Histone Modification on Parathyroid Tumors: A Review of Epigenetics
Abstract
1. Introduction
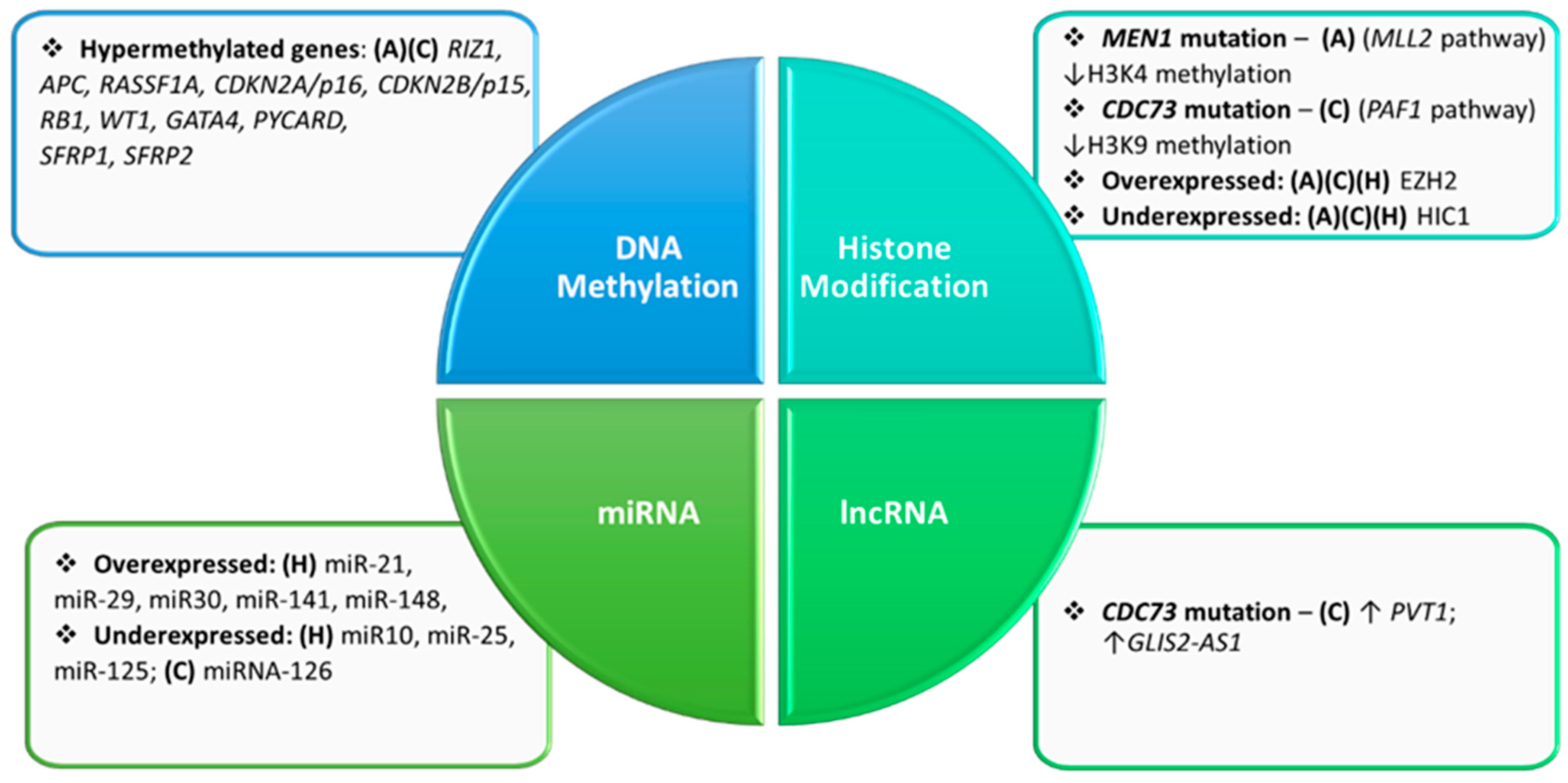
2. Epigenetics of the Parathyroid Glands
3. Potential of Histone Manipulation in Therapeutics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Owen, R. On the anatomy of the indian rhinoceros (rh. Unicornis). Trans. Zool. Soc. Lond. 1852, 4, 31–58. [Google Scholar]
- Johansson, H. The uppsala anatomist ivar sandström and the parathyroid gland. Ups. J. Med. Sci. 2015, 120, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, C. The anatomic basis of parathyroid surgery. Ann. Surg. 1976, 183, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Sarfati, E.; de Angelis, P.; D’Acrémont, B.; Casanova, D.; Dubost, C. Anatomic localization of parathyroid adenomas. Experience of 1200 cases of primary hyperparathyroidism. Minerva Chir. 1992, 47, 89–94. [Google Scholar]
- Bergson, E.J.; Heller, K.S. The clinical significance and anatomic distribution of parathyroid double adenomas. J. Am. Coll. Surg. 2004, 198, 185–189. [Google Scholar] [CrossRef]
- Abboud, B.; Sleilaty, G.; Helou, E.; Mansour, E.; Tohme, C.; Noun, R.; Sarkis, R. Existence and anatomic distribution of double parathyroid adenoma. Laryngoscope 2005, 115, 1128–1131. [Google Scholar] [CrossRef]
- Wang, F.L.; Miao, G.; Wei, J.M.; Xie, D.H.; Chen, J.; Dai, W.D.; Peng, Y.; Wu, M.X.; Chen, L.; He, S.R.; et al. Clinical and anatomic study of preserving parathyroid specific adipose attachments in total thyroidectomy. Zhonghua Wai Ke Za Zhi 2016, 54, 859–863. [Google Scholar] [CrossRef]
- Patrinos, A.; Zarokosta, M.; Piperos, T.; Tsiaoussis, J.; Noussios, G.; Mariolis-Sapsakos, T. An anatomic aberration and a surgical challenge: Mediastinal parathyroid adenoma anterior the pericardium. A case report. Int. J. Surg. Case Rep. 2019, 58, 153–156. [Google Scholar] [CrossRef]
- Conti-Freitas, C.L.; Foss-Freitas, M.C.; Lucca, L.J.; da Costa, J.A.C.; Mamede, R.C.M.; Foss, M.C. Dynamics of parathyroid hormone secretion after total parathyroidectomy and autotransplantation. World J. Surg. 2009, 33, 1403–1407. [Google Scholar] [CrossRef]
- Bilezikian, P.J.; Brandi, M.L.; Eastell, R.; Silverberg, S.J.; Udelsman, R.; Marcocci, C.; Potts, J.T. Guidelines for the management of asymptomatic primary hyperparathyroidism: Summary statement from the fourth international workshop. J. Clin. Endocrinol. Metab. 2014, 99, 3561–3569. [Google Scholar] [CrossRef]
- Yeh, W.M.; Ituarte, P.H.; Zhou, H.C.; Nishimoto, S.; Liu, I.L.; Harari, A.; Haigh, P.I.; Adams, A.L. Incidence and prevalence of primary hyperparathyroidism in a racially mixed population. J. Clin. Endocrinol. Metab. 2013, 98, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Evans, M.; Soro, M.; Barany, P.; Carrero, J.J. Secondary hyperparathyroidism and adverse health outcomes in adults with chronic kidney disease. Clin. Kidney J. 2021, 14, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Stanbury, W.S.; Lumb, G.A.; Nicholson, W.F. Elective subtotal parathyroidectomy for renal hyperparathyroidism. Lancet 1960, 1, 793–799. [Google Scholar] [CrossRef]
- Bilezikian, P.J.; Bandeira, L.; Khan, A.; Cusano, N.E. Hyperparathyroidism. Lancet 2018, 391, 168–178. [Google Scholar] [CrossRef]
