RNA Interference: Promising Approach to Combat Plant Viruses
Abstract
1. Introduction
2. Development of Resistance against Viral Infection Inside the Host Cell
2.1. Pathogen-Triggered Immunity (PTI) against Virus Infection
2.2. Effector-Triggered Immunity (ETI) against Virus Infection
2.3. Gene Silencing
2.3.1. Transcriptional Gene Silencing (TGS)
2.3.2. Post-transcriptional Gene Silencing (PTGS)/RNA Interference (RNAi)
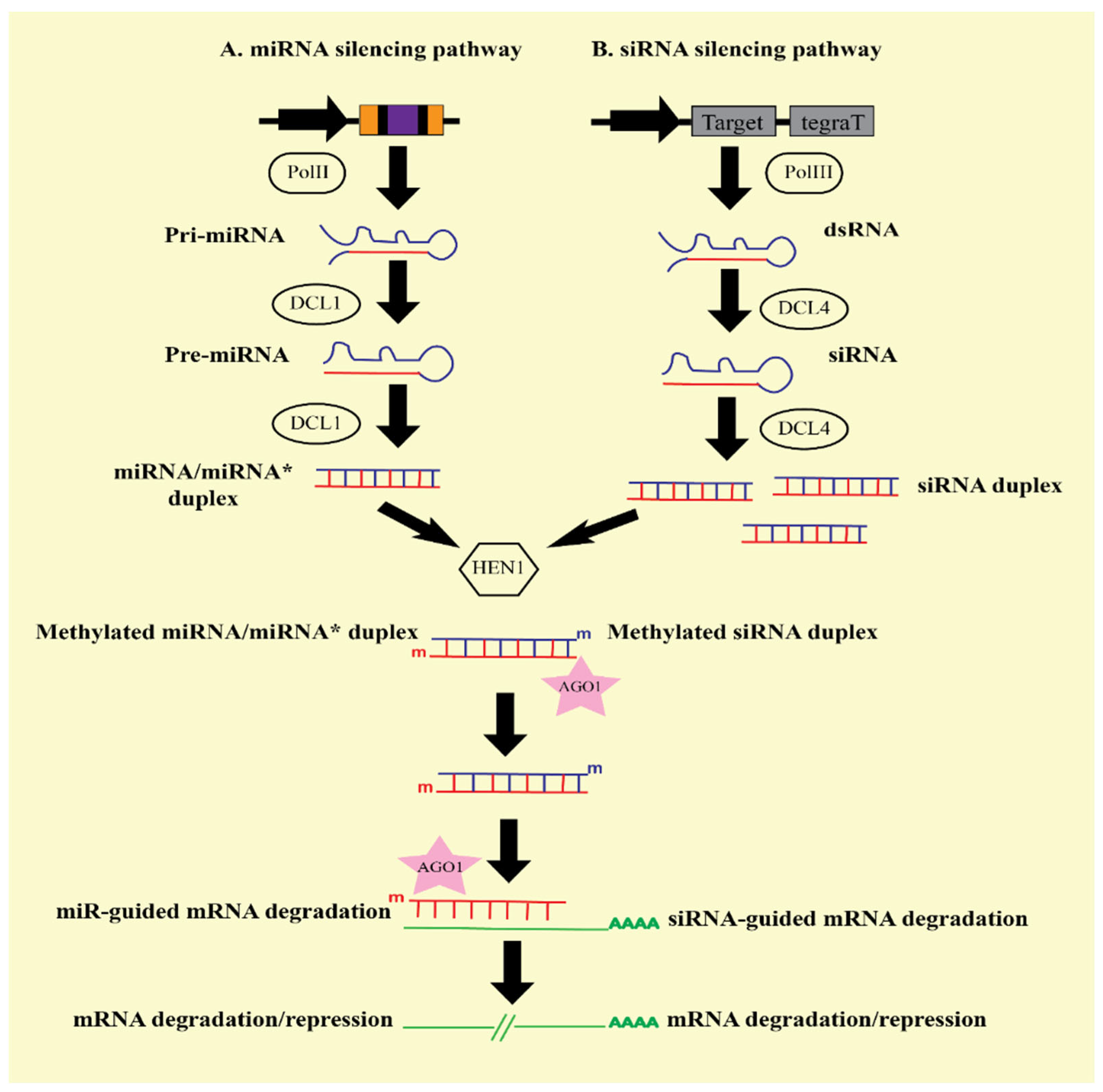
3. General Mechanism of RNA-Induced Gene Silencing
3.1. Enzymes Involved in RNA Induced Gene Silencing
3.1.1. Dicer
3.1.2. RNA-Induced Silencing Complex (RISC)
3.1.3. RNA-Dependent RNA Polymerase (RDR)
3.2. Different Types of Gene Silencing
3.2.1. Virus-Induced Gene Silencing (VIGS)
3.2.2. Host Induced Gene Silencing (HIGS)
3.2.3. Spray Induced Gene Silencing (SIGS)
3.2.4. Advantages of SIGS over HIGS and VIGS
3.3. Gene Silencing for Plant Virus Resistance
3.4. RNAi against Viral Suppressors
3.5. RNAi against Insect Vectors
3.6. HIGS against Plants Viruses
3.7. VIGS against Plant Viruses
3.8. SIGS against Plant Viruses
4. Limitations of Gene Silencing
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suweis, S.; Carr, J.A.; Maritan, A.; Rinaldo, A.; D’Odorico, P. Resilience and reactivity of global food security. Proc. Natl. Acad. Sci. USA 2015, 112, 6902–6907. [Google Scholar] [CrossRef] [PubMed]
- Sastry, K.S.; Zitter, T.A. Management of virus and viroid diseases of crops in the tropics. In Plant Virus and Viroid Diseases in the Tropics; Springer: Dordrecht, The Netherlands, 2014; pp. 149–480. [Google Scholar]
- Wale, S.J.; Platt, H.W.; Cattlin, N.D. Diseases, Pests, and Disorders of Potatoes: A Colour Handbook; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Ordon, F.; Habekuss, A.; Kastirr, U.; Rabenstein, F.; Kühne, T. Virus resistance in cereals: Sources of resistance, genetics, and breeding. J. Phytopathol. 2009, 157, 535–545. [Google Scholar] [CrossRef]
- Calvert, L.A.; Thresh, J. Utilization: The viruses and virus diseases of cassava. J. Cassava Biol. Prod. Util. 2002, 237–260. [Google Scholar]
- Thresh, J.M.; Cooter, R.J. Strategies for controlling cassava mosaic virus disease in Africa. J. Plant. Pathol. 2005, 54, 587–614. [Google Scholar] [CrossRef]
- Putra, L.K.; Kristini, A.; Achadian, E.M.A.; Damayanti, T.A. Sugarcane streak mosaic virus in Indonesia: Distribution, characterisation, yield losses, and management approaches. J. Sugar Tech. 2014, 16, 392–399. [Google Scholar] [CrossRef]
- Mahy, B.W.; Van Regenmortel, M.H. Desk Encyclopedia of Plan and Fungal Virogogy; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Leonetti, P.; Stuttmann, J.; Pantaleo, V. Regulation of plant antiviral defense genes via host RNA-silencing mechanisms. Virol. J. 2021, 18, 194. [Google Scholar] [CrossRef]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. J. Nat. 2004, 428, 764–767. [Google Scholar] [CrossRef]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D. The transcriptional innate immune response to flg22. Interplay and overlap with avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. J. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.I.; Yasuda, S. Pattern recognition receptors and signaling in plant—Microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef] [PubMed]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.F.; Zheng, H. Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. J. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Wu, J.; Lu, L.; Xu, Y.; Zhou, X. Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol. Plant 2014, 7, 691–708. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Candresse, T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, J.M. Regulation of cell death and signaling by pore-forming resistosomes. Annu. Rev. Phytopathol. 2021, 59, 239–263. [Google Scholar] [CrossRef]
