Early Life Stress Alters Expression of Glucocorticoid Stress Response Genes and Trophic Factor Transcripts in the Rodent Basal Ganglia
Abstract
1. Introduction
2. Results
2.1. Expression of Stress-Related Factors in the Substantia Nigra, Ventral Tegmental Area and Dorsal and Ventral Striatum
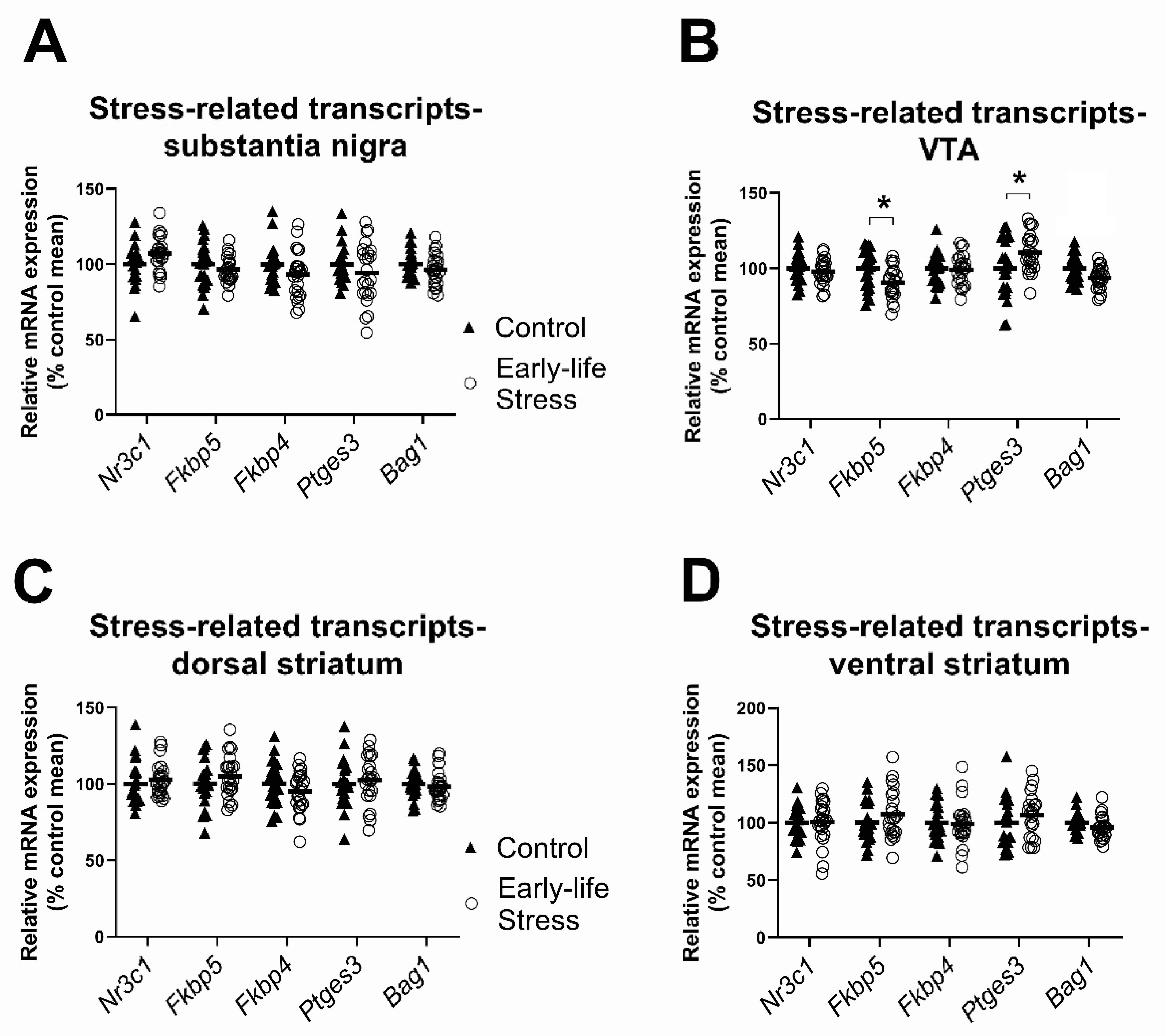
2.1.1. Effects of Early Life Stress on Expression of Stress-Related Transcripts, Seen in Both Females and Males
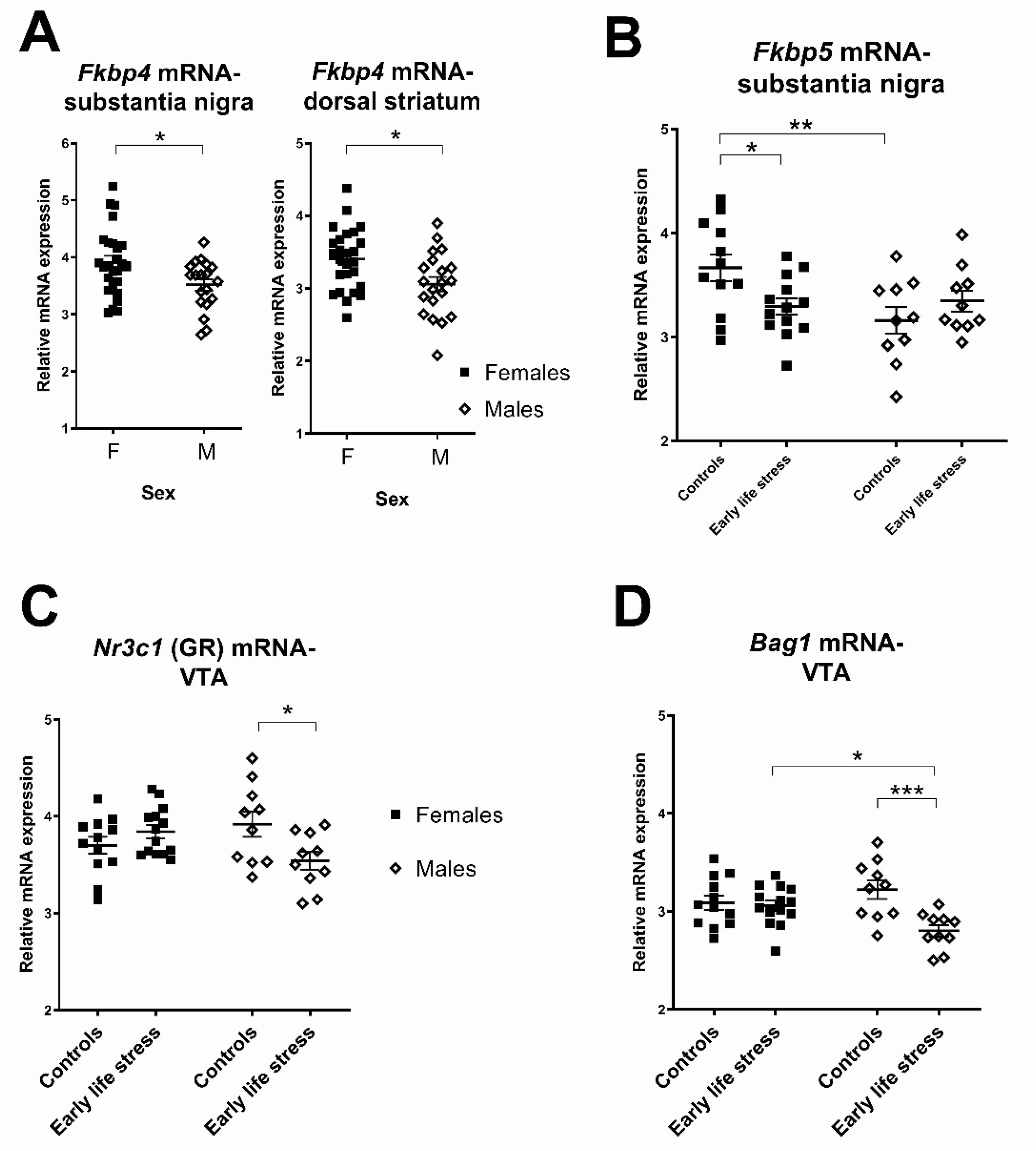
2.1.2. Sex Differences in the Effects of Early Life Stress on Expression of Stress-Related Transcripts
2.2. Expression of Bdnf Transcripts in the Substantia Nigra, Ventral Tegmental Area and Dorsal and Ventral Striatum
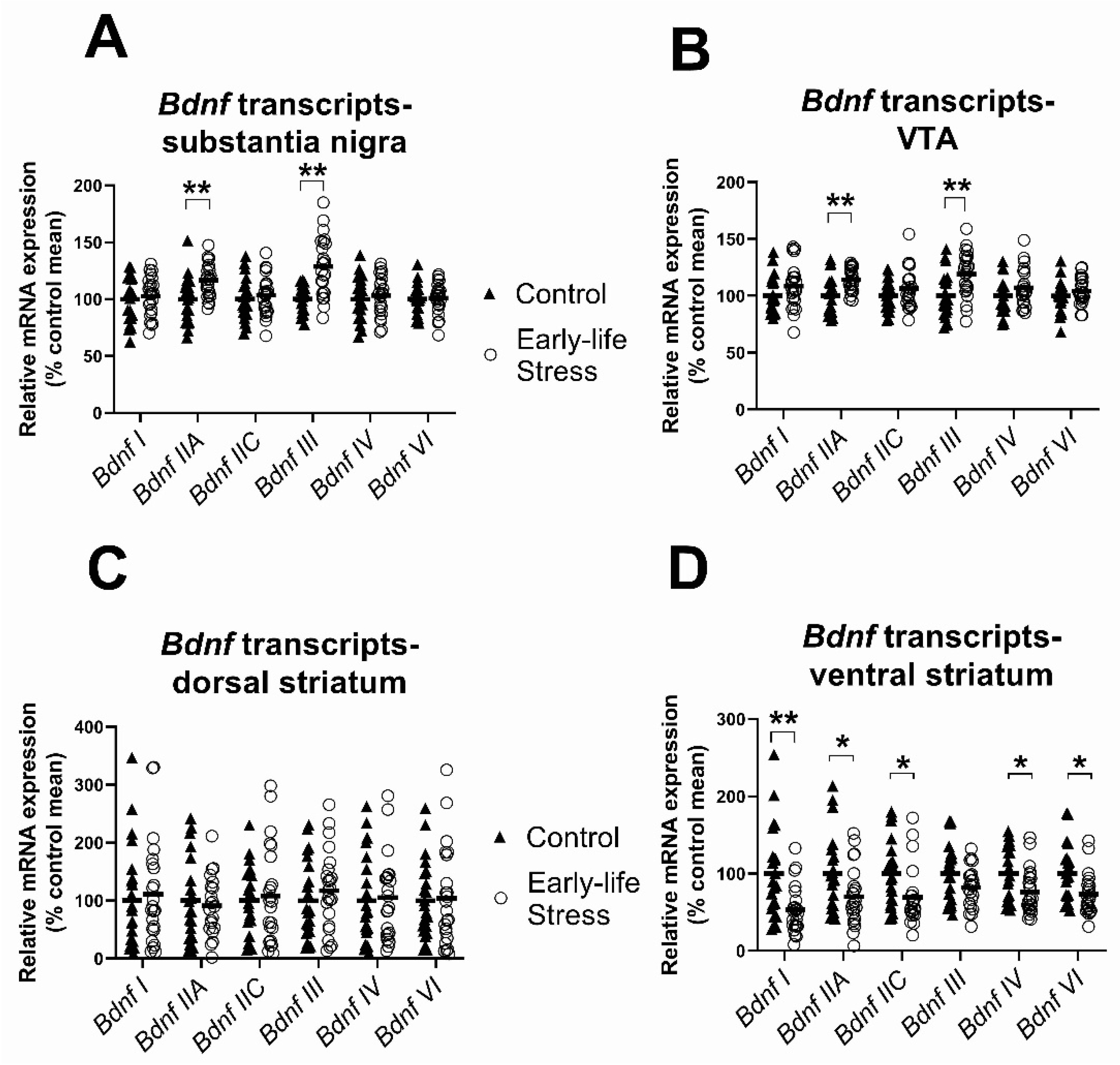
2.2.1. Effects of Early Life Stress on Expression of Bdnf Transcripts, Seen in Both Females and Males
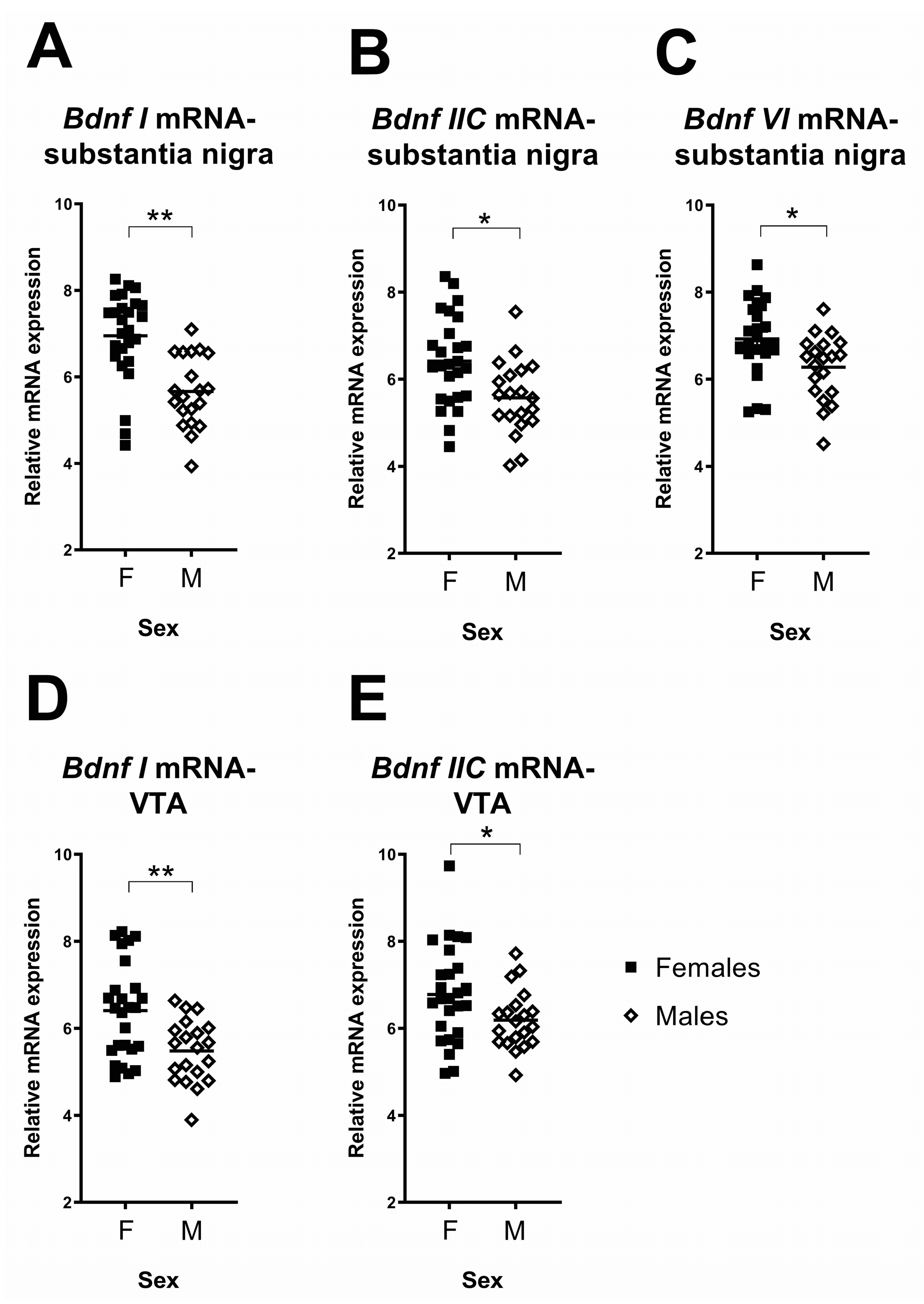
2.2.2. Sex Differences in the Effects of Early Life Stress on Expression of Bdnf Transcripts
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. RNA Extraction
4.3. cDNA Synthesis
4.4. Quantitative Polymerase Chain Reaction (qPCR)
4.5. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pacak, K.; Palkovits, M. Stressor specificity of central neuroendocrine responses: Implications for stress-related disorders. Endocr. Rev. 2001, 22, 502–548. [Google Scholar] [CrossRef]
- Lupien, S.J.; McEwen, B.S.; Gunnar, M.R.; Heim, C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 2009, 10, 434–445. [Google Scholar] [CrossRef]
- Pallarés, M.E.; Monteleone, M.C.; Pastor, V.; Grillo Balboa, J.; Alzamendi, A.; Brocco, M.A.; Antonelli, M.C. Early-Life Stress Reprograms Stress-Coping Abilities in Male and Female Juvenile Rats. Mol. Neurobiol. 2021, 58, 5837–5856. [Google Scholar] [CrossRef]
- Gondora, N.; Pople, C.B.; Tandon, G.; Robinson, M.; Solomon, E.; Beazely, M.A.; Mielke, J.G. Chronic early-life social isolation affects NMDA and TrkB receptor expression in a sex-specific manner. Neurosci. Lett. 2021, 760, 136016. [Google Scholar] [CrossRef]
- McGowan, P.O.; Sasaki, A.; D’Alessio, A.C.; Dymov, S.; Labonte, B.; Szyf, M.; Turecki, G.; Meaney, M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009, 12, 342–348. [Google Scholar] [CrossRef]
- Juszczak, G.R.; Stankiewicz, A.M. Glucocorticoids, genes and brain function. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 82, 136–168. [Google Scholar] [CrossRef]
- O’Connor, T.G.; Ben-Shlomo, Y.; Heron, J.; Golding, J.; Adams, D.; Glover, V. Prenatal anxiety predicts individual differences in cortisol in pre-adolescent children. Biol. Psychiatry 2005, 58, 211–217. [Google Scholar] [CrossRef]
- Gutteling, B.M.; de Weerth, C.; Buitelaar, J.K. Prenatal stress and children’s cortisol reaction to the first day of school. Psychoneuroendocrinology 2005, 30, 541–549. [Google Scholar] [CrossRef]
- Karlamangla, A.S.; Merkin, S.S.; Almeida, D.M.; Friedman, E.M.; Mogle, J.A.; Seeman, T.E. Early-Life Adversity and Dysregulation of Adult Diurnal Cortisol Rhythm. J. Gerontol. B Psychol. Sci. Soc. Sci. 2019, 74, 160–169. [Google Scholar] [CrossRef]
- Khashan, A.S.; Abel, K.M.; McNamee, R.; Pedersen, M.G.; Webb, R.T.; Baker, P.N.; Kenny, L.C.; Mortensen, P.B. Higher risk of offspring schizophrenia following antenatal maternal exposure to severe adverse life events. Arch. Gen. Psychiatry 2008, 65, 146–152. [Google Scholar] [CrossRef]
- Arseneault, L.; Cannon, M.; Fisher, H.L.; Polanczyk, G.; Moffitt, T.E.; Caspi, A. Childhood trauma and children’s emerging psychotic symptoms: A genetically sensitive longitudinal cohort study. Am. J. Psychiatry 2011, 168, 65–72. [Google Scholar] [CrossRef]
