Identification and Characterization of Circular RNAs in Brassica rapa in Response to Plasmodiophora brassicae
Abstract
:1. Introduction
2. Results
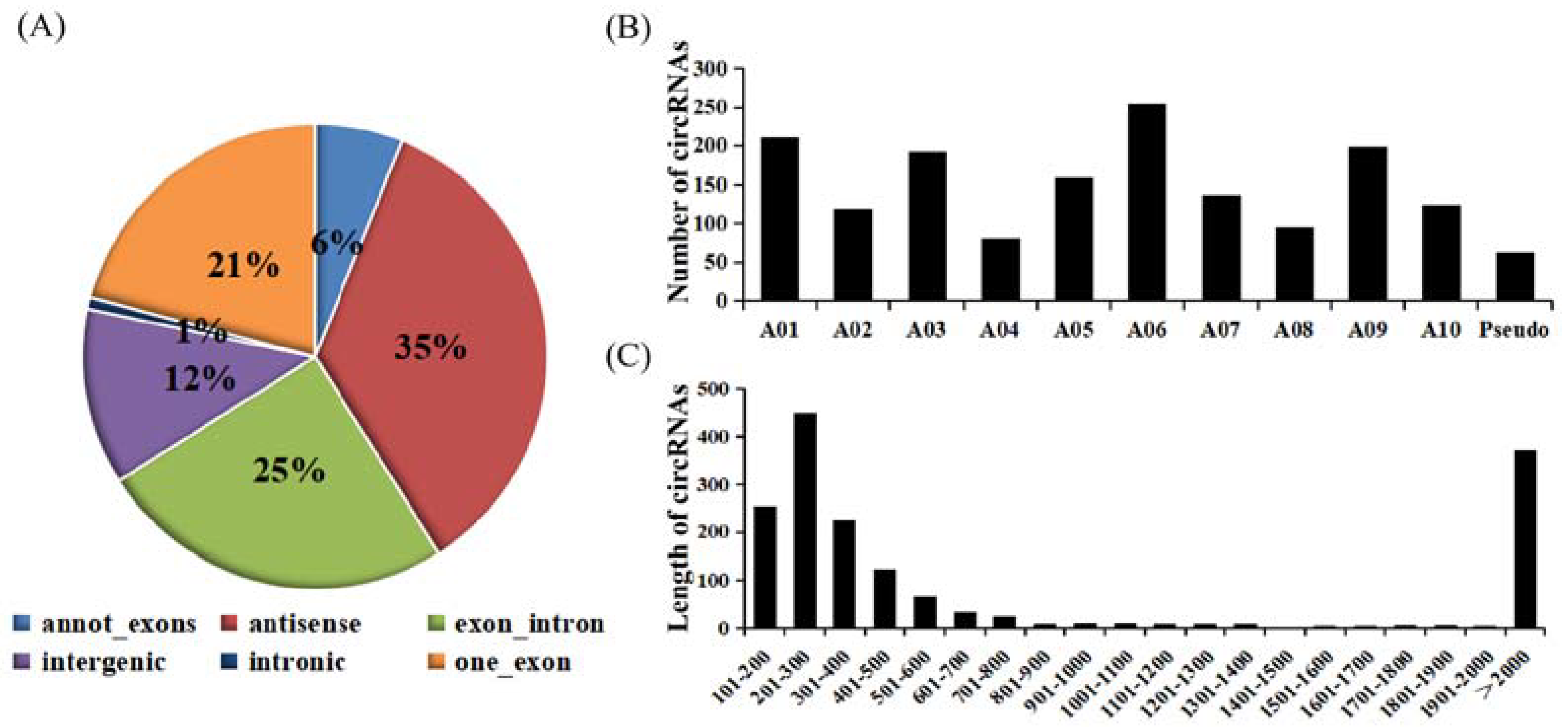
2.1. Identification and Validation of CircRNAs
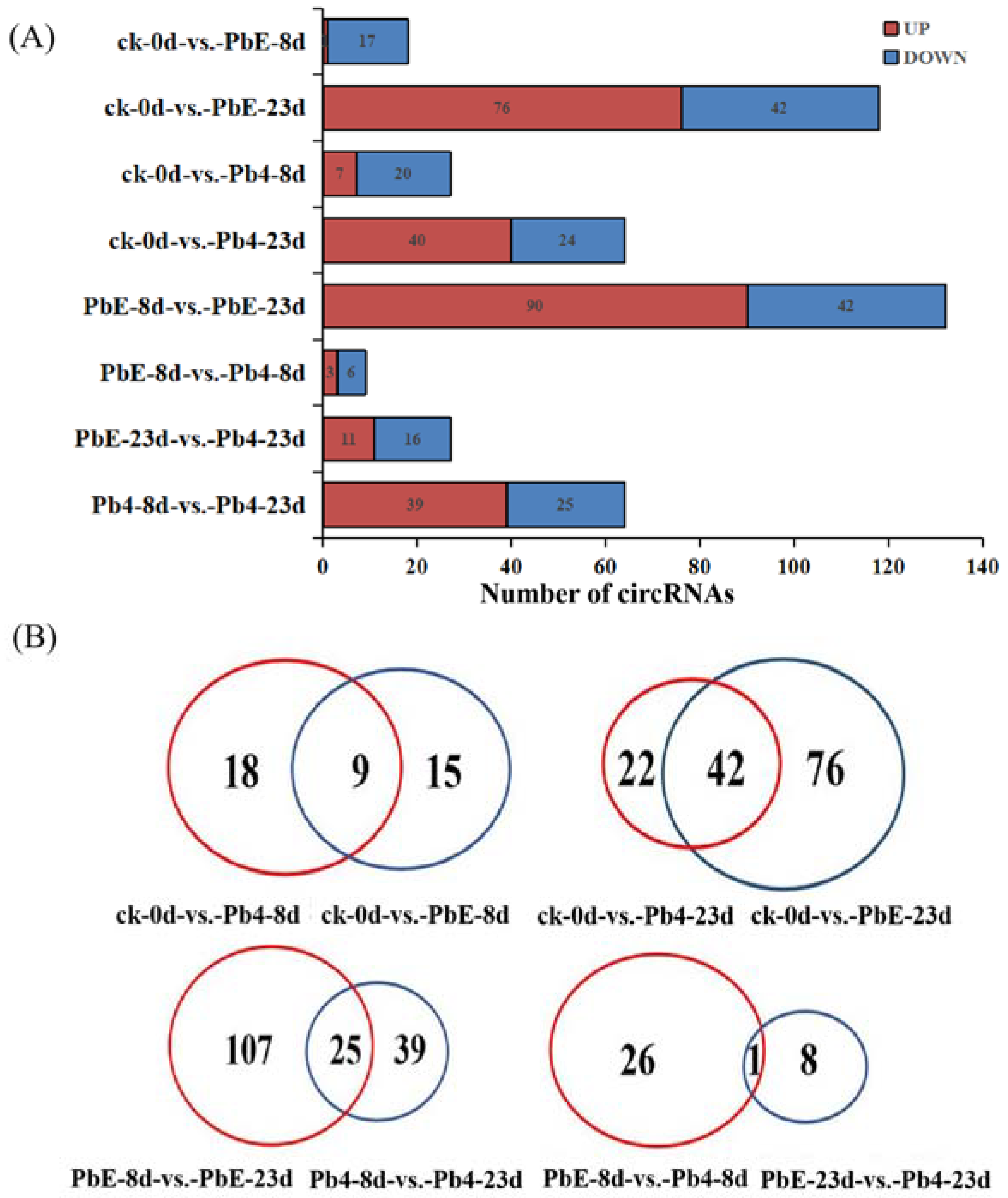
2.2. CircRNA Analysis in Response to P. brassicae
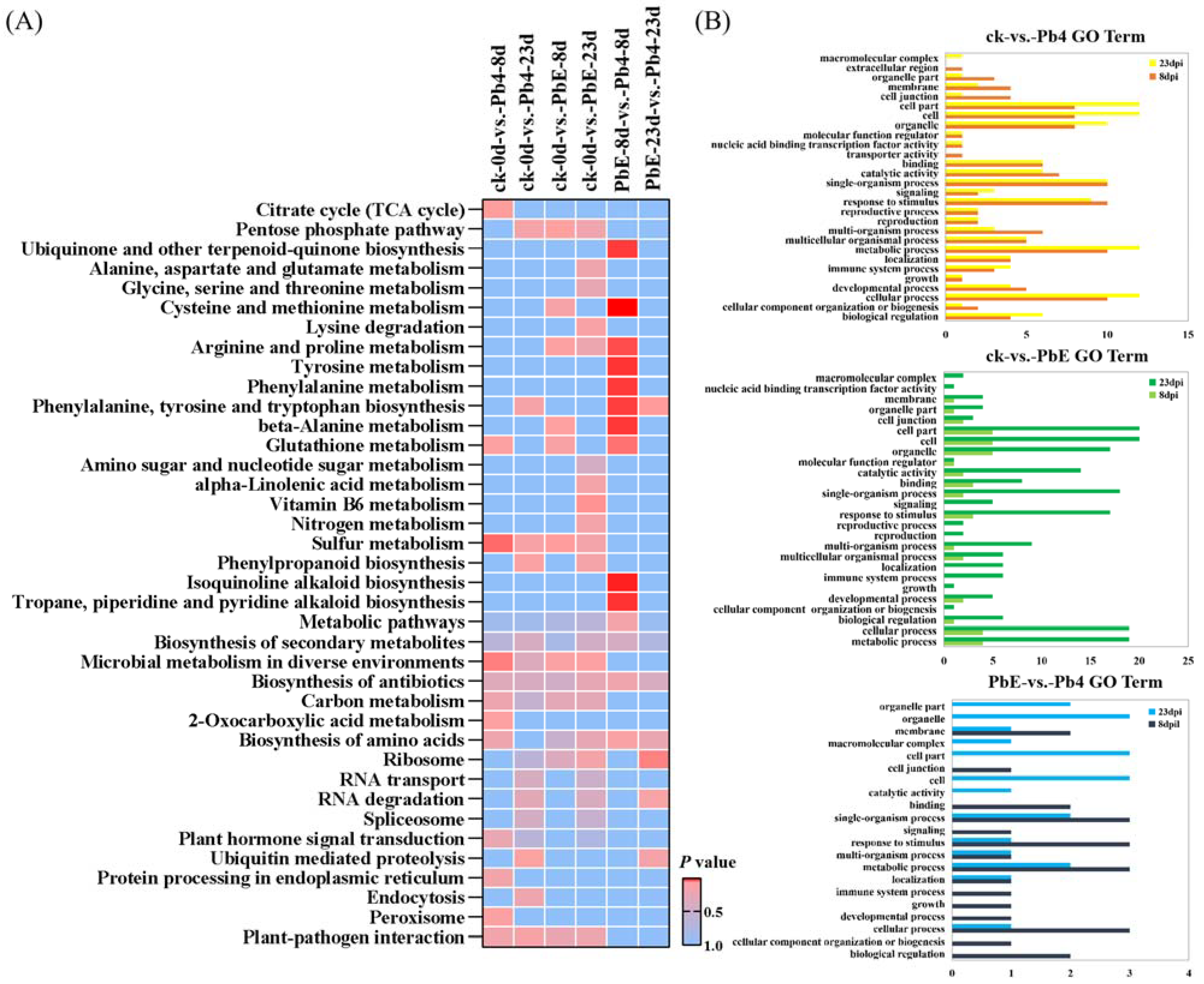
2.3. Identification of CircRNA Parental Genes
2.4. CircRNA Parental Gene Function Analysis
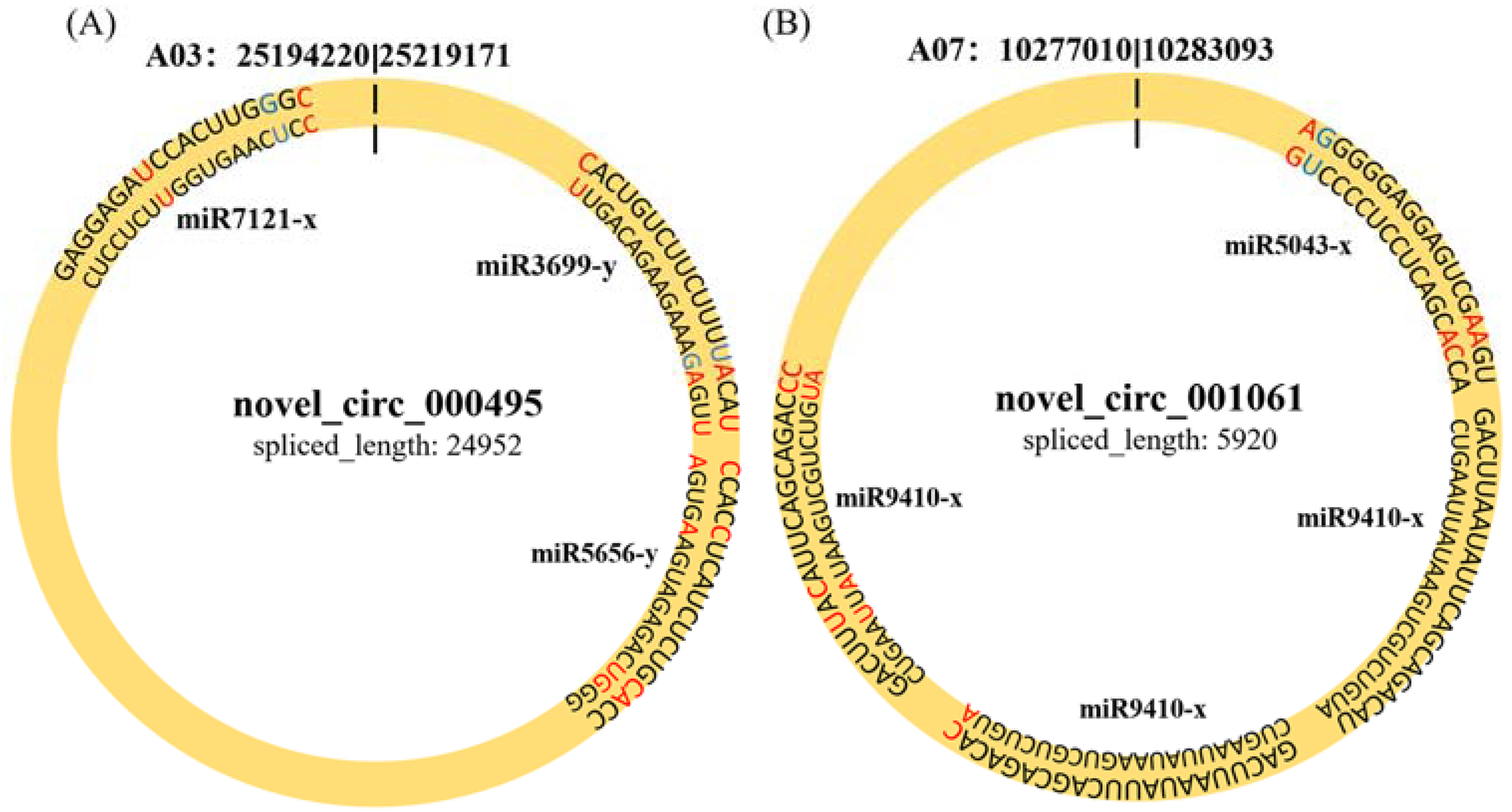
2.5. CircRNAs Acting as MiRNA Sponges
2.6. Construction of the CircRNA–MiRNA–MRNA Network
3. Discussion
3.1. CircRNA Sequencing Explains the Mechanisms of Clubroot Resistance in B. rapa
3.2. CircRNAs Act as MiRNA Sponges Affecting the Function of Target Genes
3.3. CeRNA Networks Could Provide New Insights into the Regulatory Roles of NcRNAs during P. brassicae Infection
4. Materials and Methods
4.1. Plant Materials and Inoculation with P. brassicae
4.2. RNA and DNA Isolation, Library Preparation and Sequencing
4.3. Mapping to the Reference Genome and Transcriptome Assembly
4.4. CircRNA Detection and Functional Annotation
4.5. CircRNA MiRNA-Binding Site Analysis
4.6. Integrated Analysis of CircRNAs–MiRNAs–MRNAs
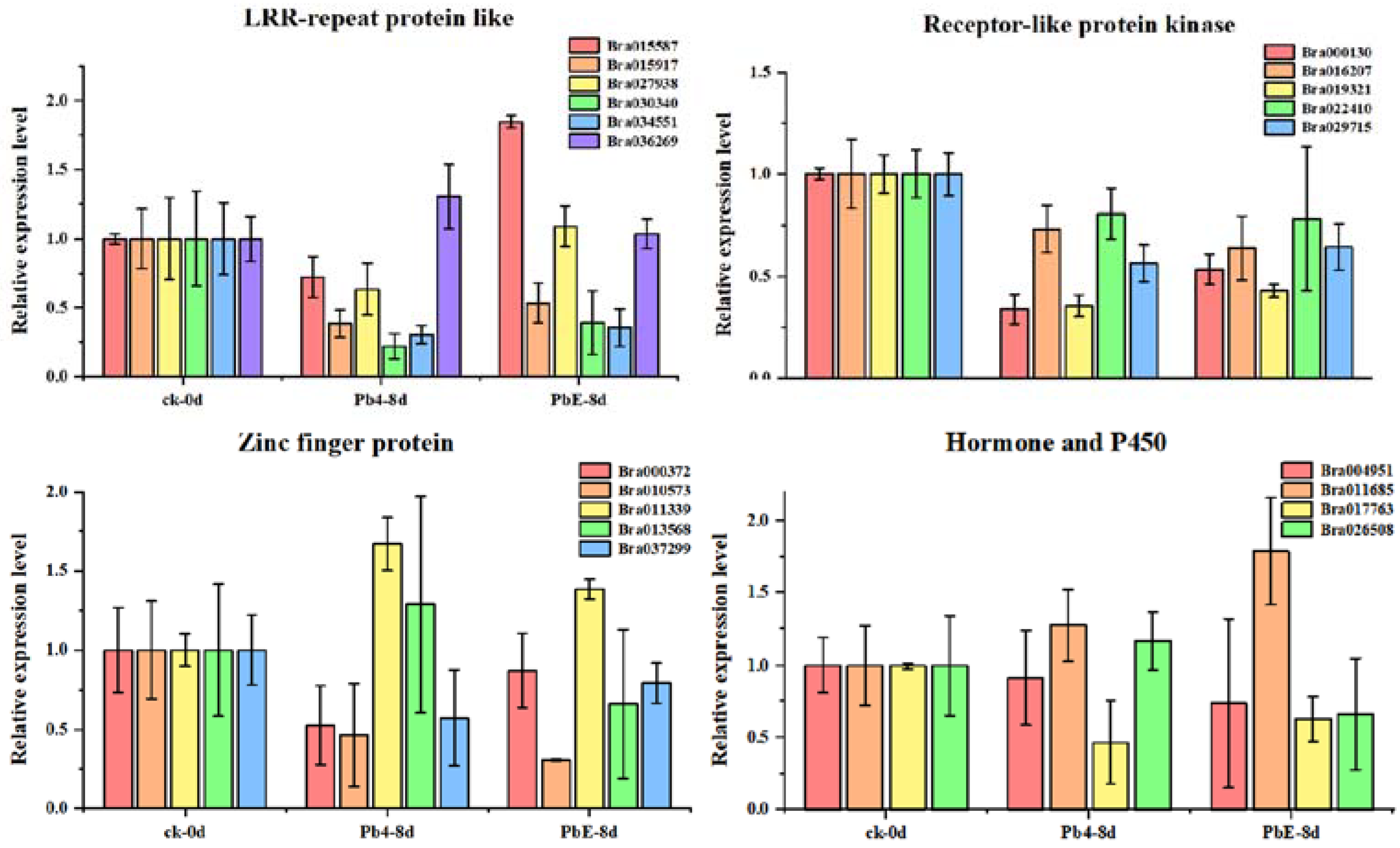
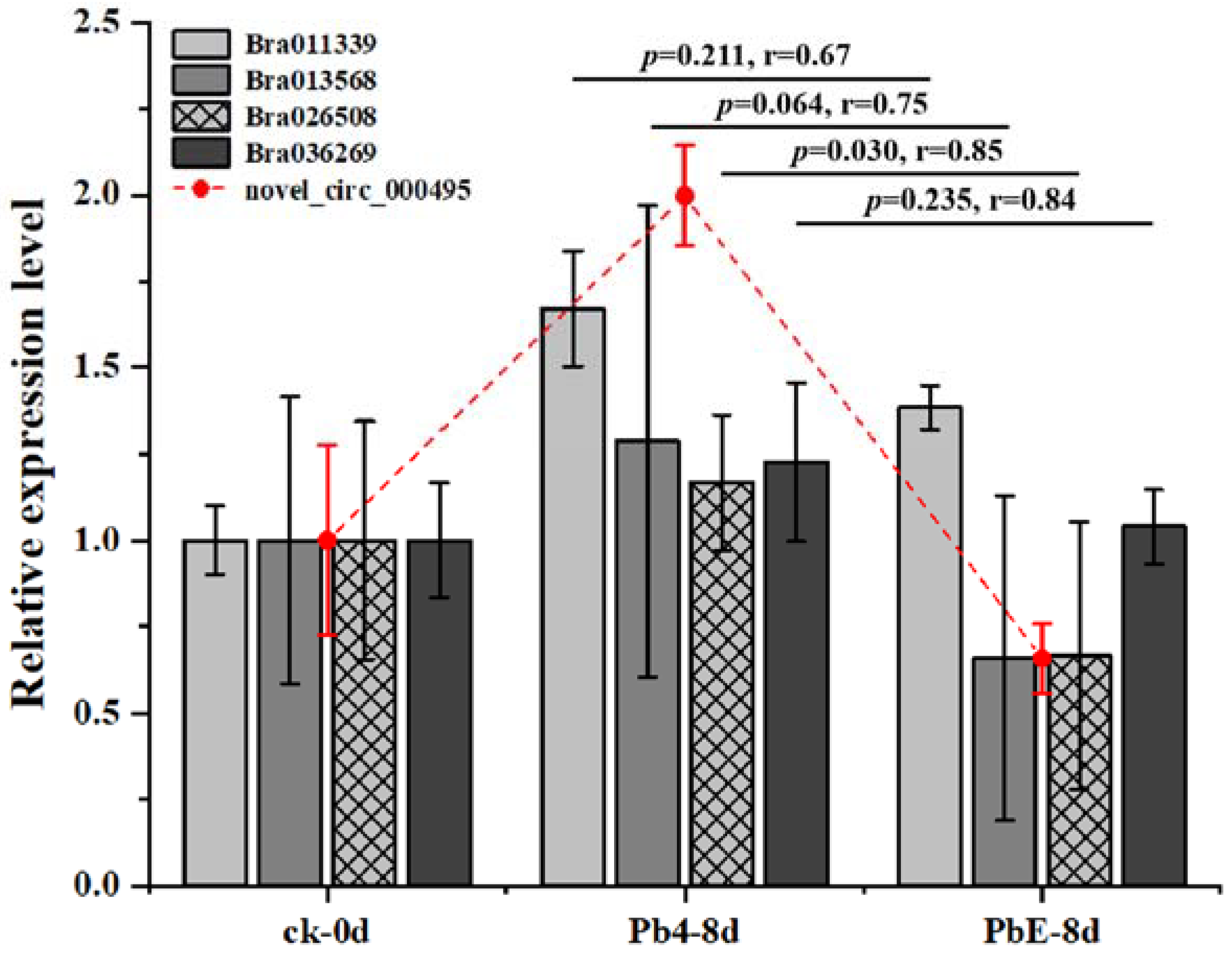
4.7. CircRNA Validation and Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and Their Roles in Stress Response in Plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Cui, J.; Wang, L.; Zhu, Y.; Lu, Z.; Jin, B.; Chen, G.; Cui, J.; Wang, L.; Zhu, Y.; et al. Genome-Wide Identification of Circular RNAs in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1678. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Gao, L.; Zhu, B.; Luo, Y.; Deng, Z.; Zuo, J. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol. Plant. 