- Lapin, D.; Bhandari, D.D.; Parker, J.E. Origins and immunity networking functions of EDS1 family proteins. Annu. Rev. Phytopathol. 2020, 58, 253–276. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Jubic, L.M.; Saile, S.; Furzer, O.J.; El Kasmi, F.; Dangl, J.L. Help wanted: Helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 2019, 50, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Bhandari, D.; von Born, P.; Bautor, J.; Guarneri, N.; Rzemieniewski, J.; Stuttmann, J.; et al. A Coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Gantner, J.; Ordon, J.; Kretschmer, C.; Guerois, R.; Stuttmann, J. An EDS1-SAG101 complex is essential for TNL-mediated immunity in nicotiana benthamiana. Plant Cell 2019, 31, 2456–2474. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Signaling in induced resistance. Adv. Virus Res. 2010, 76, 57–121. [Google Scholar]
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance—Deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Baulcombe, D.J.N. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Voinnet, O.J.N.R.G. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005, 6, 206–220. [Google Scholar] [CrossRef]
- Fagard, M.; Boutet, S.; Morel, J.B.; Bellini, C.; Vaucheret, H. AGO1, QDE-2, and RDE-1 are related proteins required for posttranscriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 2000, 97, 11650–11654. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Wang, M.B.; Lough, T.J.N. Gene silencing as an adaptive defence against viruses. Nature 2001, 411, 834–842. [Google Scholar] [CrossRef]
- Eckardt, N.A. RNA goes mobile. Am. Soc. Plant Biol. 2002, 14, 1433–1436. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing. Nature 2005, 30, 290–293. [Google Scholar]
- Tomari, Y.; Zamore, P.D.J.G. Development: Perspective: Machines for RNAi. Genes Dev. 2005, 19, 517–529. [Google Scholar] [CrossRef]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P.J.C. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T.J.G. Development: RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R.J.R. RNA interference in human cells is restricted to the cytoplasm. RNA 2002, 8, 855–860. [Google Scholar] [CrossRef]
- Haley, B.; Zamore, P.D. Biology m: Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef]
- Martinez, J.; Tuschl, T. Development: RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004, 18, 975–980. [Google Scholar] [CrossRef]
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef]
- Carmell, M.A.; Hannon, G.J. Biology m: RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004, 11, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef] [PubMed]
- Kurzynska-Kokorniak, A.; Pokornowska, M.; Koralewska, N.; Hoffmann, W.; Bienkowska-Szewczyk, K.; Figlerowicz, M. Revealing a new activity of the human dicer DUF283 domain in vitro. Sci. Rep. 2016, 6, 23989. [Google Scholar] [CrossRef] [PubMed]