- Dubose, J.; Ragsdale, T.; Morvant, J. Bodies so tiny: The history of parathyroid surgery. Curr. Surg. 2005, 62, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Giddings, E.C.; Rimmer, J.; Weir, N. History of parathyroid gland surgery: An historical case series. J. Laryngol. Otol. 2009, 123, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Wermers, A.R.; Khosla, S.; Atkinson, E.J.; Achenbach, S.J.; Oberg, A.L.; Grant, C.S.; Melton, L.J. Incidence of primary hyperparathyroidism in rochester, minnesota, 1993–2001: An update on the changing epidemiology of the disease. J. Bone Miner. Res. 2006, 21, 171–177. [Google Scholar] [CrossRef]
- Costa-Guda, J.; Arnold, A. Genetic and epigenetic changes in sporadic endocrine tumors: Parathyroid tumors. Mol. Cell. Endocrinol. 2014, 386, 46–54. [Google Scholar] [CrossRef]
- Cai, J.; Li, L.; Ye, L.; Jiang, X.H.; Shen, L.Y.; Gao, Z.B.; Fang, W.Y.; Huang, F.J.; Su, T.W.; Zhou, Y.L.; et al. Exome sequencing reveals mutant genes with low penetrance involved in men2a-associated tumorigenesis. Endocr.-Relat. Cancer 2015, 22, 23–33. [Google Scholar] [CrossRef]
- Isakov, O.; Rinella, E.S.; Olchovsky, D.; Shimon, I.; Ostrer, H.; Shomron, N.; Friedman, E. Missense mutation in the men1 gene discovered through whole exome sequencing co-segregates with familial hyperparathyroidism. Genet. Res. Camb. 2013, 95, 114–120. [Google Scholar] [CrossRef]
- Kasaian, K.; Wiseman, S.M.; Thiessen, N.; Mungall, K.L.; Corbett, R.D.; Qian, J.Q.; Nip, K.M.; He, A.; Tse, K.; Chuah, E.; et al. Complete genomic landscape of a recurring sporadic parathyroid carcinoma. J. Pathol. 2013, 230, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Park, M.H.; Woo, H.M.; Jo, H.Y.; Kim, J.H.; Choi, H.J.; Koo, S.K. Genetic analysis of parathyroid and pancreatic tumors in a patient with multiple endocrine neoplasia type 1 using whole-exome sequencing. BMC Med. Genet. 2017, 18, 106. [Google Scholar] [CrossRef] [PubMed]
- Newey, P.J.; Nesbit, M.A.; Rimmer, A.J.; Attar, M.; Head, R.T.; Christie, P.T.; Gorvin, C.M.; Stechman, M.; Gregory, L.; Mihai, R.; et al. Whole-exome sequencing studies of nonhereditary (sporadic) parathyroid adenomas. J. Clin. Endocrinol. Metab. 2012, 97, E1995–E2005. [Google Scholar] [CrossRef] [PubMed]
- Heppner, C.; Kester, M.B.; Agarwal, S.K.; Debelenko, L.V.; Emmert-Buck, M.R.; Guru, S.C.; Manickam, P.; Olufemi, S.E.; Skarulis, M.C.; Doppman, J.L.; et al. Somatic mutation of the men1 gene in parathyroid tumours. Nat. Genet. 1997, 16, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, T.M.; Valimaki, S.; Obara, T.; Gaz, R.D.; Clark, O.H.; Shoback, D.; Wierman, M.E.; Tojo, K.; Robbins, C.M.; Carpten, J.D.; et al. Somatic and germ-line mutations of the hrpt2 gene in sporadic parathyroid carcinoma. N. Engl. J. Med. 2003, 349, 1722–1729. [Google Scholar] [CrossRef]
- Tominaga, Y.; Tsuzuki, T.; Uchida, K.; Haba, T.; Otsuka, S.; Ichimori, T.; Yamada, K.; Numano, M.; Tanaka, Y.; Takagi, H. Expression of prad1 cyclin d1, retinoblastoma gene products, and ki67 in parathyroid hyperplasia caused by chronic renal failure versus primary adenoma. Kidney Int. 1999, 55, 1375–1383. [Google Scholar] [CrossRef]
- Agarwal, K.S.; Mateo, C.M.; Marx, S.J. Rare germline mutations in cyclin-dependent kinase inhibitor genes in multiple endocrine neoplasia type 1 and related states. J. Clin. Endocrinol. Metab. 2009, 94, 1826–1834. [Google Scholar] [CrossRef]