2017, 161, 311–321. [Google Scholar] [CrossRef]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Yu, X.Q.; Ottosen, C.-O.; Zhao, T.M. High Throughput Sequencing of circRNAs in Tomato Leaves Responding to Multiple Stresses of Drought and Heat. Hortic. Plant J. 2019, 6, 39–43. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Burd, C.E.; Jeck, W.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N. Expression of Linear and Novel Circular Forms of an INK4/ARF-Associated Non-Coding RNA Correlates with Atherosclerosis Risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Yu, X.Q.; Xu, L.P.; Wang, Y.L.; Zhao, L.P.; Zhao, T.M.; Yu, W.G. Genome-wide identification of circular RNAs in tomato seeds in response to high temperature. Biol. Plant. 2019, 63, 97–103. [Google Scholar] [CrossRef]
- Fan, J.; Quan, W.; Li, G.-B.; Hu, X.-H.; Wang, Q.; Wang, H.; Li, X.-P.; Luo, X.; Feng, Q.; Hu, Z.-J.; et al. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiol. 2020, 182, 272–286. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and Functional Characterization of Tomato CircRNAs Derived from Genes Involved in Fruit Pigment Accumulation. Sci. Rep. 2017, 7, 8594. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.; Yu, J.; Hou, Y.; Li, F.; Zhou, Q.; Wei, C.; Bennetzen, J.L. Circular RNA architecture and differentiation during leaf bud to young leaf development in tea (Camellia sinensis). Planta 2018, 248, 1417–1429. [Google Scholar] [CrossRef]
- Ghorbani, A.; Izadpanah, K.; Peters, J.R.; Dietzgen, R.G.; Mitter, N. Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci. 2018, 274, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Characterization and Cloning of Grape Circular RNAs Identified the Cold Resistance-Related Vv-circATS1. Plant Physiol. 2019, 180, 966–985. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Q.; Zhu, B.; Luo, Y.; Gao, L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016, 479, 132–138. [Google Scholar] [CrossRef]