- Barraud, P.; Banerjee, S.; Mohamed, W.I.; Jantsch, M.F.; Allain, F.H.T. A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1. Proc. Natl. Acad. Sci. USA 2014, 111, E1852–E1861. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Barraud, P. Functions of double-stranded RNA-binding domains in nucleocytoplasmic transport. RNA Biol. 2014, 11, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T.; Vazquez, F. RNA silencing pathways of plants: Silencing and its suppression by plant DNA viruses. Biochim. Biophys. Acta-Gene Regul. Mech. 2011, 1809, 588–600. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Schauer, S.E.; Smith, T.H.; Mallory, A.C.; Herr, J.M.J.; Roth, B.; Merchant, D.S.; Ray, A.; Bowman, L.H.; Vance, V.B. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 2005, 17, 2873–2885. [Google Scholar] [CrossRef]
- Qi, Y.; Denli, A.M.; Hannon, G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 2005, 19, 421–428. [Google Scholar] [CrossRef]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C.; Weigel, D. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef]
- Henderson, I.R.; Zhang, X.; Lu, C.; Johnson, L.; Meyers, B.C.; Green, P.J.; Jacobsen, S.E. Dissecting arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006, 38, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Coursey, T.; Regedanz, E.; Bisaro, D.M. Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J. Virol. 2018, 92, e01320-17. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.L.; Bartel, D.P.; Crété, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Himber, C.; Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005, 37, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005, 15, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Nykänen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 107, 309–321. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Parker, J.S. How to slice: Snapshots of argonaute in action. Silence 2010, 1, 3. [Google Scholar] [CrossRef]
- Simon, B.; Kirkpatrick, J.P.; Eckhardt, S.; Reuter, M.; Rocha, E.A.; Andrade-Navarro, M.A.; Sehr, P.; Pillai, R.S.; Carlomagno, T. Recognition of 2′-O-methylated 3′-end of piRNA by the PAZ domain of a piwi protein. Structure 2011, 19, 172–180. [Google Scholar] [CrossRef]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 2003, 299, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.; Sun, H.; Poethig, R.S. The arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 2003, 13, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.; Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 2010, 22, 3879–3889. [Google Scholar] [CrossRef]
- Liu, W.; Duttke, S.H.; Hetzel, J.; Groth, M.; Feng, S.; Gallego-Bartolome, J.; Zhong, Z.; Kuo, H.Y.; Wang, Z.; Zhai, J.; et al. RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in arabidopsis. Nat. Plants 2018, 4, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Lipardi, C.; Wei, Q.; Paterson, B.M. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 2001, 107, 297–307. [Google Scholar] [CrossRef]