- Schreier, A.; Wolke, D.; Thomas, K.; Horwood, J.; Hollis, C.; Gunnell, D.; Lewis, G.; Thompson, A.; Zammit, S.; Duffy, L.; et al. Prospective study of peer victimization in childhood and psychotic symptoms in a nonclinical population at age 12 years. Arch. Gen. Psychiatry 2009, 66, 527–536. [Google Scholar] [CrossRef]
- Heim, C.; Nemeroff, C.B. The role of childhood trauma in the neurobiology of mood and anxiety disorders: Preclinical and clinical studies. Biol. Psychiatry 2001, 49, 1023–1039. [Google Scholar] [CrossRef]
- Hughes, K.; Bellis, M.A.; Hardcastle, K.A.; Sethi, D.; Butchart, A.; Mikton, C.; Jones, L.; Dunne, M.P. The effect of multiple adverse childhood experiences on health: A systematic review and meta-analysis. Lancet Public Health 2017, 2, e356–e366. [Google Scholar] [CrossRef]
- de Kloet, E.R.; Joels, M.; Holsboer, F. Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 2005, 6, 463–475. [Google Scholar] [CrossRef]
- Davies, T.H.; Ning, Y.M.; Sanchez, E.R. A new first step in activation of steroid receptors: Hormone-induced switching of FKBP51 and FKBP52 immunophilins. J. Biol. Chem. 2002, 277, 4597–4600. [Google Scholar] [CrossRef]
- Morishima, Y.; Murphy, P.J.; Li, D.P.; Sanchez, E.R.; Pratt, W.B. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J. Biol. Chem. 2000, 275, 18054–18060. [Google Scholar] [CrossRef]
- Schiene-Fischer, C.; Yu, C. Receptor accessory folding helper enzymes: The functional role of peptidyl prolyl cis/trans isomerases. FEBS Lett. 2001, 495, 1–6. [Google Scholar] [CrossRef]
- Wochnik, G.M.; Ruegg, J.; Abel, G.A.; Schmidt, U.; Holsboer, F.; Rein, T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 2005, 280, 4609–4616. [Google Scholar] [CrossRef]
- Ladd, C.O.; Huot, R.L.; Thrivikraman, K.V.; Nemeroff, C.B.; Plotsky, P.M. Long-term adaptations in glucocorticoid receptor and mineralocorticoid receptor mRNA and negative feedback on the hypothalamo-pituitary-adrenal axis following neonatal maternal separation. Biol. Psychiatry 2004, 55, 367–375. [Google Scholar] [CrossRef]
- Arabadzisz, D.; Diaz-Heijtz, R.; Knuesel, I.; Weber, E.; Pilloud, S.; Dettling, A.C.; Feldon, J.; Law, A.J.; Harrison, P.J.; Pryce, C.R. Primate early life stress leads to long-term mild hippocampal decreases in corticosteroid receptor expression. Biol. Psychiatry 2010, 67, 1106–1109. [Google Scholar] [CrossRef] [PubMed]
- van der Doelen, R.H.; Calabrese, F.; Guidotti, G.; Geenen, B.; Riva, M.A.; Kozicz, T.; Homberg, J.R. Early life stress and serotonin transporter gene variation interact to affect the transcription of the glucocorticoid and mineralocorticoid receptors, and the co-chaperone FKBP5, in the adult rat brain. Front. Behav. Neurosci. 2014, 8, 355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ke, X.; Fu, Q.; Majnik, A.; Cohen, S.; Liu, Q.; Lane, R. Adverse early life environment induces anxiety-like behavior and increases expression of FKBP5 mRNA splice variants in mouse brain. Physiol. Genom. 2018, 50, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.; Fillman, S.G.; Webster, M.J.; Weickert, C.S. Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci. Rep. 2013, 3, 3539. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.; Tsai, S.Y.; Woon, H.G.; Weickert, C.S. Abnormal glucocorticoid receptor mRNA and protein isoform expression in the prefrontal cortex in psychiatric illness. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2011, 36, 2698–2709. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Sinclair, D.; O’Donnell, M.; Galletly, C.; Liu, D.; Weickert, C.S.; Weickert, T.W. Transcriptional changes in the stress pathway are related to symptoms in schizophrenia and to mood in schizoaffective disorder. Schizophr. Res. 2019, 213, 87–95. [Google Scholar] [CrossRef]
