Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993
Abstract
:1. Introduction
2. Results
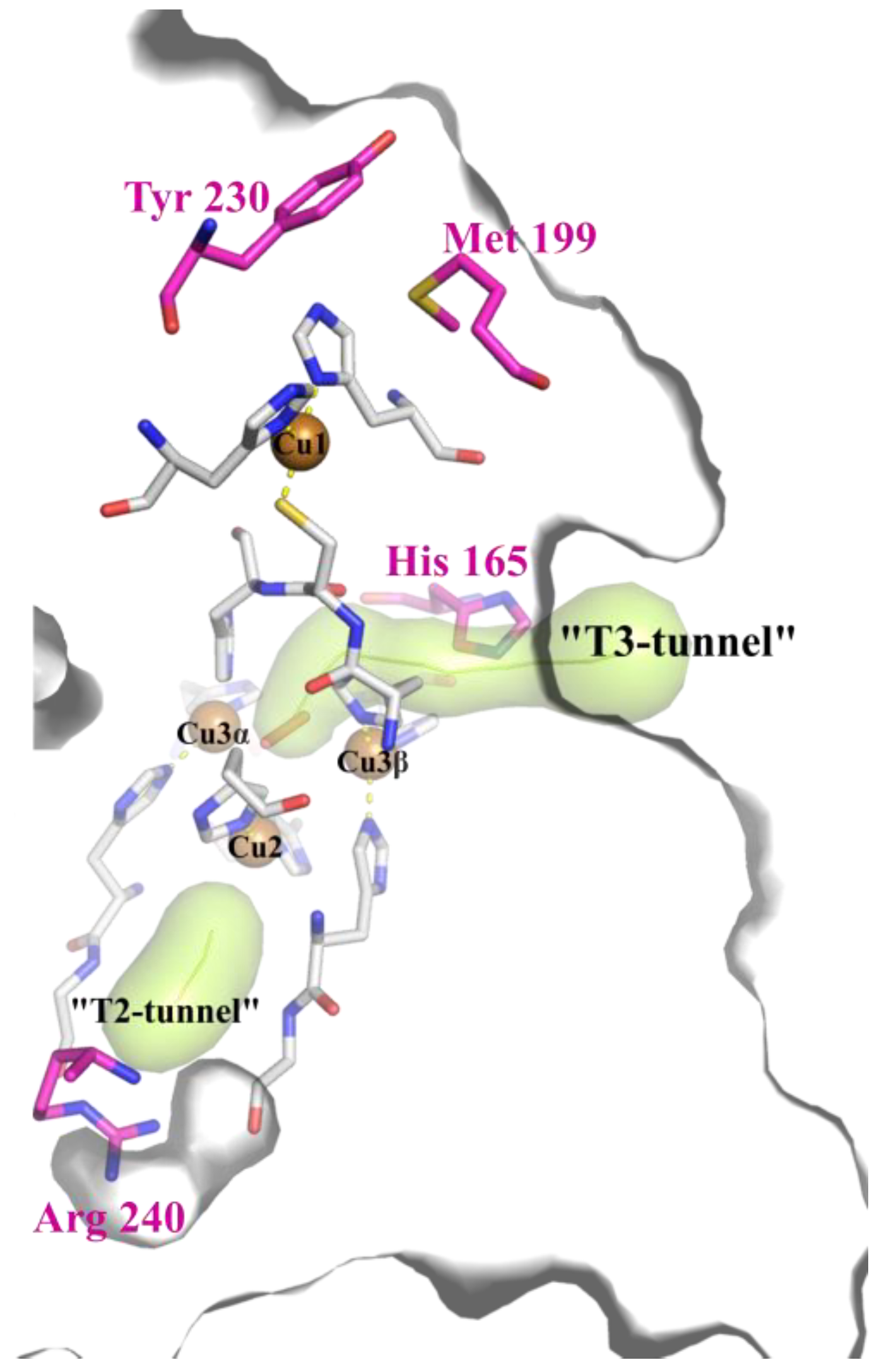
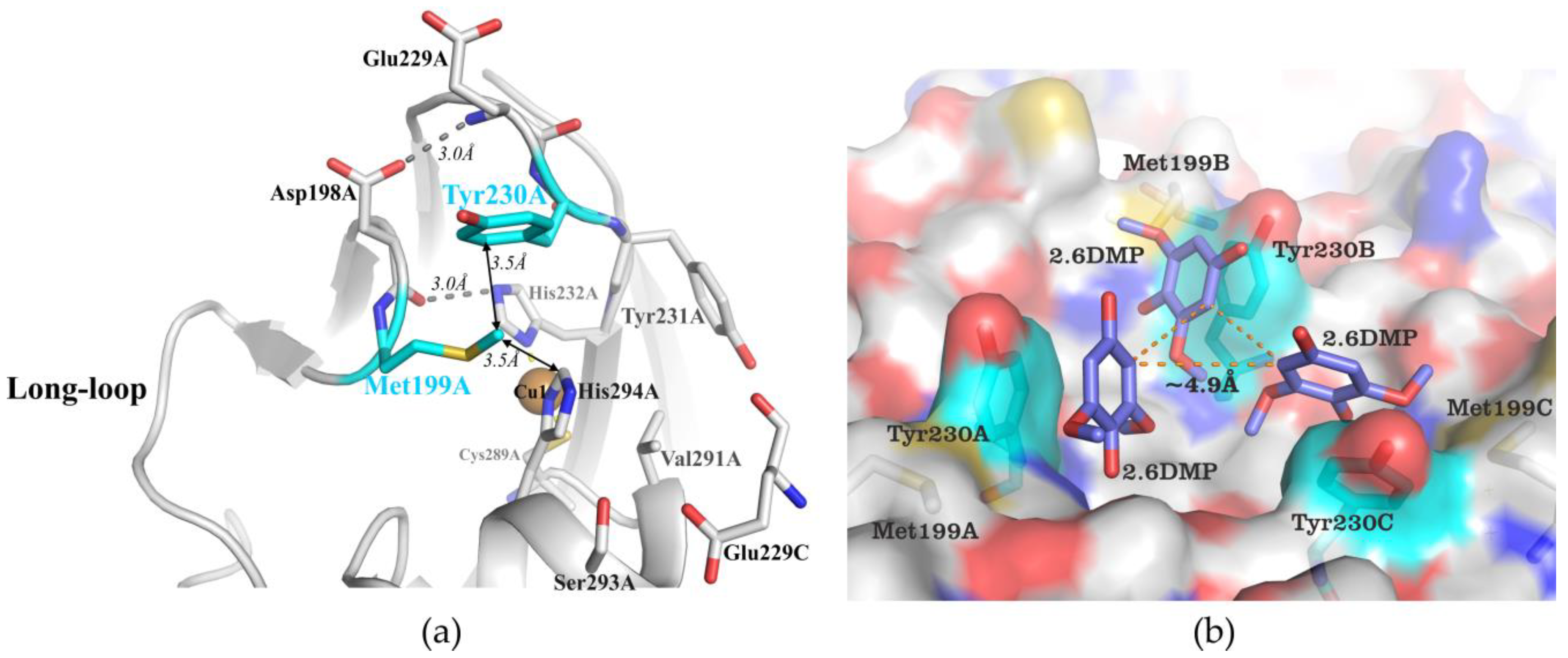