- Darbani, B.; Noeparvar, S.; Borg, S. Identification of Circular RNAs from the Parental Genes Involved in Multiple Aspects of Cellular Metabolism in Barley. Front. Plant Sci. 2016, 7, 776. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019, 98, 697–713. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Li, D.; Li, L.; Zhang, Q.; Wang, S.; Huang, H. Identification of Circular RNAs in Kiwifruit and Their Species-Specific Response to Bacterial Canker Pathogen Invasion. Front. Plant Sci. 2017, 8, 413. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Zhu, Y.; Zhao, J.; Fang, Z.; Wang, S.; Yin, J.; Chu, Z.; Ma, D. Transcriptome-Wide Identification and Characterization of Potato Circular RNAs in Response to Pectobacterium carotovorum Subspecies brasiliense Infection. Int. J. Mol. Sci. 2017, 19, 71. [Google Scholar] [CrossRef] [Green Version]
- Xiang, L.; Cai, C.; Cheng, J.; Wang, L.; Wu, C.; Shi, Y.; Luo, J.; He, L.; Deng, Y.; Zhang, X.; et al. Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq. PeerJ 2018, 6, e4500. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, A.; Liang, F.; Yao, X.; Wang, Y.; Liu, X.; Zhang, Y.; Dalelhan, J.; Zhang, B.; Qin, M.; et al. Screening of clubroot-resistant varieties and transfer of clubroot resistance genes to Brassica napus using distant hybridization. Breed. Sci. 2018, 68, 258–267. [Google Scholar] [CrossRef] [Green Version]