- Schaaf, M.J.M.; De Jong, J.; De Kloet, E.R.; Vreugdenhil, E. Downregulation of BDNF mRNA and protein in the rat hippocampus by corticosterone. Brain Res. 1998, 813, 112–120. [Google Scholar] [CrossRef]
- Smith, M.A.; Makino, S.; Kvetnansky, R.; Post, R.M. Stress and glucocorticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J. Neurosci. 1995, 15, 1768–1777. [Google Scholar] [CrossRef]
- Hofer, M.; Pagliusi, S.R.; Hohn, A.; Leibrock, J.; Barde, Y.A. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990, 9, 2459–2464. [Google Scholar] [CrossRef]
- Aid, T.; Kazantseva, A.; Piirsoo, M.; Palm, K.; Timmusk, T. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 2007, 85, 525–535. [Google Scholar] [CrossRef]
- Daskalakis, N.P.; De Kloet, E.R.; Yehuda, R.; Malaspina, D.; Kranz, T.M. Early Life Stress Effects on Glucocorticoid-BDNF Interplay in the Hippocampus. Front. Mol. Neurosci. 2015, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Jaumotte, J.D.; Wyrostek, S.L.; Zigmond, M.J. Protection of cultured dopamine neurons from MPP(+) requires a combination of neurotrophic factors. Eur. J. Neurosci. 2016, 44, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Nakaji-Hirabayashi, T.; Fujimoto, K.; Yoshikawa, C.; Kitano, H. Functional surfaces for efficient differentiation of neural stem/progenitor cells into dopaminergic neurons. J. Biomed. Mater. Res. Part A 2019, 107, 860–871. [Google Scholar] [CrossRef]
- Hyman, C.; Hofer, M.; Barde, Y.A.; Juhasz, M.; Yancopoulos, G.D.; Squinto, S.P.; Lindsay, R.M. BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature 1991, 350, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Spina, M.B.; Squinto, S.P.; Miller, J.; Lindsay, R.M.; Hyman, C. Brain-derived neurotrophic factor protects dopamine neurons against 6-hydroxydopamine and N-methyl-4-phenylpyridinium ion toxicity: Involvement of the glutathione system. J. Neurochem. 1992, 59, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in neuronal development and function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef] [PubMed]
- Hyman, C.; Juhasz, M.; Jackson, C.; Wright, P.; Ip, N.Y.; Lindsay, R.M. Overlapping and distinct actions of the neurotrophins BDNF, NT-3, and NT-4/5 on cultured dopaminergic and GABAergic neurons of the ventral mesencephalon. J. Neurosci. 1994, 14, 335–347. [Google Scholar] [CrossRef]
- Ohta, K.I.; Suzuki, S.; Warita, K.; Kaji, T.; Kusaka, T.; Miki, T. Prolonged maternal separation attenuates BDNF-ERK signaling correlated with spine formation in the hippocampus during early brain development. J. Neurochem. 2017, 141, 179–194. [Google Scholar] [CrossRef]
- Nair, A.; Vadodaria, K.C.; Banerjee, S.B.; Benekareddy, M.; Dias, B.G.; Duman, R.S.; Vaidya, V.A. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacology 2007, 32, 1504–1519. [Google Scholar] [CrossRef]
- Roceri, M.; Cirulli, F.; Pessina, C.; Peretto, P.; Racagni, G.; Riva, M.A. Postnatal repeated maternal deprivation produces age-dependent changes of brain-derived neurotrophic factor expression in selected rat brain regions. Biol. Psychiatry 2004, 55, 708–714. [Google Scholar] [CrossRef]
- Pruunsild, P.; Kazantseva, A.; Aid, T.; Palm, K.; Timmusk, T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics 2007, 90, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Cattane, N.; Begni, V.; Pariante, C.M.; Riva, M.A. The human BDNF gene: Peripheral gene expression and protein levels as biomarkers for psychiatric disorders. Transl. Psychiatry 2016, 6, e958. [Google Scholar] [CrossRef] [PubMed]
