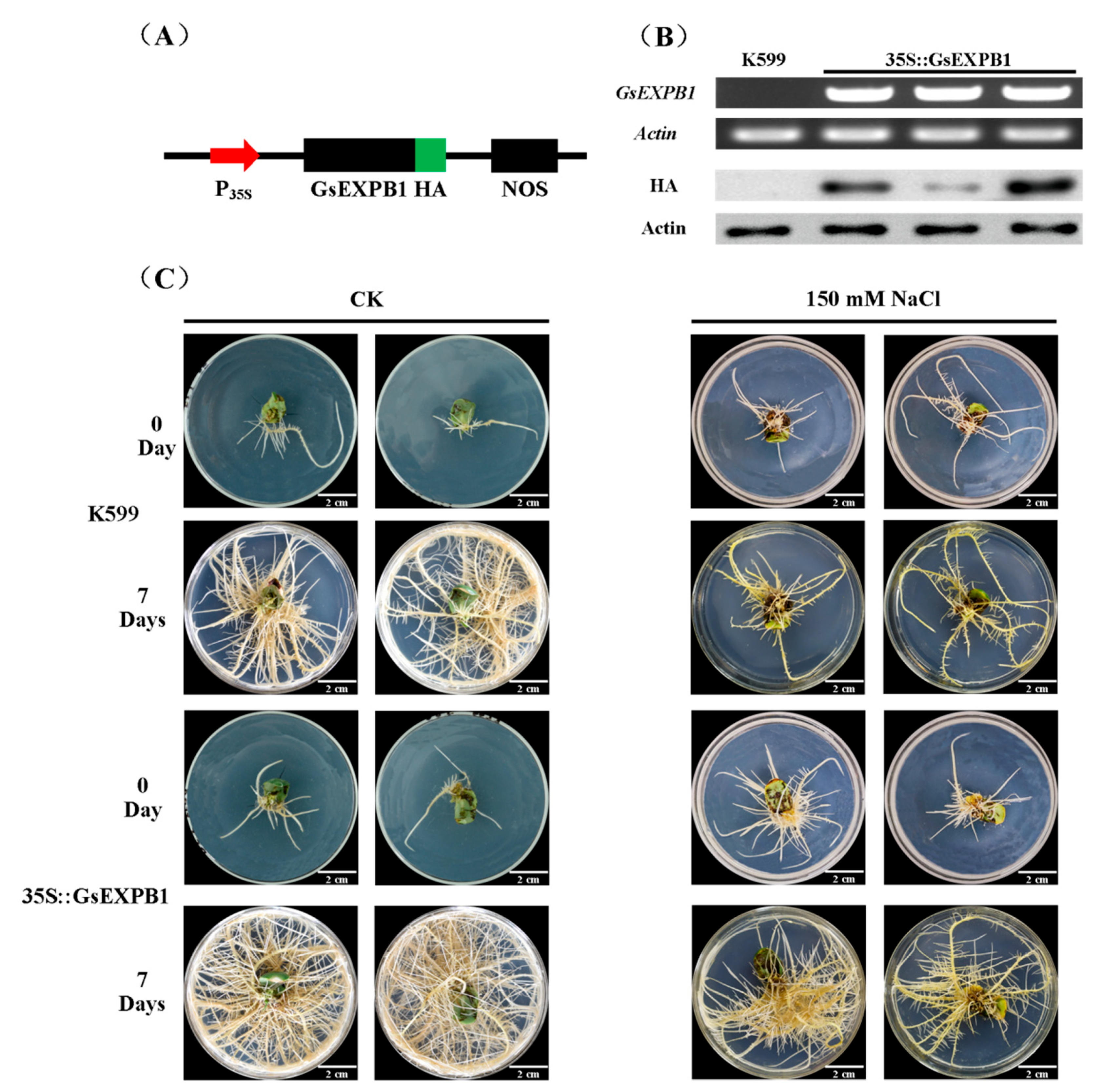
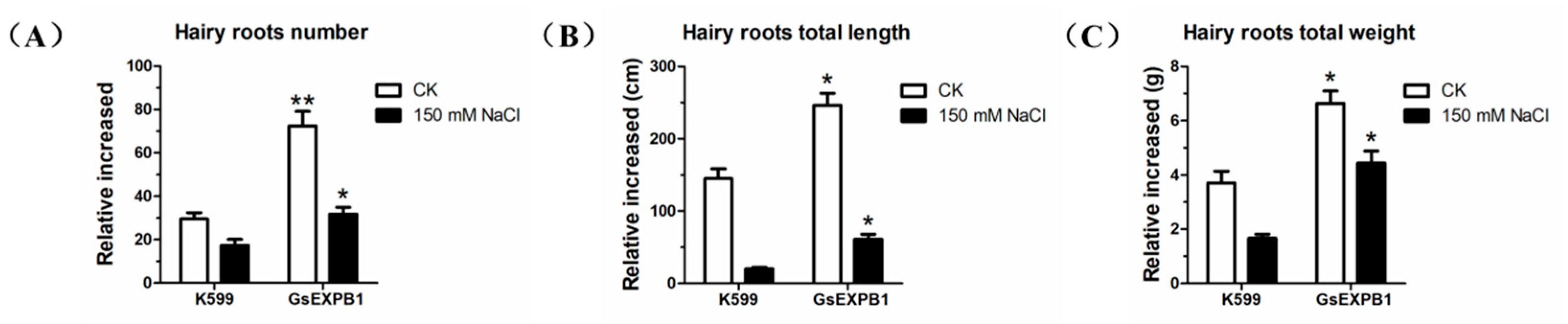
2.7. Phenotypic Observation of Soybean Hairy Roots Overexpressing GsEXPB1
3. Discussion
3.1. Characterization of the Wild Soybean Expansin Family
3.2. Potential Functions of GsEXPB1
4. Materials and Methods
4.1. Genome-Wide Identification of Wild Soybean Expansin Genes
4.2. Determination of the Basic Transcriptome in Nutritional Organs
4.3. qRT–PCR
4.4. Construction of the GsEXPB1 Amino Acid Evolutionary Tree and Its Subcellular Localization
4.5. Overexpression of GsEXPB1 through Soybean Hairy Roots
4.6. Phenotypic Observation of Hairy Roots from Soybean Overexpressing GsEXPB1
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Kofsky, J.; Zhang, H.; Song, B.H. The Untapped Genetic Reservoir: The Past, Current, and Future Applications of the Wild Soybean (Glycine soja). Front. Plant Sci. 2018, 9, 949. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Liu, Y.; Minh, T.N.; Lu, H.; Zhang, P.; Li, W.; Xiao, J.; Ding, X.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chung, C.Y.; Li, M.W.; Wong, F.L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.; Wong, T.H.; Tong, S.W.; et al. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.C.; Kim, S.H.; Lee, H.S.; Adeva, C.; Jeon, Y.A.; Luong, N.H.; Kim, W.J.; Akhtamov, M.; Park, Y.J.; Ahn, S.N. Characterization of a New qLTG3-1 Allele for Low-temperature Germinability in Rice from the Wild Species Oryza Rufipogon. Rice 2020, 13, 10. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Narváez-Barragán, D.A.; Tovar-Herrera, O.E.; Segovia, L.; Serrano, M.; Martinez-Anaya, C. Expansin-related proteins: Biology, microbe-plant interactions and associated plant-defense responses. Microbiology 2020, 166, 1007–1018. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Chen, F.; Bradford, K.J. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000, 124, 1265–1274. [Google Scholar] [CrossRef]
- Morris, K.; Linkies, A.; Müller, K.; Oracz, K.; Wang, X.; Lynn, J.R.; Leubner-Metzger, G.; Finch-Savage, W.E. Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: A global tissue-specific transcript analysis. Plant Physiol. 2011, 155, 1851–1870. [Google Scholar] [CrossRef]
- Kwasniewski, M.; Szarejko, I. Molecular cloning and characterization of beta-expansin gene related to root hair formation in barley. Plant Physiol. 2006, 141, 1149–1158. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Zhou, S.; Han, Y.Y.; Chen, Y.; Kong, X.; Wang, W. The involvement of expansins in response to water stress during leaf development in wheat. J. Plant Physiol. 2015, 183, 64–74. [Google Scholar] [CrossRef]