- Cromer, K.M.; Starker, L.F.; Choi, M.; Udelsman, R.; Nelson-Williams, C.; Lifton, R.P.; Carling, T. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J. Clin. Endocrinol. Metab. 2012, 97, E1774–E1781. [Google Scholar] [CrossRef]
- Bird, A. Dna methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A.; Patel, S.G. The genetic and epigenetic landscape of early-onset colorectal cancer. Colorectal Cancer 2020, 9, 14. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Mazzio, E.A.; Soliman, K.F. Basic concepts of epigenetics: Impact of environmental signals on gene expression. Epigenetics 2012, 7, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Minucci, S.; Pelicci, P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 2006, 6, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Vedamurthy, B.M.; Choudhari, R.; Ostwal, Y.B.; Mantelingu, K.; Kodaganur, G.S.; Kundu, T.K. Nitric oxide-mediated histone hyperacetylation in oral cancer: Target for a water-soluble hat inhibitor, ctk7a. Chem. Biol. 2010, 17, 903–913. [Google Scholar] [CrossRef]
- Juhlin, C.C.; Kiss, N.B.; Villablanca, A.; Haglund, F.; Nordenstrom, J.; Hoog, A.; Larsson, C. Frequent promoter hypermethylation of the apc and rassf1a tumour suppressors in parathyroid tumours. PLoS ONE 2010, 5, e9472. [Google Scholar] [CrossRef]
- Salcuni, A.S.; Cetani, F.; Guarnieri, V.; Nicastro, V.; Romagnoli, E.; de Martino, D.; Scillitani, A.; Cole, D.E.C. Parathyroid carcinoma. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 877–889. [Google Scholar] [CrossRef]
- Guarnieri, V.; Muscarella, L.A.; Verdelli, C.; Corbetta, S. Alterations of dna methylation in parathyroid tumors. Mol. Cell. Endocrinol. 2018, 469, 60–69. [Google Scholar] [CrossRef]
- Hewitt, K.M.; Sharma, P.K.; Samowitz, W.; Hobbs, M. Aberrant methylation of the hrpt2 gene in parathyroid carcinoma. Ann. Otol. Rhinol. Laryngol. 2007, 116, 928–933. [Google Scholar] [CrossRef]
- Starker, L.F.; Svedlund, J.; Udelsman, R.; Dralle, H.; Akerstrom, G.; Westin, G.; Lifton, R.P.; Bjorklund, P.; Carling, T. The dna methylome of benign and malignant parathyroid tumors. Genes Chromosomes Cancer 2011, 50, 735–745. [Google Scholar] [CrossRef]
- Varshney, S.; Bhadada, S.K.; Sachdeva, N.; Arya, A.K.; Saikia, U.N.; Behera, A.; Rao, S.D. Methylation status of the cpg islands in vitamin d and calcium-sensing receptor gene promoters does not explain the reduced gene expressions in parathyroid adenomas. J. Clin. Endocrinol. Metab. 2013, 98, E1631–E1635. [Google Scholar] [CrossRef] [PubMed]
- Hofman-Bang, J.; Gravesen, E.; Olgaard, K.; Lewin, E. Epigenetic methylation of parathyroid car and vdr promoters in experimental secondary hyperparathyroidism. Int. J. Nephrol. 2012, 2012, 123576. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landgraf, P.; Ludwig, J.; Rice, A.; Ojo, T.; Lin, C.; Holoch, D.; Lim, C.; Tuschl, T. Identification of micrornas and other small regulatory rnas using cdna library sequencing. Methods 2008, 44, 3–12. [Google Scholar] [CrossRef]
- Kilav-Levin, R.; Hassan, A.; Nechama, M.; Shilo, V.; Silver, J.; Ben-Dov, I.Z.; Naveh-Many, T. Post-transcriptional mechanisms regulating parathyroid hormone gene expression in secondary hyperparathyroidism. FEBS J. 2020, 287, 2903–2913. [Google Scholar] [CrossRef]