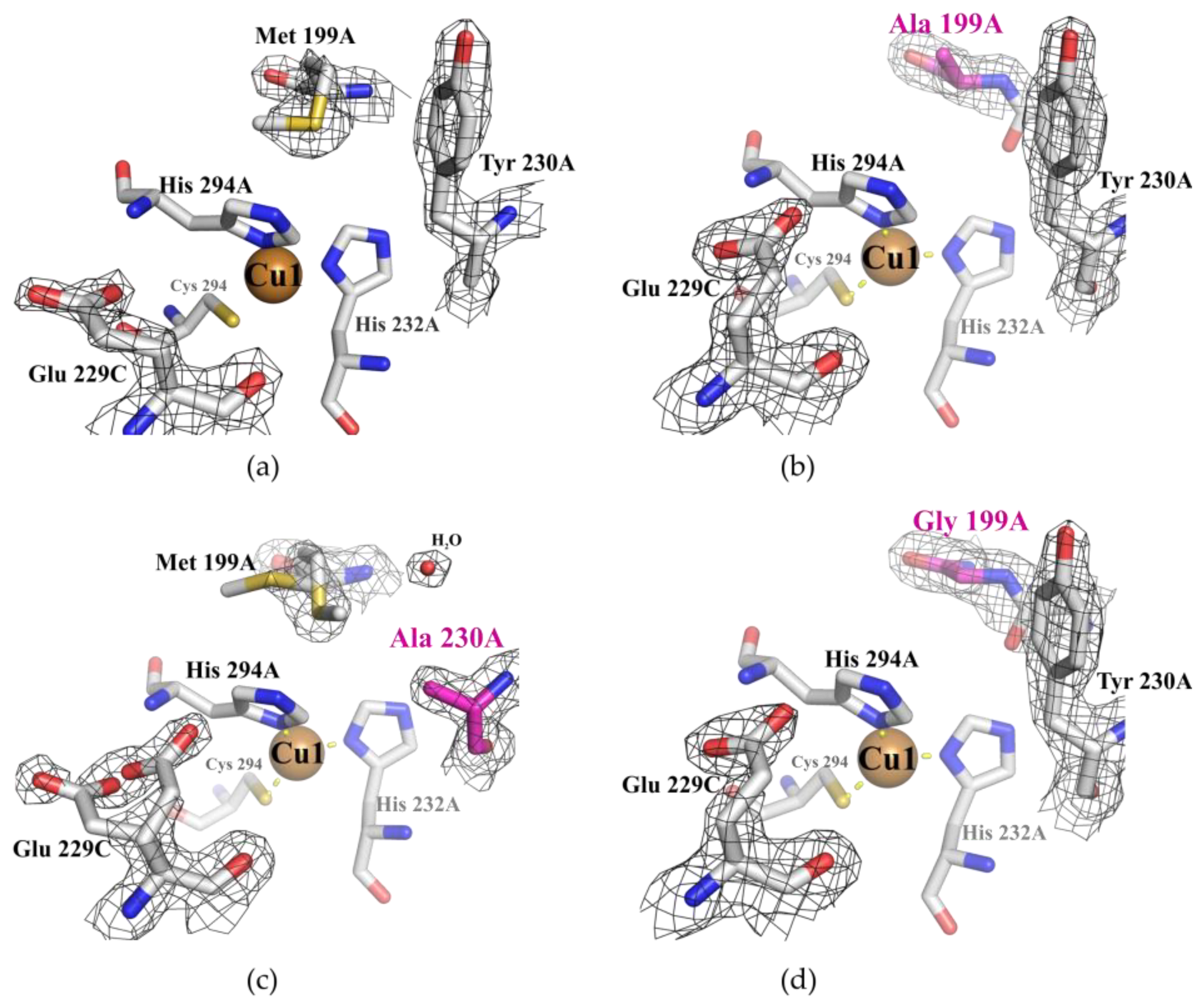
2.1. Structural Analysis of the Wild Type and Mutant Enzymes
2.2. Kinetic Analysis of the Wild Type and Mutant Enzymes
2.3. Inhibition Assays Using Sodium Azide