- Zong, J.; Yao, X.; Yin, J.; Zhang, D.; Ma, H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 2009, 447, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, K.; Wang, J.; Chen, X.; Chen, Z.; Cai, R.; Xiang, Y. Comprehensive analysis of dicer-like, argonaute, and RNA-dependent RNA polymerase gene families in grapevine (Vitis vinifera). J. Plant Growth Regul. 2015, 34, 108–121. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.-S.; Chen, W.-T.; Mao, Z.-C.; Kang, H.-X.; Chen, G.-H.; Yang, Y.-H.; Xie, B.-Y. Genome-wide identification of dicer-like, argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Pandey, S.P.; Baldwin, I.T. RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of nicotiana attenuata to herbivore attack in nature. Plant J. 2007, 50, 40–53. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Q.; Wu, B.; Ai, T.; Guo, X. NgRDR1, an RNA-dependent RNA polymerase isolated from Nicotiana glutinosa, was involved in biotic and abiotic stresses. Plant Physiol. Biochem. 2009, 47, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Hunter, L.J.; Westwood, J.H.; Heath, G.; Macaulay, K.; Smith, A.G.; MacFarlane, S.A.; Palukaitis, P.; Carr, J.P. Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS ONE 2013, 8, e66530. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Kulkarni, K.; Souret, F.F.; MuthuValliappan, R.; Tej, S.S.; Poethig, R.S.; Henderson, I.R.; Jacobsen, S.E.; Wang, W.; Green, P.J.; et al. MicroRNAs and other small RNAs enriched in the arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006, 16, 1276–1288. [Google Scholar] [CrossRef]
- Nuthikattu, S.; McCue, A.D.; Panda, K.; Fultz, D.; DeFraia, C.; Thomas, E.N.; Slotkin, R.K. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013, 162, 116–131. [Google Scholar] [CrossRef]
- Voinnet, O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001, 17, 449–459. [Google Scholar] [CrossRef]
- Liu, H.; Reavy, B.; Swanson, M.; MacFarlane, S.A. Functional replacement of the tobacco rattle virus cysteine-rich protein by pathogenicity proteins from unrelated plant viruses. Virology 2002, 298, 232–239. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Parre, E.; Ghars, M.A.; Leprince, A.S.; Thiery, L.; Lefebvre, D.; Bordenave, M.; Richard, L.; Mazars, C.; Abdelly, C.; Savouré, A. Calcium signaling via phospholipase C is essential for proline accumulation upon ionic but not nonionic hyperosmotic stresses in Arabidopsis. Plant Physiol. 2007, 144, 503–512. [Google Scholar] [CrossRef]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-dependent RNA polymerase gene in arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000, 101, 543–553. [Google Scholar] [CrossRef]
- Morel, J.B.; Godon, C.; Mourrain, P.; Béclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 2002, 14, 629–639. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011, 16, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, M.H.; Donson, J.; Della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Kjemtrup, S.; Sampson, K.S.; Peele, C.G.; Nguyen, L.V.; Conkling, M.A.; Thompson, W.F.; Robertson, D. Gene silencing from plant DNA carried by a geminivirus. Plant J. 1998, 14, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.T.; Voinnet, O.; Baulcombe, D.C. Initiation and maintenance of virus-induced gene silencing. Plant Cell 1998, 10, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Liou, M.R.; Huang, Y.W.; Hu, C.C.; Lin, N.S.; Hsu, Y.H. A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 2014, 12, 330–343. [Google Scholar] [CrossRef]
- Valentine, T.; Shaw, J.; Blok, V.C.; Phillips, M.S.; Oparka, K.J.; Lacomme, C. Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol. 2004, 136, 3999–4009. [Google Scholar] [CrossRef]