- Berton, O.; McClung, C.A.; Dileone, R.J.; Krishnan, V.; Renthal, W.; sRusso, S.J.; Graham, D.; Tsankova, N.M.; Bolanos, C.A.; Rios, M.; et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 2006, 311, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Wook Koo, J.; Labonté, B.; Engmann, O.; Calipari, E.S.; Juarez, B.; Lorsch, Z.; Walsh, J.J.; Friedman, A.K.; Yorgason, J.T.; Han, M.H.; et al. Essential Role of Mesolimbic Brain-Derived Neurotrophic Factor in Chronic Social Stress-Induced Depressive Behaviors. Biol. Psychiatry 2016, 80, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Webster, M.J.; Cassano, H.; Weickert, C.S. Changes in alternative brain-derived neurotrophic factor transcript expression in the developing human prefrontal cortex. Eur. J. Neurosci. 2009, 29, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Kheirbek, M.A.; Drew, L.J.; Burghardt, N.S.; Costantini, D.O.; Tannenholz, L.; Ahmari, S.E.; Zeng, H.; Fenton, A.A.; Hen, R. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 2013, 77, 955–968. [Google Scholar] [CrossRef]
- Gasbarri, A.; Verney, C.; Innocenzi, R.; Campana, E.; Pacitti, C. Mesolimbic dopaminergic neurons innervating the hippocampal formation in the rat: A combined retrograde tracing and immunohistochemical study. Brain Res. 1994, 668, 71–79. [Google Scholar] [CrossRef]
- Howes, O.; Bose, S.; Turkheimer, F.; Valli, I.; Egerton, A.; Stahl, D.; Valmaggia, L.; Allen, P.; Murray, R.; McGuire, P. Progressive increase in striatal dopamine synthesis capacity as patients develop psychosis: A PET study. Mol. Psychiatry 2011, 16, 885–886. [Google Scholar] [CrossRef]
- Howes, O.D.; Montgomery, A.J.; Asselin, M.C.; Murray, R.M.; Valli, I.; Tabraham, P.; Bramon-Bosch, E.; Valmaggia, L.; Johns, L.; Broome, M.; et al. Elevated striatal dopamine function linked to prodromal signs of schizophrenia. Arch. Gen. Psychiatry 2009, 66, 13–20. [Google Scholar] [CrossRef]
- Mizrahi, R.; Addington, J.; Rusjan, P.M.; Suridjan, I.; Ng, A.; Boileau, I.; Pruessner, J.C.; Remington, G.; Houle, S.; Wilson, A.A. Increased stress-induced dopamine release in psychosis. Biol. Psychiatry 2012, 71, 561–567. [Google Scholar] [CrossRef]
- Egerton, A.; Chaddock, C.A.; Winton-Brown, T.T.; Bloomfield, M.A.; Bhattacharyya, S.; Allen, P.; McGuire, P.K.; Howes, O.D. Presynaptic striatal dopamine dysfunction in people at ultra-high risk for psychosis: Findings in a second cohort. Biol. Psychiatry 2013, 74, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Wiesel, F.A.; Sedvall, G. Effect of antipsychotic drugs on homovanillic acid levels in striatum and olfactory tubercle of the rat. Eur. J. Pharmacol. 1975, 30, 364–367. [Google Scholar] [CrossRef]
- Bjorklund, A.; Dunnett, S.B. Dopamine neuron systems in the brain: An update. Trends Neurosci. 2007, 30, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Haber, S.N. The place of dopamine in the cortico-basal ganglia circuit. Neuroscience 2014, 282, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Holly, E.N.; Miczek, K.A. Ventral tegmental area dopamine revisited: Effects of acute and repeated stress. Psychopharmacology 2016, 233, 163–186. [Google Scholar] [CrossRef]
- Vaessen, T.; Hernaus, D.; Myin-Germeys, I.; van Amelsvoort, T. The dopaminergic response to acute stress in health and psychopathology: A systematic review. Neurosci. Biobehav. Rev. 2015, 56, 241–251. [Google Scholar] [CrossRef]
- Sinclair, D.; Purves-Tyson, T.D.; Allen, K.M.; Weickert, C.S. Impacts of stress and sex hormones on dopamine neurotransmission in the adolescent brain. Psychopharmacology 2014, 231, 1581–1599. [Google Scholar] [CrossRef]