- Devi, M.J.; Taliercio, E.W.; Sinclair, T.R. Leaf expansion of soybean subjected to high and low atmospheric vapour pressure deficits. J. Exp. Bot. 2015, 66, 1845–1850. [Google Scholar] [CrossRef]
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef]
- Wei, P.C.; Zhang, X.Q.; Zhao, P.; Wang, X.C. Regulation of stomatal opening by the guard cell expansin AtEXPA1. Plant Signal. Behav. 2011, 6, 740–742. [Google Scholar] [CrossRef]
- Zenoni, S.; Reale, L.; Tornielli, G.B.; Lanfaloni, L.; Porceddu, A.; Ferrarini, A.; Moretti, C.; Zamboni, A.; Speghini, A.; Ferranti, F.; et al. Downregulation of the Petunia hybrida alpha-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell 2004, 16, 295–308. [Google Scholar] [CrossRef]
- Minoia, S.; Boualem, A.; Marcel, F.; Troadec, C.; Quemener, B.; Cellini, F.; Petrozza, A.; Vigouroux, J.; Lahaye, M.; Carriero, F.; et al. Induced mutations in tomato SlExp1 alter cell wall metabolism and delay fruit softening. Plant Sci. 2016, 242, 195–202. [Google Scholar] [CrossRef]
- Palapol, Y.; Kunyamee, S.; Thongkhum, M.; Ketsa, S.; Ferguson, I.B.; van Doorn, W.G. Expression of expansin genes in the pulp and the dehiscence zone of ripening durian (Durio zibethinus) fruit. J. Plant Physiol. 2015, 182, 33–39. [Google Scholar] [CrossRef]
- Xu, B.; Gou, J.Y.; Li, F.G.; Shangguan, X.X.; Zhao, B.; Yang, C.Q.; Wang, L.J.; Yuan, S.; Liu, C.J.; Chen, X.Y. A cotton BURP domain protein interacts with α-expansin and their co-expression promotes plant growth and fruit production. Mol. Plant 2013, 6, 945–958. [Google Scholar] [CrossRef]
- Li, Y.; Tu, L.; Pettolino, F.A.; Ji, S.; Hao, J.; Yuan, D.; Deng, F.; Tan, J.; Hu, H.; Wang, Q.; et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol. J. 2016, 14, 951–963. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, Y.; Zhang, G.; An, J.; Yang, J.; Wang, Y.; Wang, W. Overexpression of the wheat expansin gene TaEXPA2 improves oxidative stress tolerance in transgenic Arabidopsis plants. Plant Physiol. Biochem. 2018, 124, 190–198. [Google Scholar] [CrossRef]
- Calderini, D.F.; Castillo, F.M.; Arenas-M, A.; Molero, G.; Reynolds, M.P.; Craze, M.; Bowden, S.; Milner, M.J.; Wallington, E.J.; Dowle, A.; et al. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef]