- Martín-Hernández, A.M.; Baulcombe, D.C. Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J. Virol. 2008, 82, 4064–4071. [Google Scholar] [CrossRef]
- Csorba, T.; Pantaleo, V.; Burgyán, J. RNA silencing: An antiviral mechanism. Adv. Virus Res. 2009, 75, 35–71. [Google Scholar]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Niu, Y.; Zhang, K.; Liu, Y.; Zhou, X. Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virol. J. 2011, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-induced gene silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef] [PubMed]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2012, 10, 150–163. [Google Scholar] [CrossRef]
- Akbar, S.; Tahir, M.; Wang, M.B.; Liu, Q. Expression analysis of hairpin RNA carrying sugarcane mosaic virus (SCMV) derived sequences and transgenic resistance development in a model rice plant. Biomed. Res. Int. 2017, 2017, 1646140. [Google Scholar] [CrossRef]
- Sivamani, E.; Brey, C.W.; Talbert, L.E.; Young, M.A.; Dyer, W.E.; Kaniewski, W.K.; Qu, R. Resistance to wheat streak mosaic virus in transgenic wheat expressing the viral replicase (NIb) gene. Mol. Breed. 2000, 6, 469–477. [Google Scholar] [CrossRef]
- Sivamani, E.; Brey, C.W.; Talbert, L.E.; Young, M.A.; Dyer, W.E.; Kaniewski, W.K.; Qu, R. Resistance to wheat streak mosaic virus in transgenic wheat engineered with the viral coat protein gene. Transgenic Res. 2002, 11, 31–41. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Berger, P.H. Transgene silencing in wheat transformed with the WSMV-CP gene. Biotechnology 2005, 4, 62–68. [Google Scholar]
- Chaloner, T.; van Kan, J.A.; Grant-Downton, R.T. RNA ‘Information warfare’in pathogenic and mutualistic interactions. J. Trends Plant Sci. 2016, 21, 738–748. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- Vetukuri, R.R.; Dubey, M.; Kalyandurg, P.B.; Carlsson, A.S.; Whisson, S.C.; Ortiz, R. Spray-induced gene silencing: An innovative strategy for plant trait improvement and disease control. Crop Breed. Appl. Biotechnol. 2021, 21. [Google Scholar] [CrossRef]
- Tenllado, F.; Dıaz-Ruız, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically modified organism-free RNA interference: Exogenous application of RNA molecules in plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of exogenous dsRNAs-induced RNAi in agriculture: Challenges and triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Sammons, R.D.; Ivashuta, S.; Liu, H.; Wang, D.; Feng, P.C.; Kouranov, A.Y.; Andersen, S.E. Polynucleotide Molecules for Gene Regulation in Plants. U.S. Patent 20110296556A1, 1 September 2015. [Google Scholar]
- Numata, K.; Ohtani, M.; Yoshizumi, T.; Demura, T.; Kodama, Y. Local gene silencing in plants via synthetic ds RNA and carrier peptide. Plant Biotechnol. J. 2014, 12, 1027–1034. [Google Scholar] [CrossRef]
- Lau, S.E.; Schwarzacher, T.; Othman, R.Y.; Harikrishna, J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a dendrobium hybrid. BMC Plant Biol. 2015, 15, 194. [Google Scholar] [CrossRef]
- van Schie, C.C.; Takken, F.L. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Sanchez, J.N.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021, 19, 1756. [Google Scholar] [CrossRef]
- Baulcombe, D.C. VIGS, HIGS and FIGS: Small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 2015, 26, 141–146. [Google Scholar] [CrossRef]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef]