- Wallenhammar, A.-C.; Wallenhammar, A. Prevalence of Plasmodiophora brassicae in a spring oilseed rape growing area in central Sweden and factors influencing soil infestation levels. Plant Pathol. 1996, 45, 710–719. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone Crosstalk in Plant Disease and Defense: More than Just JASMONATE-SALICYLATE Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Pinot, F.; Skrabs, M.; Compagnon, V.; Salaün, J.P.; Benveniste, I.; Schreiber, L.; Durst, F. omega-Hydroxylation of epoxy- and hydroxy-fatty acids by CYP94A1: Possible involvement in plant defence. Biochem. Soc. Trans. 2000, 28, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Chételat, A.; Caldelari, D.; Farmer, E.E. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 1999, 11, 485–494. [Google Scholar] [CrossRef]
- Summanwar, A.; Basu, U.; Rahman, H.; Kav, N. Identification of lncRNAs Responsive to Infection by Plasmodiophora brassicae in Clubroot-Susceptible and -Resistant Brassica napus Lines Carrying Resistance Introgressed from Rutabaga. Mol. Plant-Microbe Interact. 2019, 32, 1360–1377. [Google Scholar] [CrossRef]
- Verma, S.S.; Rahman, M.H.; Deyholos, M.K.; Basu, U.; Kav, N.N.V. Differential Expression of miRNAs in Brassica napus Root following Infection with Plasmodiophora brassicae. PLoS ONE 2014, 9, e86648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016, 7, 12060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Glawischnig, E. Camalexin. Phytochemistry 2007, 68, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Katagiri, F.; Mindrinos, M.; Glazebrook, J. Use of Arabidopsis thaliana defense-related mutants to dissect the plant response to pathogens. Proc. Natl. Acad. Sci. USA 1995, 92, 4189–4196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Levrel, A.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci. 2015, 6, 539. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Ju, H.-W.; Min, J.-H.; Zhang, X.; Kim, S.-H.; Yang, K.-Y.; Kim, C.S. Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 2013, 203–204, 98–106. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.-H.; Bindereif, A. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Szabo, L.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Erratum to: Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2016, 17, 263. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yu, Y.; Zhang, X.; Liu, C.; Ye, C.; Fan, L. PcircRNA_finder: A software for circRNA prediction in plants. Bioinformatics 2016, 32, 3528–3529. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Wang, L.Y.; Li, S.; Xu, M.; Guan, X.Y.; Zhou, B.L. Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors. FEBS Lett. 2017, 591, 3660–3669. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Sun, X.; Liu, Y.; Li, H.; Deng, G.; Lin, H.; Wang, S. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol. Biol. 2018, 96, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.-F.; Zhou, J.-J.; Hu, C.-G.; Zhang, J.-Z. Transcriptome-wide identification and functional prediction of novel and flowering-related circular RNAs from trifoliate orange (Poncirus trifoliata L. Raf.). Planta 2018, 247, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; Zhang, P.Y.; Guo, M.R.; Chen, K.S. Secondary metabolites and plant defence against pathogenic disease. Plant Physiol. J. 2012, 48, 429–434. [Google Scholar] [CrossRef]
- Chevilly, S.; Dolz-Edo, L.; López-Nicolás, J.M.; Morcillo, L.; Vilagrosa, A.; Yenush, L.; Mulet, J.M. Physiological and Molecular Characterization of the Differential Response of Broccoli (Brassica oleracea var. Italica) Cultivars Reveals Limiting Factors for Broccoli Tolerance to Drought Stress. J. Agric. Food Chem. 2021, 69, 10394–10404. [Google Scholar] [CrossRef]