- Li, F.; Xing, S.; Guo, Q.; Zhao, M.; Zhang, J.; Gao, Q.; Wang, G.; Wang, W. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J. Plant Physiol. 2011, 168, 960–966. [Google Scholar] [CrossRef]
- Li, F.; Han, Y.; Feng, Y.; Xing, S.; Zhao, M.; Chen, Y.; Wang, W. Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 2013, 163, 281–291. [Google Scholar] [CrossRef]
- Chen, Y.; Han, Y.; Kong, X.; Kang, H.; Ren, Y.; Wang, W. Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+ /K+ and antioxidant competence. Physiol. Plant. 2017, 159, 161–177. [Google Scholar] [CrossRef]
- Jadamba, C.; Kang, K.; Paek, N.C.; Lee, S.I.; Yoo, S.C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F.; Hu, B. TaEXPB7-B, a β-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.N.; Xu, Y.Q.; Wang, X.; Feng, X.; Zhao, Q.Q.; Feng, S.S.; Zhao, Z.Y.; Hu, B.Z.; Li, F.L. Overexpression of paralogues of the wheat expansin gene TaEXPA8 improves low-temperature tolerance in Arabidopsis. Plant Biol. 2019, 21, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chen, Y.; An, J.; Zhao, Z.; Zhang, G.; Wang, Y.; Wang, W. Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to Cd toxicity. Plant Sci. 2018, 270, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef]
- de Lucas, M.; Davière, J.M.; Rodríguez-Falcón, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blázquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef]
- Zhang, J.F.; Xu, Y.Q.; Dong, J.M.; Peng, L.N.; Feng, X.; Wang, X.; Li, F.; Miao, Y.; Yao, S.K.; Zhao, Q.Q.; et al. Genome-wide identification of wheat (Triticum aestivum) expansins and expansin expression analysis in cold-tolerant and cold-sensitive wheat cultivars. PLoS ONE 2018, 13, e0195138. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, H.; Chen, W.; Liu, J.; Jiang, C.; Jiang, H.; Zhu, S.; Cheng, B. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol. Genet. Genom. 2014, 289, 1061–1074. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Ding, A.; Marowa, P.; Kong, Y. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol. Genet. Genom. 2016, 291, 1891–1907. [Google Scholar] [CrossRef]
- Guimaraes, L.A.; Mota, A.; Araujo, A.; de Alencar Figueiredo, L.F.; Pereira, B.M.; de Passos Saraiva, M.A.; Silva, R.B.; Danchin, E.; Guimaraes, P.M.; Brasileiro, A. Genome-wide analysis of expansin superfamily in wild Arachis discloses a stress-responsive expansin-like B gene. Plant Mol. Biol. 2017, 94, 79–96. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, L.; Wang, X.; Han, Z.; Ouyang, B.; Zhang, J.; Li, H. Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol. Genet. Genom. 2016, 291, 597–608. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, R.; Gao, Z.; Chen, C.; Jiang, Z.; Shu, H. A genome-wide analysis of the expansin genes in Malus × Domestica. Mol. Genet. Genom. 2014, 289, 225–236. [Google Scholar] [CrossRef]