- Reyes, C.A.; De Francesco, A.; Peña, E.J.; Costa, N.; Plata, M.I.; Sendin, L.; Castagnaro, A.P.; García, M.L. Resistance to citrus psorosis virus in transgenic sweet orange plants is triggered by coat protein–RNA silencing. J. Biotechnol. 2011, 151, 151–158. [Google Scholar] [CrossRef]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Kumar, S.; Tanti, B.; Patil, B.L.; Mukherjee, S.K.; Sahoo, L. RNAi-derived transgenic resistance to mungbean yellow mosaic India virus in cowpea. PLoS ONE 2017, 12, e0186786. [Google Scholar]
- Ntui, V.O.; Kong, K.; Khan, R.S.; Igawa, T.; Janavi, G.J.; Rabindran, R.; Nakamura, I.; Mii, M. Resistance to Sri Lankan cassava mosaic virus (SLCMV) in genetically engineered cassava cv. KU50 through RNA silencing. PLoS ONE 2015, 10, e0120551. [Google Scholar] [CrossRef]
- Bau, H.J.; Cheng, Y.H.; Yu, T.A.; Yang, J.S.; Yeh, S.D. Broad-spectrum resistance to different geographic strains of Papaya ringspot virus in coat protein gene transgenic papaya. Phytopathology 2003, 93, 112–120. [Google Scholar] [CrossRef]
- Ye, C.; Li, H. 20 years of transgenic research in China for resistance to papaya ringspot virus. Transgenic Plant J. 2010, 4, 58–63. [Google Scholar]
- Soler, N.; Plomer, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 2012, 10, 597–608. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Mangwende, T.; Martin, D.P.; Bezuidenhout, M.; Thomson, J.A.; Rybicki, E.P. Inhibition of maize streak virus (MSV) replication by transient and transgenic expression of MSV replication-associated protein mutants. J. Gen. Virol. 2007, 88, 325–336. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Wang, Y.G.; Shen, X.J.; Li, L.; Zhou, S.F.; Li, W.C.; Fu, F.L. RNA interference-mediated resistance to maize dwarf mosaic virus. Plant Cell Tissue Organ. Cult. 2013, 113, 571–578. [Google Scholar] [CrossRef]
- Gao, L.; Ding, X.; Li, K.; Liao, W.; Zhong, Y.; Ren, R.; Liu, Z.; Adhimoolam, K.; Zhi, H. Genetics A: Characterization of soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 2015, 128, 1489–1505. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020, 18, 328. [Google Scholar] [CrossRef]
- Kundu, J.K.; Briard, P.; Hily, J.M.; Ravelonandro, M.; Scorza, R. Role of the 25–26 nt siRNA in the resistance of transgenic prunus domestica graft inoculated with plum pox virus. Virus Genes 2008, 36, 215–220. [Google Scholar] [CrossRef]
- Song, G.Q.; Sink, K.C.; Walworth, A.E.; Cook, M.A.; Allison, R.F.; Lang, G.A. Engineering cherry rootstocks with resistance to P runus necrotic ring spot virus through RNA i-mediated silencing. Plant Biotechnol. J. 2013, 11, 702–708. [Google Scholar] [CrossRef]
- Shekhawat, U.K.; Ganapathi, T.R.; Hadapad, A.B. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. J. Gen. Virol. 2012, 93, 1804–1813. [Google Scholar] [CrossRef]
- Duan, C.G.; Fang, Y.Y.; Zhou, B.J.; Zhao, J.H.; Hou, W.N.; Zhu, H.; Ding, S.W.; Guo, H.S. Suppression of arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the cucumber mosaic virus 2b protein. Plant Cell 2012, 24, 259–274. [Google Scholar] [CrossRef]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef]
- Wang, M.B.; Abbott, D.C.; Waterhouse, P.M. A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol. Plant Pathol. 2000, 1, 347–356. [Google Scholar] [CrossRef]