- Shilo, V.; Levi, I.M.Y.; Abel, R.; Mihailovic, A.; Wasserman, G.; Naveh-Many, T.; Ben-Dov, I.Z. Let-7 and microrna-148 regulate parathyroid hormone levels in secondary hyperparathyroidism. J. Am. Soc. Nephrol. 2017, 28, 2353–2363. [Google Scholar] [CrossRef]
- Esau, C.C. Inhibition of microrna with antisense oligonucleotides. Methods 2008, 44, 55–60. [Google Scholar] [CrossRef]
- Shilo, V.; Silver, J.; Naveh-Many, T. Micro-rnas in the parathyroid: A new portal in understanding secondary hyperparathyroidism. Curr. Opin. Nephrol. Hypertens. 2016, 25, 271–277. [Google Scholar] [CrossRef]
- Sun, Y.Q.; Bai, Y.F.; Zhang, F.; Wang, Y.; Guo, Y.; Guo, L.L. Mir-126 inhibits non-small cell lung cancer cells proliferation by targeting egfl7. Biochem. Biophys. Res. Commun. 2010, 391, 1483–1489. [Google Scholar] [CrossRef]
- Guo, C.G.; Sah, J.F.; Beard, L.; Willson, J.K.V.; Markowitz, S.D.; Guda, K. The noncoding rna, mir-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47, 939–946. [Google Scholar] [CrossRef]
- Juhlin, C.C.; Erickson, L.A. Genomics and epigenomics in parathyroid neoplasia: From bench to surgical pathology practice. Endocr. Pathol. 2020, 32, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Y.; Wang, M.Y.; Zhang, R.H.; Wang, P.P.; Cui, M.; Su, Z.; Gao, X.; Liao, Q.; Zhao, Y.P. Profiling analysis of long non-coding rna and mrna in parathyroid carcinoma. Endocr.-Relat. Cancer 2019, 26, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.Q.; Hardin, H.; Chu, Y.H.; Rehrauer, W.; Lloyd, R.V. Parathyroid neoplasms: Immunohistochemical characterization and long noncoding rna (lncrna) expression. Endocr. Pathol. 2019, 30, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Farber, L.J.; Kort, E.J.; Wang, P.F.; Chen, J.D.; Teh, B.T. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mrna. Mol. Carcinog. 2010, 49, 215–223. [Google Scholar] [CrossRef]
- Mueller, C.L.; Porter, S.E.; Hoffman, M.G.; Jaehning, J.A. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 2004, 14, 447–456. [Google Scholar] [CrossRef]
- Pokholok, D.K.; Hannett, N.M.; Young, R.A. Exchange of rna polymerase ii initiation and elongation factors during gene expression in vivo. Mol. Cell 2002, 9, 799–809. [Google Scholar] [CrossRef]
- Verdelli, C.; Corbetta, S. Epigenetic alterations in parathyroid cancers. Int. J. Mol. Sci. 2017, 18, 310. [Google Scholar] [CrossRef]
- Lindberg, D.; Akerstrom, G.; Westin, G. Evaluation of cdkn2c/p18, cdkn1b/p27 and cdkn2b/p15 mrna expression, and cpg methylation status in sporadic and men1-associated pancreatic endocrine tumours. Clin. Endocrinol. 2008, 68, 271–277. [Google Scholar] [CrossRef]
- Yokoyama, A.; Somervaille, T.C.P.; Smith, K.S.; Rozenblatt-Rosen, O.; Meyerson, M.; Cleary, M.L. The menin tumor suppressor protein is an essential oncogenic cofactor for mll-associated leukemogenesis. Cell 2005, 123, 207–218. [Google Scholar] [CrossRef]
- Wu, X.; Hua, X. Menin, histone h3 methyltransferases, and regulation of cell proliferation: Current knowledge and perspective. Curr. Mol. Med. 2008, 8, 805–815. [Google Scholar] [CrossRef]
- Karnik, S.K.; Hughes, C.M.; Gu, X.Y.; Rozenblatt-Rosen, O.; McLean, G.W.; Xiong, Y.; Meyerson, M.; Kim, S.K. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27(kip1) and p18(ink4c). Proc. Natl. Acad. Sci. USA 2005, 102, 14659–14664. [Google Scholar] [CrossRef] [PubMed]