- Jin, K.M.; Zhuo, R.Y.; Xu, D.; Wang, Y.J.; Fan, H.J.; Huang, B.Y.; Qiao, G.R. Genome-Wide Identification of the Expansin Gene Family and Its Potential Association with Drought Stress in Moso Bamboo. Int. J. Mol. Sci. 2020, 21, 9491. [Google Scholar] [CrossRef]
- Lv, L.M.; Zuo, D.Y.; Wang, X.F.; Cheng, H.L.; Zhang, Y.P.; Wang, Q.L.; Song, G.L.; Ma, Z.Y. Genome-wide identification of the expansin gene family reveals that expansin genes are involved in fibre cell growth in cotton. BMC Plant Biol. 2020, 20, 223. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, Y.; Zhi, J.; Li, X.; Liu, H.; Xu, J. Over-expression of the poplar expansin gene PtoEXPA12 in tobacco plants enhanced cadmium accumulation. Int. J. Biol. Macromol. 2018, 116, 676–682. [Google Scholar] [CrossRef]
- Chen, S.; Luo, Y.; Wang, G.; Feng, C.; Li, H. Genome-wide identification of expansin genes in Brachypodium distachyon and functional characterization of BdEXPA27. Plant Sci. 2020, 296, 110490. [Google Scholar] [CrossRef]
- Won, S.K.; Choi, S.B.; Kumari, S.; Cho, M.; Lee, S.H.; Cho, H.T. Root hair-specific EXPANSIN B genes have been selected for Graminaceae root hairs. Mol. Cells 2010, 30, 369–376. [Google Scholar] [CrossRef]
- Zou, X.; Liu, L.; Hu, Z.; Wang, X.; Zhu, Y.; Zhang, J.; Li, X.; Kang, Z.; Lin, Y.; Yin, C. Salt-induced inhibition of rice seminal root growth is mediated by ethylene-jasmonate interaction. J. Exp. Bot. 2021, 72, 5656–5672. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Kende, H.; Bradford, K.; Brummell, D.; Cho, H.T.; Cosgrove, D.; Fleming, A.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef]
- He, X.; Zeng, J.; Cao, F.; Ahmed, I.M.; Zhang, G.; Vincze, E.; Wu, F. HvEXPB7, a novel β-expansin gene revealed by the root hair transcriptome of Tibetan wild barley, improves root hair growth under drought stress. J. Exp. Bot. 2015, 66, 7405–7419. [Google Scholar] [CrossRef]
- ZhiMing, Y.; Bo, K.; XiaoWei, H.; ShaoLei, L.; YouHuang, B.; WoNa, D.; Ming, C.; Hyung-Taeg, C.; Ping, W. Root hair-specific expansins modulate root hair elongation in rice. Plant J. 2011, 66, 725–734. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Expanding wheat yields with expansin. New Phytol. 2021, 230, 403–405. [Google Scholar] [CrossRef]