- Yang, Y.; Sherwood, T.A.; Patte, C.P.; Hiebert, E.; Polston, J.E. Use of tomato yellow leaf curl virus (TYLCV) rep gene sequences to engineer TYLCV resistance in tomato. Phytopathology 2004, 94, 490–496. [Google Scholar] [CrossRef][Green Version]
- Tabassum, B.; Sher, Z.; Tariq, M.; Khan, A.; Shahid, N.; Bilal, M.; Ramzan, M.; Shehzad, I.M.; Nasir, I.A.; Husnain, T. Overview of acquired virus resistance in transgenic plants. Exp. Agricult. Hortic. 2013, 2, 12–28. [Google Scholar]
- Tyagi, H.; Rajasubramaniam, S.; Rajam, M.V.; Dasgupta, I. RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants. Transgenic Res. 2008, 17, 897–904. [Google Scholar] [CrossRef]
- Anandalakshmi, R.; Pruss, G.J.; Ge, X.; Marathe, R.; Mallory, A.C.; Smith, T.H.; Vance, V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 13079–13084. [Google Scholar] [CrossRef]
- Buchmann, R.C.; Asad, S.; Wolf, J.N.; Mohannath, G.; Bisaro, D.M. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 2009, 83, 5005–5013. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, A. The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 2017, 91, e01478-16. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef]
- Li, F.; Huang, C.; Li, Z.; Zhou, X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014, 10, e1003921. [Google Scholar] [CrossRef]
- Varanda, C.M.; Materatski, P.; Campos, M.D.; Clara, M.I.E.; Nolasco, G.; Félix, M.D.R. Olive mild mosaic virus coat protein and P6 are suppressors of RNA silencing, and their silencing confers resistance against OMMV. J. Viruses 2018, 10, 416. [Google Scholar] [CrossRef]
- Senshu, H.; Yamaji, Y.; Minato, N.; Shiraishi, T.; Maejima, K.; Hashimoto, M.; Miura, C.; Neriya, Y.; Namba, S. A dual strategy for the suppression of host antiviral silencing: Two distinct suppressors for viral replication and viral movement encoded by potato virus M. J. Virol. 2011, 85, 10269–10278. [Google Scholar] [CrossRef]
- Brault, V.; Uzest, M.; Monsion, B.; Jacquot, E.; Blanc, S. Aphids as transport devices for plant viruses. J. Comptes. Rendus. Biol. 2010, 333, 524–538. [Google Scholar] [CrossRef]
- Heck, M.; Brault, V. Targeted disruption of aphid transmission: A vision for the management of crop diseases caused by Luteoviridae members. Curr. Opin. Virol. 2018, 33, 24–32. [Google Scholar] [CrossRef]
- Deshoux, M.; Monsion, B.; Uzest, M. Insect cuticular proteins and their role in transmission of phytoviruses. Curr. Opin. Virol. 2018, 33, 137–143. [Google Scholar] [CrossRef]
- Pitino, M.; Coleman, A.D.; Maffei, M.E.; Ridout, C.J.; Hogenhout, S.A. Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE 2011, 6, e25709. [Google Scholar] [CrossRef]
- Mutti, N.S.; Louis, J.; Pappan, L.K.; Pappan, K.; Begum, K.; Chen, M.S.; Park, Y.; Dittmer, N.; Marshall, J.; Reese, J.C.; et al. A protein from the salivary glands of the pea aphid, acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA 2008, 105, 9965–9969. [Google Scholar] [CrossRef]
- Guo, H.; Song, X.; Wang, G.; Yang, K.; Wang, Y.; Niu, L.; Chen, X.; Fang, R. Plant-generated artificial small RNAs mediated aphid resistance. PLoS ONE 2014, 9, e97410. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, J.; Francis, F.; Chen, J. Molecular characterization and gene silencing of Laccase 1 in the grain aphid, sitobion avenae. Arch. Insect Biochem. Physiol. 2018, 97, e21446. [Google Scholar] [CrossRef]