- Aleman, A.; Kahn, R.S.; Selten, J.P. Sex differences in the risk of schizophrenia: Evidence from meta-analysis. Arch. Gen. Psychiatry 2003, 60, 565–571. [Google Scholar] [CrossRef]
- McGrath, J.; Saha, S.; Welham, J.; El Saadi, O.; MacCauley, C.; Chant, D. A systematic review of the incidence of schizophrenia: The distribution of rates and the influence of sex, urbanicity, migrant status and methodology. BMC Med. 2004, 2, 13. [Google Scholar] [CrossRef]
- Bian, Y.; Yang, L.; Wang, Z.; Wang, Q.; Zeng, L.; Xu, G. Repeated Three-Hour Maternal Separation Induces Depression-Like Behavior and Affects the Expression of Hippocampal Plasticity-Related Proteins in C57BL/6N Mice. Neural Plast. 2015, 2015, 627837. [Google Scholar] [CrossRef]
- Maniam, J.; Morris, M.J. Voluntary exercise and palatable high-fat diet both improve behavioural profile and stress responses in male rats exposed to early life stress: Role of hippocampus. Psychoneuroendocrinology 2010, 35, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Maniam, J.; Morris, M.J. Palatable cafeteria diet ameliorates anxiety and depression-like symptoms following an adverse early environment. Psychoneuroendocrinology 2010, 35, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Pryce, C.R.; Feldon, J. Long-term neurobehavioural impact of the postnatal environment in rats: Manipulations, effects and mediating mechanisms. Neurosci. Biobehav. Rev. 2003, 27, 57–71. [Google Scholar] [CrossRef]
- Tatro, E.T.; Everall, I.P.; Kaul, M.; Achim, C.L. Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: Implications for major depressive disorder. Brain Res. 2009, 1286, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Spivey, J.M.; Shumake, J.; Colorado, R.A.; Conejo-Jimenez, N.; Gonzalez-Pardo, H.; Gonzalez-Lima, F. Adolescent female rats are more resistant than males to the effects of early stress on prefrontal cortex and impulsive behavior. Dev. Psychobiol. 2009, 51, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, K.D.; Demady, D.R.; Stancato, L.F.; Krishna, P.; Pratt, W.B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor.hsp90 heterocomplexes formed by hsp90.p60.hsp70. J. Biol. Chem. 1997, 272, 21213–21220. [Google Scholar] [CrossRef]
- Maurel, O.M.; Torrisi, S.A.; Barbagallo, C.; Purrello, M.; Salomone, S.; Drago, F.; Ragusa, M.; Leggio, G.M. Dysregulation of miR-15a-5p, miR-497a-5p and miR-511-5p Is Associated with Modulation of BDNF and FKBP5 in Brain Areas of PTSD-Related Susceptible and Resilient Mice. Int. J. Mol. Sci. 2021, 22, 5157. [Google Scholar] [CrossRef]
- Kwon, J.; Kim, Y.J.; Choi, K.; Seol, S.; Kang, H.J. Identification of stress resilience module by weighted gene co-expression network analysis in Fkbp5-deficient mice. Mol. Brain 2019, 12, 99. [Google Scholar] [CrossRef]
- Miller, O.; Shakespeare-Finch, J.; Bruenig, D.; Mehta, D. DNA methylation of NR3C1 and FKBP5 is associated with posttraumatic stress disorder, posttraumatic growth, and resilience. Psychol. Trauma Theory Res. Pract. Policy 2020, 12, 750–755. [Google Scholar] [CrossRef]
- Murer, M.G.; Yan, Q.; Raisman-Vozari, R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog. Neurobiol. 2001, 63, 71–124. [Google Scholar] [CrossRef]
- Shepard, R.D.; Gouty, S.; Kassis, H.; Berenji, A.; Zhu, W.; Cox, B.M.; Nugent, F.S. Targeting histone deacetylation for recovery of maternal deprivation-induced changes in BDNF and AKAP150 expression in the VTA. Exp. Neurol. 2018, 309, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Koppel, I.; Aid-Pavlidis, T.; Jaanson, K.; Sepp, M.; Pruunsild, P.; Palm, K.; Timmusk, T. Tissue-specific and neural activity-regulated expression of human BDNF gene in BAC transgenic mice. BMC Neurosci. 2009, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.C.; Rodríguez, G.; Asede, D.; Jung, K.; Hwang, I.W.; Ogelman, R.; Bolton, M.M.; Kwon, H.B. Dysregulation of the mesoprefrontal dopamine circuit mediates an early-life stress-induced synaptic imbalance in the prefrontal cortex. Cell Rep. 2021, 35, 109074. [Google Scholar] [CrossRef] [PubMed]