- Chen, L.J.; Zou, W.S.; Fei, C.Y.; Wu, G.; Li, X.Y.; Lin, H.H.; Xi, D.H. α-Expansin EXPA4 Positively Regulates Abiotic Stress Tolerance but Negatively Regulates Pathogen Resistance in Nicotiana tabacum. Plant Cell Physiol. 2018, 59, 2317–2330. [Google Scholar] [CrossRef]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef]
- Chen, Y.; Han, Y.; Zhang, M.; Zhou, S.; Kong, X.; Wang, W. Overexpression of the Wheat Expansin Gene TaEXPA2 Improved Seed Production and Drought Tolerance in Transgenic Tobacco Plants. PLoS ONE 2016, 11, e0153494. [Google Scholar] [CrossRef]
- Li, C.; Zhang, H.; Wang, X.; Liao, H. A comparison study of Agrobacterium-mediated transformation methods for root-specific promoter analysis in soybean. Plant Cell Rep. 2014, 33, 1921–1932. [Google Scholar] [CrossRef]


| Species (Scientific Name) | Total Number | EXPA | EXPB | EXLA | EXLB |
|---|---|---|---|---|---|
| Gossypium hirsutum | 93 | 67 | 8 | 6 | 12 |
| Triticum aestivum | 241 | 121 | 104 | 16 | 0 |
| Moso Bamboo | 82 | 45 | 7 | 1 | 29 |
| Brachypodium distachyon | 38 | 30 | 4 | 3 | 1 |
| Gossypium hirsutum | 93 | 67 | 8 | 6 | 12 |
| Ziziphus jujuba | 30 | 19 | 3 | 1 | 7 |
| Glycine max | 75 | 49 | 9 | 2 | 15 |
| Nicotiana tabacum | 52 | 36 | 6 | 3 | 7 |
| Solanum lycopersicum | 38 | 25 | 8 | 1 | 4 |
| Glycine soja | 75 | 50 | 9 | 2 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Li, C.; He, F.; Xu, Y.; Li, L.; Wang, X.; Chen, Q.; Li, F. Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root. Int. J. Mol. Sci. 2022, 23, 5407. https://doi.org/10.3390/ijms23105407
Feng X, Li C, He F, Xu Y, Li L, Wang X, Chen Q, Li F. Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root. International Journal of Molecular Sciences. 2022; 23(10):5407. https://doi.org/10.3390/ijms23105407
Chicago/Turabian StyleFeng, Xu, Cuiting Li, Fumeng He, Yongqing Xu, Li Li, Xue Wang, Qingshan Chen, and Fenglan Li. 2022. "Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root" International Journal of Molecular Sciences 23, no. 10: 5407. https://doi.org/10.3390/ijms23105407
APA StyleFeng, X., Li, C., He, F., Xu, Y., Li, L., Wang, X., Chen, Q., & Li, F. (2022). Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root. International Journal of Molecular Sciences, 23(10), 5407. https://doi.org/10.3390/ijms23105407