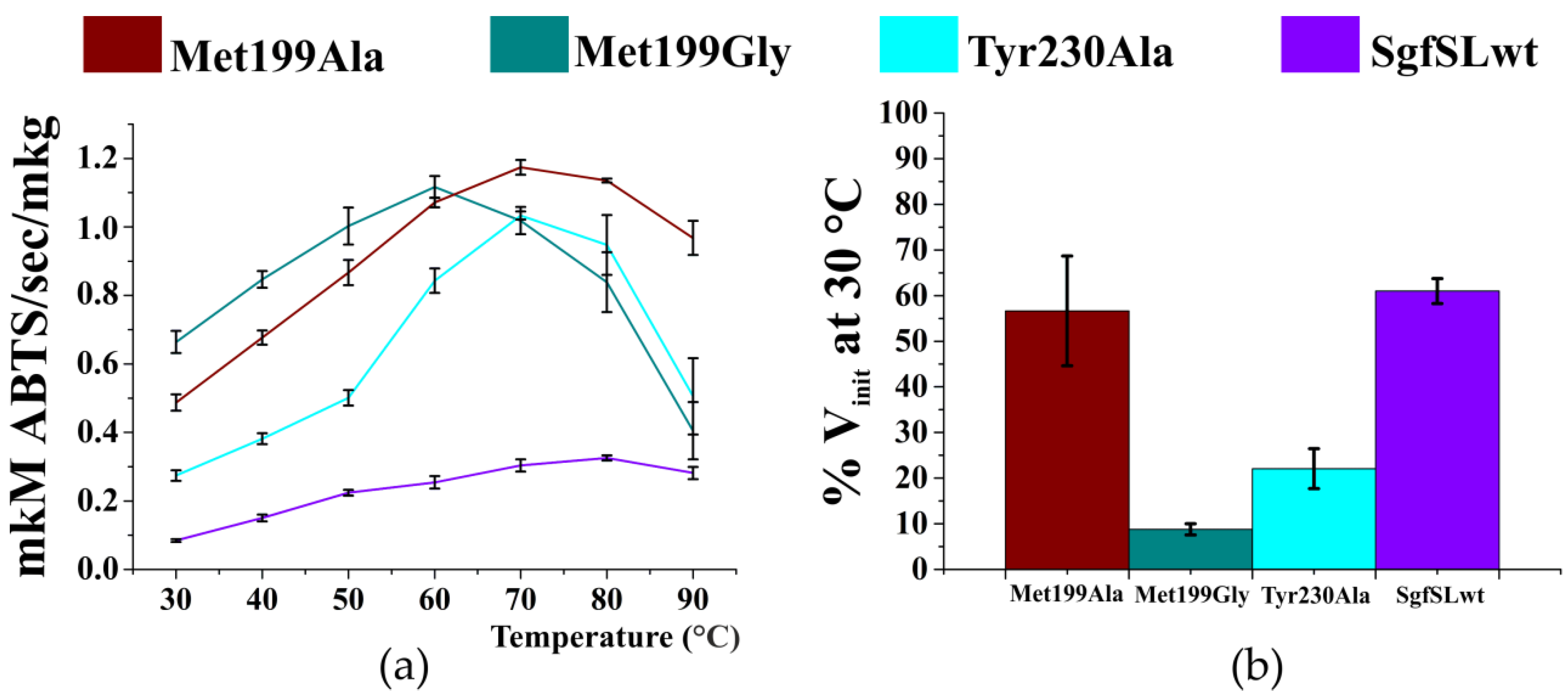
2.4. Optimal Temperature and Thermal Stability of SgfSL Variants
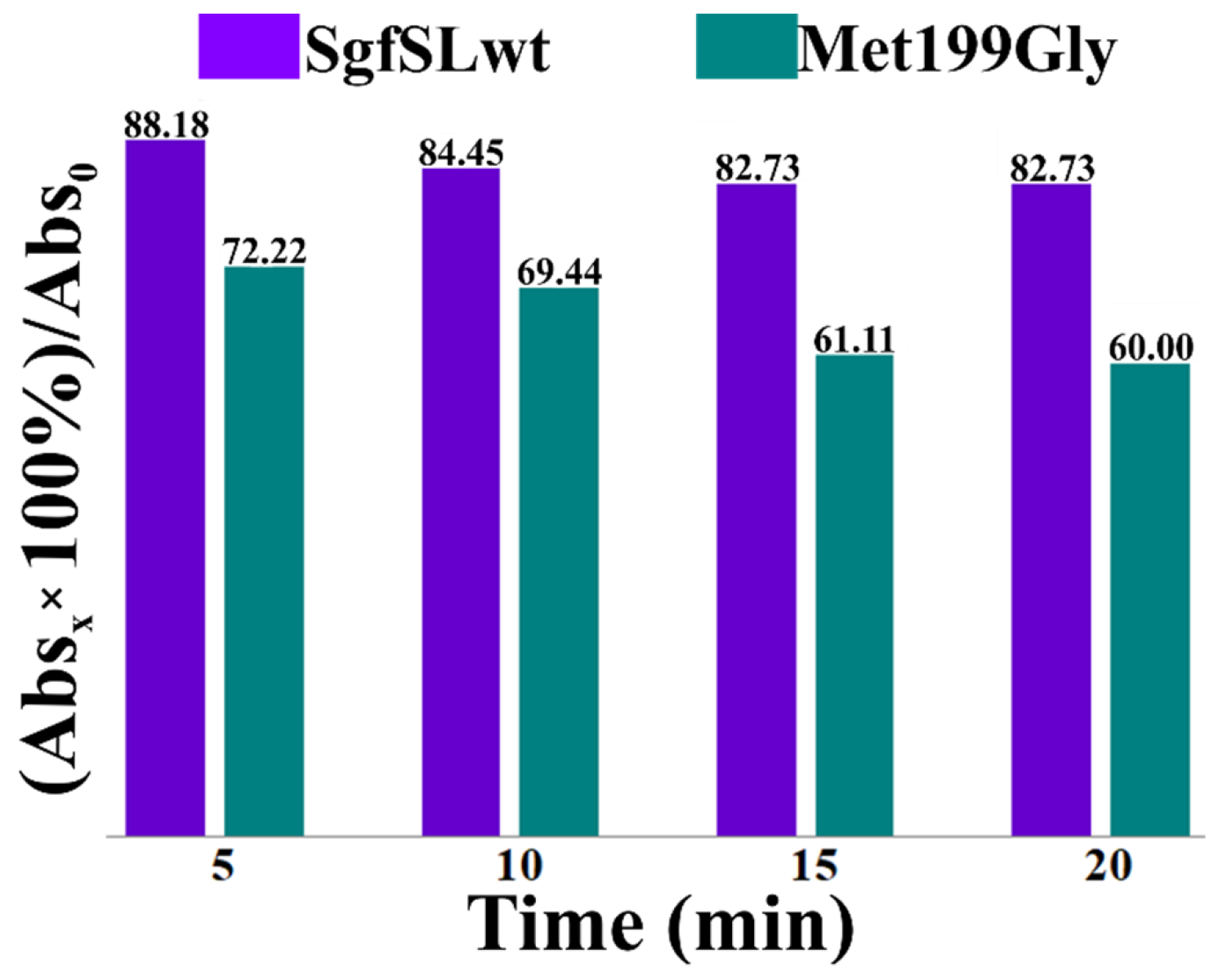
2.5. Comparative Analysis of the Changes in Intensities of Cu1 