- Neeley, E.W.; Berger, R.; Koenig, J.I.; Leonard, S. Prenatal stress differentially alters brain-derived neurotrophic factor expression and signaling across rat strains. Neuroscience 2011, 187, 24–35. [Google Scholar] [CrossRef]
- Mallei, A.; Ieraci, A.; Popoli, M. Chronic social defeat stress differentially regulates the expression of BDNF transcripts and epigenetic modifying enzymes in susceptible and resilient mice. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 2019, 20, 555–566. [Google Scholar] [CrossRef]
- Zafra, F.; Hengerer, B.; Leibrock, J.; Thoenen, H.; Lindholm, D. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. Embo J. 1990, 9, 3545–3550. [Google Scholar] [CrossRef]
- Kikusui, T.; Ichikawa, S.; Mori, Y. Maternal deprivation by early weaning increases corticosterone and decreases hippocampal BDNF and neurogenesis in mice. Psychoneuroendocrinology 2009, 34, 762–772. [Google Scholar] [CrossRef]
- Viveros, M.P.; Diaz, F.; Mateos, B.; Rodriguez, N.; Chowen, J.A. Maternal deprivation induces a rapid decline in circulating leptin levels and sexually dimorphic modifications in hypothalamic trophic factors and cell turnover. Horm. Behav. 2010, 57, 405–414. [Google Scholar] [CrossRef]
- Luft, C.; Levices, I.P.; da Costa, M.S.; de Oliveira, J.R.; Donadio, M.V.F. Effects of running before pregnancy on long-term memory and hippocampal alterations induced by prenatal stress. Neurosci. Lett. 2021, 746, 135659. [Google Scholar] [CrossRef]
- Lazic, S.E.; Essioux, L. Improving basic and translational science by accounting for litter-to-litter variation in animal models. BMC Neurosci. 2013, 14, 37. [Google Scholar] [CrossRef]
- Ellenbroek, B.A.; Cools, A.R. The Long-Term Effects of Maternal Deprivation Depend on the Genetic Background. Neuropsychopharmacology 2000, 23, 99–106. [Google Scholar] [CrossRef]
- Ellenbroek, B.A.; de Bruin, N.M.; van Den Kroonenburg, P.T.; van Luijtelaar, E.L.; Cools, A.R. The effects of early maternal deprivation on auditory information processing in adult Wistar rats. Biol. Psychiatry 2004, 55, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Ellenbroek, B.A.; van den Kroonenberg, P.T.; Cools, A.R. The effects of an early stressful life event on sensorimotor gating in adult rats. Schizophr. Res. 1998, 30, 251–260. [Google Scholar] [CrossRef]
- Lehmann, J.; Pryce, C.R.; Feldon, J. Lack of effect of an early stressful life event on sensorimotor gating in adult rats. Schizophr. Res. 2000, 41, 365–371. [Google Scholar] [CrossRef]
- Bodensteiner, K.J.; Christianson, N.; Siltumens, A.; Krzykowski, J. Effects of Early Maternal Separation on Subsequent Reproductive and Behavioral Outcomes in Male Rats. J. Gen. Psychol. 2014, 141, 228–246. [Google Scholar] [CrossRef]
- Carlyle, B.C.; Duque, A.; Kitchen, R.R.; Bordner, K.A.; Coman, D.; Doolittle, E.; Papademetris, X.; Hyder, F.; Taylor, J.R.; Simen, A.A. Maternal separation with early weaning: A rodent model providing novel insights into neglect associated developmental deficits. Dev. Psychopathol. 2012, 24, 1401–1416. [Google Scholar] [CrossRef]
- Nylander, I.; Roman, E. Is the rodent maternal separation model a valid and effective model for studies on the early-life impact on ethanol consumption? Psychopharmacology 2013, 229, 555–569. [Google Scholar] [CrossRef]