- Verdelli, C.; Forno, I.; Vaira, V.; Corbetta, S. Epigenetic alterations in human parathyroid tumors. Endocrine 2015, 49, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Verdelli, C.; Corbetta, S. Epigenetics of human parathyroid tumors. Int. J. Endocr. Oncol. 2017, 4, 103–111. [Google Scholar] [CrossRef]
- Chrun, E.S.; Modolo, F.; Daniel, F.I. Histone modifications: A review about the presence of this epigenetic phenomenon in carcinogenesis. Pathol. Res. Pract. 2017, 213, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Naveh-Many, T.; Volovelsky, O. Parathyroid cell proliferation in secondary hyperparathyroidism of chronic kidney disease. Int. J. Mol. Sci. 2020, 21, 4332. [Google Scholar] [CrossRef] [PubMed]
- Svedlund, J.; Edblom, S.K.; Marquez, V.E.; Akerstrom, G.; Bjorklund, P.; Westin, G. Hypermethylated in cancer 1 (hic1), a tumor suppressor gene epigenetically deregulated in hyperparathyroid tumors by histone h3 lysine modification. J. Clin. Endocrinol. Metab. 2012, 97, E1307–E1315. [Google Scholar] [CrossRef]
- Cetani, F.; Pardi, E.; Marcocci, C. Parathyroid carcinoma: A clinical and genetic perspective. Minerva Endocrinol. 2018, 43, 144–155. [Google Scholar] [CrossRef]
- Fleuriel, C.; Touka, M.; Boulay, G.; Guerardel, C.; Rood, B.R.; Leprince, D. Hic1 (hypermethylated in cancer 1) epigenetic silencing in tumors. Int. J. Biochem. Cell Biol. 2009, 41, 26–33. [Google Scholar] [CrossRef]
- Jenal, M.; Britschgi, C.; Fey, M.F.; Tschan, M.P. Inactivation of the hypermethylated in cancer 1 tumour suppressor—Not just a question of promoter hypermethylation? Swiss Med. Wkly. 2011, 141, 28–33. [Google Scholar] [CrossRef]
- Westin, G. Molecular genetics and epigenetics of nonfamilial (sporadic) parathyroid tumours. J. Intern. Med. 2016, 280, 551–558. [Google Scholar] [CrossRef]
- Svedlund, J.; Barazeghi, E.; Stalberg, P.; Hellman, P.; Akerstrom, G.; Bjorklund, P.; Westin, G. The histone methyltransferase ezh2, an oncogene common to benign and malignant parathyroid tumors. Endocr.-Relat. Cancer 2014, 21, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Silva-Figueroa, A.M.; Perrier, N.D. Epigenetic processes in sporadic parathyroid neoplasms. Mol. Cell. Endocrinol. 2018, 469, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Bhadada, S.K.; Dahiya, D.; Arya, A.K.; Saikia, U.N.; Sachdeva, N.; Kaur, J.; Brandi, M.L.; Rao, S.D. Reduced calcium sensing receptor (casr) expression is epigenetically deregulated in parathyroid adenomas. J. Clin. Endocrinol. Metab. 2020, 105, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Brewer, K.; Costa-Guda, J.; Arnold, A. Molecular genetic insights into sporadic primary hyperparathyroidism. Endocr.-Relat. Cancer 2019, 26, R53–R72. [Google Scholar] [CrossRef]
- Chiariello, A.M.; Bianco, S.; Esposito, A.; Fiorillo, L.; Conte, M.; Irani, E.; Musella, F.; Abraham, A.; Prisco, A.; Nicodemi, M. Physical mechanisms of chromatin spatial organization. Febs J. 2022, 289, 1180–1190. [Google Scholar] [CrossRef]
- Feng, Y.L.; Liu, X.G.; Pauklin, S. 3d chromatin architecture and epigenetic regulation in cancer stem cells. Protein Cell 2021, 12, 440–454. [Google Scholar] [CrossRef]
- Mangelinck, A.; Mann, C. Dna methylation and histone variants in aging and cancer. Int. Rev. Cell Mol. Biol. 2021, 364, 1–110. [Google Scholar]