- Gao, X.; Luo, J.; Zhu, X.; Wang, L.; Ji, J.; Zhang, L.; Zhang, S.; Cui, J. Growth and fatty acid metabolism of aphis gossypii parasitized by the parasitic wasp lysiphlebia japonica. J. Agric. Food Chem. 2019, 67, 8756–8765. [Google Scholar] [CrossRef]
- Shoup Rupp, J.L.; Cruz, L.F.; Trick, H.N.; Fellers, J.P. RNAi-mediated, stable resistance to triticum mosaic virus in wheat. Crop Sci. 2016, 56, 1602–1610. [Google Scholar] [CrossRef]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Methods Mol. Biol. 2013, 975, 1–14. [Google Scholar]
- Zhang, H.; Wang, X.; Giroux, M.J.; Huang, L. A wheat COP9 subunit 5-like gene is negatively involved in host response to leaf rust. J. Mol. Plant Pathol 2017, 18, 125–133. [Google Scholar] [CrossRef]
- Gomariz-Fernández, A.; Sánchez-Gerschon, V.; Fourquin, C.; Ferrándiz, C. The role of SHI/STY/SRS genes in organ growth and carpel development is conserved in the distant eudicot species arabidopsis thaliana and nicotiana benthamiana. Front. Plant Sci. 2017, 8, 814. [Google Scholar] [CrossRef]
- Li, R.; Reed, D.W.; Liu, E.; Nowak, J.; Pelcher, L.E.; Page, J.E.; Covello, P.S. Functional genomic analysis of alkaloid biosynthesis in Hyoscyamus niger reveals a cytochrome P450 involved in littorine rearrangement. Chem. Biol. 2006, 13, 513–520. [Google Scholar] [CrossRef]
- Deng, X.; Elomaa, P.; Nguyen, C.X.; Hytönen, T.; Valkonen, J.P.; Teeri, T.H. Virus-induced gene silencing for asteraceae—A reverse genetics approach for functional genomics in Gerbera hybrida. Plant Biotechnol. J. 2012, 10, 970–978. [Google Scholar] [CrossRef]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M.; et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. J. Virol. 2009, 386, 407–416. [Google Scholar] [CrossRef]
- Sasaki, S.; Yamagishi, N.; Yoshikawa, N. Efficient virus-induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors. Plant Methods 2011, 7, 15. [Google Scholar] [CrossRef]
- Agüero, J.; del Carmen Vives, M.; Velázquez, K.; Pina, J.A.; Navarro, L.; Moreno, P.; Guerri, J. Effectiveness of gene silencing induced by viral vectors based on Citrus leaf blotch virus is different in Nicotiana benthamiana and citrus plants. J. Virol. 2014, 460, 154–164. [Google Scholar] [CrossRef]
- Nagamatsu, A.; Masuta, C.; Senda, M.; Matsuura, H.; Kasai, A.; Hong, J.S.; Kitamura, K.; Abe, J.; Kanazawa, A. Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol. J. 2007, 5, 778–790. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Mei, Y.; Zhang, C.; Kernodle, B.M.; Hill, J.H.; Whitham, S.A. A foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiol. 2016, 171, 760–772. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, S.; Wei, Y.; Zhang, M.-Q. RNA Interference: Promising Approach to Combat Plant Viruses. Int. J. Mol. Sci. 2022, 23, 5312. https://doi.org/10.3390/ijms23105312
Akbar S, Wei Y, Zhang M-Q. RNA Interference: Promising Approach to Combat Plant Viruses. International Journal of Molecular Sciences. 2022; 23(10):5312. https://doi.org/10.3390/ijms23105312
Chicago/Turabian StyleAkbar, Sehrish, Yao Wei, and Mu-Qing Zhang. 2022. "RNA Interference: Promising Approach to Combat Plant Viruses" International Journal of Molecular Sciences 23, no. 10: 5312. https://doi.org/10.3390/ijms23105312
APA StyleAkbar, S., Wei, Y., & Zhang, M.-Q. (2022). RNA Interference: Promising Approach to Combat Plant Viruses. International Journal of Molecular Sciences, 23(10), 5312. https://doi.org/10.3390/ijms23105312