- Ploj, K.; Roman, E.; Nylander, I. Long-term effects of short and long periods of maternal separation on brain opioid peptide levels in male Wistar rats. Neuropeptides 2003, 37, 149–156. [Google Scholar] [CrossRef]
- Ploj, K.; Roman, E.; Nylander, I. Effects of maternal separation on brain nociceptin/orphanin FQ peptide levels in male Wistar rats. Pharmacol. Biochem. Behav. 2002, 73, 123–129. [Google Scholar] [CrossRef]
- Ploj, K.; Roman, E.; Nylander, I. Long-term effects of maternal separation on ethanol intake and brain opioid and dopamine receptors in male Wistar rats. Neuroscience 2003, 121, 787–799. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]

| Transcript | Outliers (Removed from Analysis) | N (after Removing Outliers) | ANOVA (Early Life Stress) | ANOVA (Sex) | ANOVA (Early Life Stress × Sex Interaction) |
|---|---|---|---|---|---|
| Substantia Nigra | |||||
| Nr3c1 (GR) | - | 46 | F(1,42) = 3.78, p = 0.06 | F(1,42) = 0.17, p = 0.68 | F(1,42) = 0.03, p = 0.87 |
| Fkbp5 | Stress (1) | 45 | F(1,41) = 0.70, p = 0.41 | F(1,41) = 4.17, p = 0.05 | F(1,41) = 6.33, p = 0.02 |
| Fkbp4 | Control (1) | 45 | F(1,41) = 2.70, p = 0.11 | F(1,41) = 6.38, p = 0.02 | F(1,41) = 0.12, p = 0.73 |
| Ptges3 | Control (3) | 43 | F(1,39) = 1.41, p = 0.24 | F(1,39) = 2.33, p = 0.14 | F(1,39) = 0.40, p = 0.53 |
| Bag1 | - | 46 | F(1,42) = 2.18, p = 0.15 | F(1,42) = 1.16, p = 0.29 | F(1,42) = 1.19, p = 0.28 |
| VTA | |||||
| Nr3c1 (GR) | - | 46 | F(1,42) = 1.64, p = 0.21 | F(1,42) = 0.19, p = 0.67 | F(1,42) = 7.87, p= 0.008 |
| Fkbp5 | Stress (2) | 44 | F(1,40) = 7.27, p = 0.01 | F(1,40) = 0.40, p = 0.53 | F(1,40) = 0.16, p = 0.69 |
| Fkbp4 | Stress (1) | 45 | F(1,41) = 0.11, p = 0.75 | F(1,41) = 0.03, p = 0.85 | F(1,41) = 0.02, p = 0.90 |
| Ptges3 | - | 46 | F(1,42) = 4.24, p = 0.05 | F(1,42) = 1.20, p = 0.28 | F(1,42) = 1.13, p = 0.29 |
| Bag1 | - | 46 | F(1,42) = 10.25, p = 0.003 | F(1,42) = 0.82, p = 0.37 | F(1,42) = 7.66, p = 0.008 |
| Dorsal Striatum | |||||
| Nr3c1 (GR) | - | 46 | F(1,42) = 0.71, p = 0.40 | F(1,42) = 0.007, p = 0.93 | F(1,42) = 0.90, p = 0.35 |
| Fkbp5 | - | 46 | F(1,42) = 1.28, p = 0.27 | F(1,42) = 0.36, p = 0.55 | F(1,42) = 0.03, p = 0.86 |
| Fkbp4 | - | 46 | F(1,42) = 1.80, p = 0.19 | F(1,42) = 7.55, p = 0.009 | F(1,42) = 0.23, p = 0.63 |
| Ptges3 | Stress (1) | 45 | F(1,41) = 0.28, p = 0.60 | F(1,41) = 0.001, p = 0.98 | F(1,41) = 0.03, p = 0.86 |
| Bag1 * | - | 46 | F(1,42) = 0.23, p = 0.64 | F(1,42) = 0.08, p = 0.78 | F(1,42) = 2.07, p = 0.16 |
| Ventral Striatum | |||||
| Nr3c1 (GR) | - | 46 | F(1,42) = 0.009, p= 0.93 | F(1,42) = 0.04, p = 0.85 | F(1,42) = 2.78, p = 0.10 |
| Fkbp5 | - | 46 | F(1,42) = 1.39, p = 0.25 | F(1,42) = 0.76, p = 0.39 | F(1,42) = 0.14, p = 0.72 |
| Fkbp4 | Control (1) | 45 | F(1,41) = 0.001, p = 0.98 | F(1,41) = 1.77, p = 0.19 | F(1,41) = 1.78, p = 0.19 |
| Ptges3 | Stress (2) | 44 | F(1,40) = 1.23, p = 0.28 | F(1,40) = 0.67, p = 0.42 | F(1,40) = 0.11, p = 0.74 |
| Bag1 | - | 46 | F(1,42) = 2.60, p = 0.11 | F(1,42) = 0.25, p = 0.62 | F(1,42) = 0.32, p = 0.57 |
| Transcripts | Outliers (Removed from Analyses) | N (after Removing Outliers) | ANOVA (Early Life Stress) | ANOVA (Sex) | ANOVA (Early Life Stress × Sex Interaction) |
|---|---|---|---|---|---|
| Substantia Nigra | |||||
| Bdnf I | - | 46 | F(1,42) = 0.34, p = 0.56 | F(1,42) = 21.51, p < 0.001 | F(1,42) = 1.86, p = 0.18 |
| Bdnf IIA | - | 46 | F(1,42) = 10.93, p = 0.002 | F(1,42) = 1.88, p = 0.18 | F(1,42) = 0.45, p = 0.50 |
| Bdnf IIC | - | 46 | F(1,42) = 0.32, p = 0.58 | F(1,42) = 8.52, p = 0.006 | F(1,42) = 0.005, p = 0.94 |