- Chevilly, S.; Dolz-Edo, L.; Morcillo, L.; Vilagrosa, A.; López-Nicolás, J.M.; Yenush, L.; Mulet, J.M. Identification of distinctive physiological and molecular responses to salt stress among tolerant and sensitive cultivars of broccoli (Brassica oleracea var. Italica). BMC Plant Biol. 2021, 21, 488. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a Viral Suppressor of RNA Silencing, Interferes with Arabidopsis Development and miRNA Function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Navarro, L.; Jay, F.; Nomura, K.; He, S.Y.; Voinnet, O. Suppression of the MicroRNA Pathway by Bacterial Effector Proteins. Science 2008, 321, 964–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, C.; Wang, J.; Zhao, J.-H.; Fang, Y.-Y.; He, X.-F.; Guo, H.-S.; Duan, C.-G. A Brassica miRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-F.; Zhang, Y.-C.; Chen, Y.-Q.; Yu, Y. Circular RNAs roll into the regulatory network of plants. Biochem. Biophys. Res. Commun. 2017, 488, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Chen, G.; Shi, T. Identifying and Characterizing the Circular RNAs during the Lifespan of Arabidopsis Leaves. Front. Plant Sci. 2017, 8, 1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of Circular RNAs and Their Targets in Leaves of Triticum aestivum L. under Dehydration Stress. Front. Plant Sci. 2017, 7, 2024. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Chen, Y. Identification of Potentially Functional CircRNA-miRNA-mRNA Regulatory Network in Hepatocellular Carcinoma by Integrated Microarray Analysis. Med Sci. Monit. Basic Res. 2018, 24, 70–78. [Google Scholar] [CrossRef]
- Summanwar, A.; Basu, U.; Kav, N.N.; Rahman, H. Identification of lncRNAs in response to infection by Plasmodiophora brassicae in Brassica napus and development of lncRNA-based SSR markers. Genome 2021, 64, 547–566. [Google Scholar] [CrossRef]