- Qi, Y.; Zhang, B. Predicting three-dimensional genome organization with chromatin states. PLoS Comput. Biol. 2019, 15, e1007024. [Google Scholar] [CrossRef]
- Johnstone, S.E.; Reyes, A.; Qi, Y.; Adriaens, C.; Hegazi, E.; Pelka, K.; Chen, J.H.; Zou, L.S.; Drier, Y.; Hecht, V.; et al. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell 2020, 182, 1474–1489.e23. [Google Scholar] [CrossRef]
- Fang, S.H.; Guidroz, J.A.; O’Malley, Y.; Lal, G.; Sugg, S.L.; Howe, J.R.; Jensen, C.S.; Weigel, R.J. Expansion of a cell population expressing stem cell markers in parathyroid glands from patients with hyperparathyroidism. Ann. Surg. 2010, 251, 107–113. [Google Scholar] [CrossRef]
- Hoffmeyer, K.; Raggioli, A.; Rudloff, S.; Anton, R.; Hierholzer, A.; del Valle, I.; Hein, K.; Vogt, R.; Kemler, R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012, 336, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.B.; Lim, J.J.; Chow, E.K.H. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Castilho, R.M.; Squarize, C.H.; Almeida, L.O. Epigenetic modifications and head and neck cancer: Implications for tumor progression and resistance to therapy. Int. J. Mol. Sci. 2017, 18, 1506. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Wittekind, C.; Tannapfel, A. Genetic and epigenetic alterations of 9p21 gene products in benign and malignant tumors of the head and neck. Pathol. Res. Pract. 2003, 199, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.L.; Hormaeche, I.; Licht, J.D. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene 2007, 26, 6697–6714. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, T.; Bosserhoff, A.K. Epigenetic events in malignant melanoma. Pigment. Cell Res. 2007, 20, 92–111. [Google Scholar] [CrossRef]
- Gazdzicka, J.; Golabek, K.; Strzelczyk, J.K.; Ostrowska, Z. Epigenetic modifications in head and neck cancer. Biochem. Genet. 2020, 58, 213–244. [Google Scholar] [CrossRef]
- Furlan, C.; Polesel, J.; Barzan, L.; Franchin, G.; Sulfaro, S.; Romeo, S.; Colizzi, F.; Rizzo, A.; Baggio, V.; Giacomarra, V.; et al. Prognostic significance of line-1 hypomethylation in oropharyngeal squamous cell carcinoma. Clin. Epigenetics 2017, 9. [Google Scholar] [CrossRef]
- Allameh, A.; Moazeni-Roodi, A.; Harirchi, I.; Ravanshad, M.; Motiee-Langroudi, M.; Garajei, A.; Hamidavi, A.; Mesbah-Namin, S.A. Promoter dna methylation and mrna expression level of p16 gene in oral squamous cell carcinoma: Correlation with clinicopathological characteristics. Pathol. Oncol. Res. 2019, 25, 1535–1543. [Google Scholar] [CrossRef]
- Sushma, P.S.; Jamil, K.; Kumar, P.U.; Satyanarayana, U.; Ramakrishna, M.; Triveni, B. Pten and p16 genes as epigenetic biomarkers in oral squamous cell carcinoma (oscc): A study on south indian population. Tumor Biol. 2016, 37, 7625–7632. [Google Scholar] [CrossRef]
- Choudhury, J.H.; Ghosh, S.K. Promoter hypermethylation profiling identifies subtypes of head and neck cancer with distinct viral, environmental, genetic and survival characteristics. PLoS ONE 2015, 10, e0129808. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.H.; Wang, H.D.; Zhong, Z.H. Associations of rassf1a, rar, and cdh1 promoter hypermethylation with oral cancer risk: A prisma-compliant meta-analysis. Medicine 2018, 97, e9971. [Google Scholar] [CrossRef] [PubMed]