| Bdnf III | - | 46 | F(1,42) = 22.54, p < 0.001 | F(1,42) = 0.41, p = 0.53 | F(1,42) = 0.41, p = 0.52 |
| Bdnf IV | - | 46 | F(1,42) = 0.51, p = 0.48 | F(1,42) = 3.11, p = 0.09 | F(1,42) = 2.28, p = 0.14 |
| Bdnf VI | - | 46 | F(1,42) = 0.06, p = 0.82 | F(1,42) = 7.12, p = 0.01 | F(1,42) = 0.27, p = 0.60 |
| (VTA | |||||
| Bdnf I * | - | 46 | F(1,42) = 2.02, p = 0.16 | F(1,42) = 10.46, p = 0.002 | F(1,42) = 1.64, p = 0.21 |
| Bdnf IIA | - | 46 | F(1,42) = 11.61, p = 0.001 | F(1,42) = 0.65, p = 0.43 | F(1,42) = 0.91, p = 0.34 |
| Bdnf IIC | - | 46 | F(1,42) = 1.51, p = 0.23 | F(1,42) = 4.08, p = 0.05 | F(1,42) = 1.05, p = 0.31 |
| Bdnf III | - | 46 | F(1,42) = 11.18, p = 0.002 | F(1,42) = 1.25, p = 0.27 | F(1,42) = 0.17, p = 0.69 |
| Bdnf IV | Stress (1) | 45 | F(1,41) = 1.59, p = 0.21 | F(1,41) = 1.79, p = 0.19 | F(1,41) = 2.58, p = 0.12 |
| Bdnf VI | Stress (1) | 45 | F(1,41) = 0.89, p = 0.35 | F(1,41) = 0.41, p = 0.53 | F(1,41) = 3.17, p = 0.08 |
| Dorsal Striatum | |||||
| Bdnf I * | Stress (1) | 45 | F(1,41) = 0.27, p = 0.61 | F(1,41) = 0.19, p = 0.67 | F(1,41) = 1.02, p = 0.32 |
| Bdnf IIA | Stress (3), Control (1) | 42 | F(1,38) = 0.22, p = 0.65 | F(1,38) = 0.09, p = 0.77 | F(1,38) = 0.32, p = 0.57 |
| Bdnf IIC | Stress (1), Control (1) | 44 | F(1,40) = 0.05, p = 0.83 | F(1,40) = 1.97, p = 0.17 | F(1,40) = 0.87, p = 0.36 |
| Bdnf III | Stress (1) | 45 | F(1,41) = 0.45, p = 0.51 | F(1,41) = 1.20, p = 0.28 | F(1,41) = 1.61, p = 0.21 |
| Bdnf IV | Stress (2) | 44 | F(1,40) = 0.02, p = 0.89 | F(1,40) = 1.19, p = 0.28 | F(1,40) = 0.98, p = 0.33 |
| Bdnf VI* | Stress (1) | 45 | F(1,41) = 0.28, p = 0.60 | F(1,41) = 2.58, p = 0.12 | F(1,41) = 0.57, p = 0.45 |
| Ventral Striatum | |||||
| Bdnf I | Stress (2) | 44 | F(1,40) = 10.08, p = 0.003 | F(1,40) = 1.36, p = 0.25 | F(1,40) = 1.22, p = 0.28 |
| Bdnf IIA * | Stress (1) | 45 | F(1,41) = 5.48, p = 0.02 | F(1,41) < 0.001, p = 0.98 | F(1,41) = 0.14, p = 0.72 |
| Bdnf IIC | Stress (1) | 45 | F(1,41) = 6.48, p = 0.02 | F(1,41) = 0.18, p = 0.68 | F(1,41) = 0.05, p = 0.83 |
| Bdnf III | Stress (2) | 44 | F(1,40) = 3.11, p = 0.09 | F(1,40) = 0.79, p = 0.38 | F(1,40) = 0.07, p = 0.80 |
| Bdnf IV * | - | 46 | F(1,42) = 6.26, p = 0.02 | F(1,42) = 2.09, p = 0.16 | F(1,42) = 0.24, p = 0.63 |
| Bdnf VI | Stress (1) | 45 | F(1,41) = 7.43, p = 0.009 | F(1,41) = 0.76, p = 0.39 | F(1,41) = 0.84, p = 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, C.H.; Shannon Weickert, C.; Weickert, T.W.; Sinclair, D. Early Life Stress Alters Expression of Glucocorticoid Stress Response Genes and Trophic Factor Transcripts in the Rodent Basal Ganglia. Int. J. Mol. Sci. 2022, 23, 5333. https://doi.org/10.3390/ijms23105333
Tran CH, Shannon Weickert C, Weickert TW, Sinclair D. Early Life Stress Alters Expression of Glucocorticoid Stress Response Genes and Trophic Factor Transcripts in the Rodent Basal Ganglia. International Journal of Molecular Sciences. 2022; 23(10):5333. https://doi.org/10.3390/ijms23105333
Chicago/Turabian StyleTran, Cynthia Haidee, Cynthia Shannon Weickert, Thomas Wesley Weickert, and Duncan Sinclair. 2022. "Early Life Stress Alters Expression of Glucocorticoid Stress Response Genes and Trophic Factor Transcripts in the Rodent Basal Ganglia" International Journal of Molecular Sciences 23, no. 10: 5333. https://doi.org/10.3390/ijms23105333
APA StyleTran, C. H., Shannon Weickert, C., Weickert, T. W., & Sinclair, D. (2022). Early Life Stress Alters Expression of Glucocorticoid Stress Response Genes and Trophic Factor Transcripts in the Rodent Basal Ganglia. International Journal of Molecular Sciences, 23(10), 5333. https://doi.org/10.3390/ijms23105333