- Field, B.; Osbourn, A.E. Metabolic Diversification—Independent Assembly of Operon-Like Gene Clusters in Different Plants. Science 2008, 320, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.X.; Liang, Y.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Development of a Sinitic Clubroot Differential Set for the Pathotype Classification of Plasmodiophora brassicae. Front. Plant Sci. 2020, 11, 568771. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.Y.; Piao, Z.Y.; Zhan, Z.X.; Zhao, Y.Z.; Pang, W.X.; Li, X.N.; Piao, Z. Transcriptome Arofile of Brassica rapa L. Reveals the Involvement of Jasmonic Acid, Ethylene, and Brassinosteroid Signaling Pathways in Clubroot Resistance. Agronomy 2019, 9, 589. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Sample | Treatment | Replicate |
|---|---|---|
| ck-0d-1 | control | biological replicate 1 |
| ck-0d-2 | control | biological replicate 2 |
| ck-0d-3 | control | biological replicate 3 |
| Pb4-8d-1 | inoculated with Pb4 | biological replicate 1 |
| Pb4-8d-2 | inoculated with Pb4 | biological replicate 2 |
| Pb4-8d-3 | inoculated with Pb4 | biological replicate 3 |
| PbE-8d-1 | inoculated with PbE | biological replicate 1 |
| PbE-8d-2 | inoculated with PbE | biological replicate 2 |
| PbE-8d-3 | inoculated with PbE | biological replicate 3 |
| Pb4-23d-1 | inoculated with Pb4 | biological replicate 1 |
| Pb4-23d-2 | inoculated with Pb4 | biological replicate 2 |
| Pb4-23d-3 | inoculated with Pb4 | biological replicate 3 |
| PbE-23d-1 | inoculated with PbE | biological replicate 1 |
| PbE-23d-2 | inoculated with PbE | biological replicate 2 |
| PbE-23d-3 | inoculated with PbE | biological replicate 3 |
| Group | CircRNA ID | Source Gene ID | log2 (FC) | p Value | Chr | Strand |
|---|---|---|---|---|---|---|