- Prystowsky, M.B.; Adomako, A.; Smith, R.V.; Kawachi, N.; McKimpson, W.; Atadja, P.; Chen, Q.; Schlecht, N.F.; Parish, J.L.; Childs, G.; et al. The histone deacetylase inhibitor lbh589 inhibits expression of mitotic genes causing g2/m arrest and cell death in head and neck squamous cell carcinoma cell lines. J. Pathol. 2009, 218, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Uzawa, K.; Miyamoto, I.; Kimura, Y.; Kasama, H.; Ishige, S.; Yamamoto, A.; Saito, Y.; Shimizu, T.; Tanzawa, H. Inactivation of dermatopontin via histone deacetylation in human oral cancer. J. Oral Maxillofac. Surg. Med. Pathol. 2017, 29, 400–404. [Google Scholar] [CrossRef]
- Rosato, R.R.; Almenara, J.A.; Grant, S. The histone deacetylase inhibitor ms-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21(cip1/waf1). Cancer Res. 2003, 63, 3637–3645. [Google Scholar]
- Giudice, F.S.; Pinto, D.S.; Nor, J.E.; Squarize, C.H.; Castilho, R.M. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE 2013, 8, e58672. [Google Scholar] [CrossRef]
- Webber, L.P.; Wagner, V.P.; Curra, M.; Vargas, P.A.; Meurer, L.; Carrard, V.C.; Squarize, C.H.; Castilho, R.M.; Martins, M.D. Hypoacetylation of acetyl-histone h3 (h3k9ac) as marker of poor prognosis in oral cancer. Histopathology 2017, 71, 278–286. [Google Scholar] [CrossRef]
- Almeida, L.O.; Guimaraes, D.M.; Martins, M.D.; Martins, M.A.T.; Warner, K.A.; Nor, J.E.; Castilho, R.M.; Squarize, C.H. Unlocking the chromatin of adenoid cystic carcinomas using hdac inhibitors sensitize cancer stem cells to cisplatin and induces tumor senescence. Stem Cell Res. 2017, 21, 94–105. [Google Scholar] [CrossRef]
- Rodriguez-Rodero, S.; Delgado-Alvarez, E.; Fernandez, A.F.; Fernandez-Morera, J.L.; Menendez-Torre, E.; Fraga, M.F. Epigenetic alterations in endocrine-related cancer. Endocr.-Relat. Cancer 2014, 21, R319–R330. [Google Scholar] [CrossRef]
- Sponziello, M.; Durante, C.; Boichard, A.; Dima, M.; Puppin, C.; Verrienti, A.; Tamburrano, G.; di Rocco, G.; Redler, A.; Lacroix, L.; et al. Epigenetic-related gene expression profile in medullary thyroid cancer revealed the overexpression of the histone methyltransferases ezh2 and smyd3 in aggressive tumours. Mol. Cell. Endocrinol. 2014, 392, 8–13. [Google Scholar] [CrossRef]
- Puppin, C.; Passon, N.; Lavarone, E.; di Loreto, C.; Frasca, F.; Vella, V.; Vigneri, R.; Damante, G. Levels of histone acetylation in thyroid tumors. Biochem. Biophys. Res. Commun. 2011, 411, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Watanabe, M.; Suzuki, M.; Toyota, M.; Sekita, N.; Hirokawa, Y.; Mizokami, A.; Ito, H.; Yatani, R.; Shiraishi, T. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab. Investig. 2000, 80, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Fialova, B.; Trtkova, K.S.; Paskova, L.; Langova, K.; Kolar, Z. Effect of histone deacetylase and dna methyltransferase inhibitors on the expression of the androgen receptor gene in androgen-independent prostate cancer cell lines. Oncol. Rep. 2013, 29, 2039–2045. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, F.Q.; Ye, B.; Hong, L.S.; Xu, H.D.; Fishbein, M.C. Epigenetic modifications of histone h4 in lung neuroendocrine tumors. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at lys16 and trimethylation at lys20 of histone h4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H. Hdac and hdac inhibitor: From cancer to cardiovascular diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef]
- Hu, Z.C.; Rong, Y.C.; Li, S.T.; Qu, S.Q.; Huang, S.B. Upregulated histone deacetylase 6 associates with malignant progression of melanoma and predicts the prognosis of patients. Cancer Manag. Res. 2020, 12, 12993–13001. [Google Scholar] [CrossRef]
- Mastoraki, A.; Schizas, D.; Vlachou, P.; Melissaridou, N.M.; Charalampakis, N.; Fioretzaki, R.; Kole, C.; Savvidou, O.; Vassiliu, P.; Pikoulis, E. Assessment of synergistic contribution of histone deacetylases in prognosis and therapeutic management of sarcoma. Mol. Diagn. Ther. 2020, 24, 557–569. [Google Scholar] [CrossRef]
- Cao, L.L.; Song, X.X.; Pei, L.; Liu, L.H.; Wang, H.; Jia, M. Histone deacetylase hdac1 expression correlates with the progression and prognosis of lung cancer a meta-analysis. Medicine 2017, 96, e7663. [Google Scholar] [CrossRef]
- Chervona, Y.; Costa, M. Histone modifications and cancer: Biomarkers of prognosis? Am. J. Cancer Res. 2012, 2, 589–597. [Google Scholar]
- Giaginis, C.; Alexandrou, P.; Delladetsima, I.; Giannopoulou, I.; Patsouris, E.; Theocharis, S. Clinical significance of histone deacetylase (hdac)-1, hdac-2, hdac-4, and hdac-6 expression in human malignant and benign thyroid lesions. Tumor Biol. 2014, 35, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Movafagh, S.; Munson, A. Histone deacetylase inhibitors in cancer prevention and therapy. Epigenetics Cancer Prev. 2019, 8, 75–105. [Google Scholar] [CrossRef]
- Ho, E.; Clarke, J.D.; Dashwood, R.H. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J. Nutr. 2009, 139, 2393–2396. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Lin, Y.; Tseng, R.; Shun, C.; Wu, M. Prevention of colitis and colitis-associated colorectal cancer by a novel polypharmacological histone deacetylase inhibitor. Clin. Cancer Res. 2018, 24, 499. [Google Scholar] [CrossRef]
- Kalin, J.H.; Eroglu, A.; Liu, H.; Holtzclaw, W.D.; Leigh, I.; Proby, C.M.; Fahey, J.W.; Cole, P.A.; Dinkova-Kostova, A.T. Investigation into the use of histone deacetylase inhibitor ms-275 as a topical agent for the prevention and treatment of cutaneous squamous cell carcinoma in an skh-1 hairless mouse model. PLoS ONE 2019, 14, e0213095. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conti de Freitas, L.C.; Castilho, R.M.; Squarize, C.H. Histone Modification on Parathyroid Tumors: A Review of Epigenetics. Int. J. Mol. Sci. 2022, 23, 5378. https://doi.org/10.3390/ijms23105378
Conti de Freitas LC, Castilho RM, Squarize CH. Histone Modification on Parathyroid Tumors: A Review of Epigenetics. International Journal of Molecular Sciences. 2022; 23(10):5378. https://doi.org/10.3390/ijms23105378
Chicago/Turabian StyleConti de Freitas, Luiz C., Rogerio M. Castilho, and Cristiane H. Squarize. 2022. "Histone Modification on Parathyroid Tumors: A Review of Epigenetics" International Journal of Molecular Sciences 23, no. 10: 5378. https://doi.org/10.3390/ijms23105378
APA StyleConti de Freitas, L. C., Castilho, R. M., & Squarize, C. H. (2022). Histone Modification on Parathyroid Tumors: A Review of Epigenetics. International Journal of Molecular Sciences, 23(10), 5378. https://doi.org/10.3390/ijms23105378