| ck-0d-vs.-Pb4-8d | novel_circ_000064 | Bra013579 | 4.246827502 | 0.004744418 | A01 | − |
| novel_circ_000079 | Bra013579 | 2.893516888 | 0.009588829 | A01 | − | |
| novel_circ_000086 | Bra013579 | −2.84065774 | 0.028360819 | A01 | + | |
| novel_circ_000264 | Bra039746 | −19.12803312 | 0.04701868 | A02 | + | |
| ck-0d-vs.-PbE-8d | novel_circ_000074 | Bra013579 | 19.51979677 | 0.011219066 | A01 | − |
| novel_circ_000086 | Bra013579 | −3.947713015 | 0.012980797 | A01 | + | |
| PbE-8d-vs.-Pb4-8d | novel_circ_001061 | Bra012389 | −4.222569626 | 0.038710775 | A07 | − |
| novel_circ_000495 | Bra019293 | 4.995712702 | 0.001038162 | A03 | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Nwafor, C.C.; Piao, Y.; Li, X.; Zhan, Z.; Piao, Z. Identification and Characterization of Circular RNAs in Brassica rapa in Response to Plasmodiophora brassicae. Int. J. Mol. Sci. 2022, 23, 5369. https://doi.org/10.3390/ijms23105369
Liu H, Nwafor CC, Piao Y, Li X, Zhan Z, Piao Z. Identification and Characterization of Circular RNAs in Brassica rapa in Response to Plasmodiophora brassicae. International Journal of Molecular Sciences. 2022; 23(10):5369. https://doi.org/10.3390/ijms23105369
Chicago/Turabian StyleLiu, Huishan, Chinedu Charles Nwafor, Yinglan Piao, Xiaonan Li, Zongxiang Zhan, and Zhongyun Piao. 2022. "Identification and Characterization of Circular RNAs in Brassica rapa in Response to Plasmodiophora brassicae" International Journal of Molecular Sciences 23, no. 10: 5369. https://doi.org/10.3390/ijms23105369
APA StyleLiu, H., Nwafor, C. C., Piao, Y., Li, X., Zhan, Z., & Piao, Z. (2022). Identification and Characterization of Circular RNAs in Brassica rapa in Response to Plasmodiophora brassicae. International Journal of Molecular Sciences, 23(10), 5369. https://doi.org/10.3390/ijms23105369