Absorption Peaks of SgfSLwt and Met199Gly after Heating at 80 °C
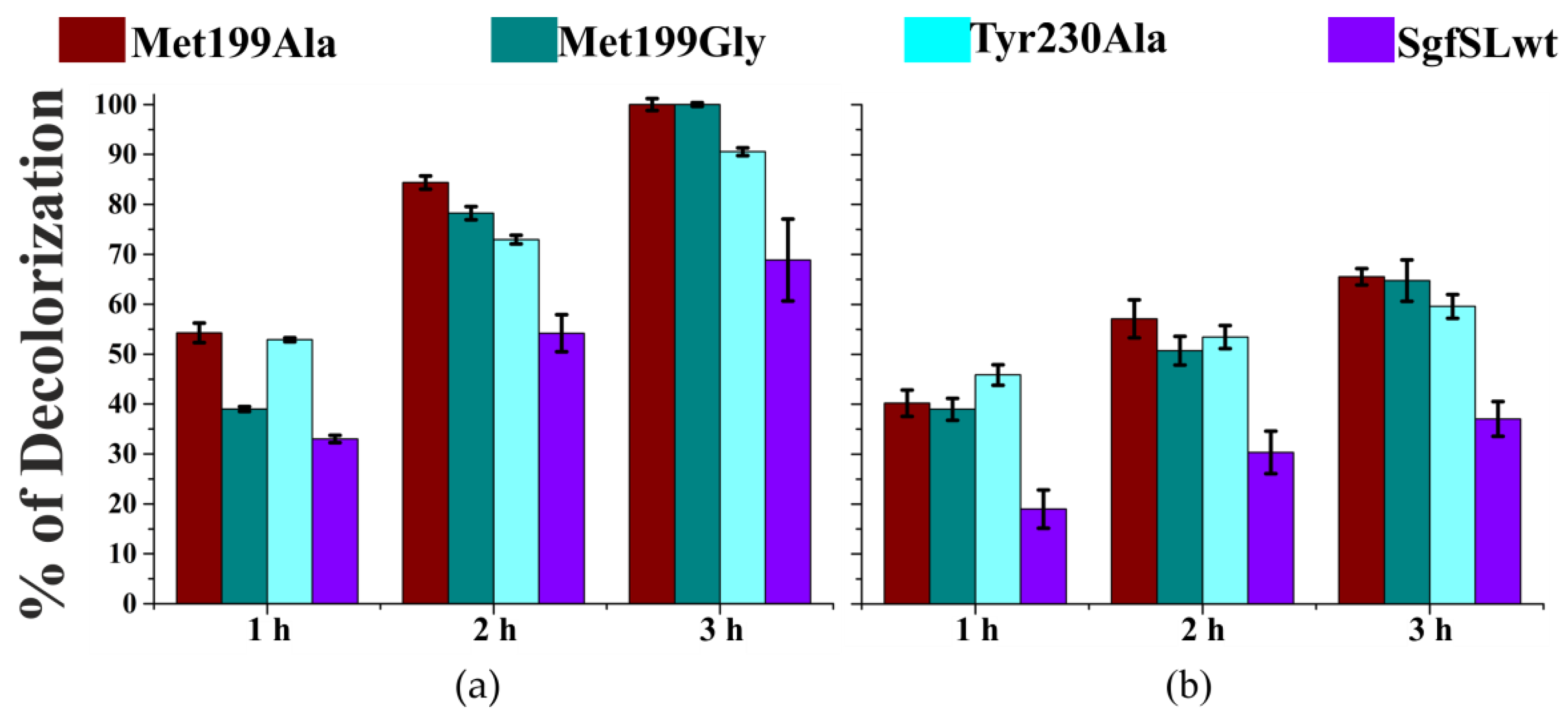
2.6. Decolorization of Industrial Dyes by SgfSLwt and Mutant Forms with Replacements in SBP
3. Discussion
3.1. Mutations in the Substrate-Binding Pocket of the 2D Laccase SgfSL Affect Its Activity toward Different Substrates
3.2. Changing the Geometry of the Substrate-Binding Pocket by Met199Gly and Tyr230Ala Mutations Reduces the Activity of SgfSL after High-Temperature Incubation
3.3. The Revision of the Role of His165 Located in the T3 Tunnel in the 2D Laccases Functioning
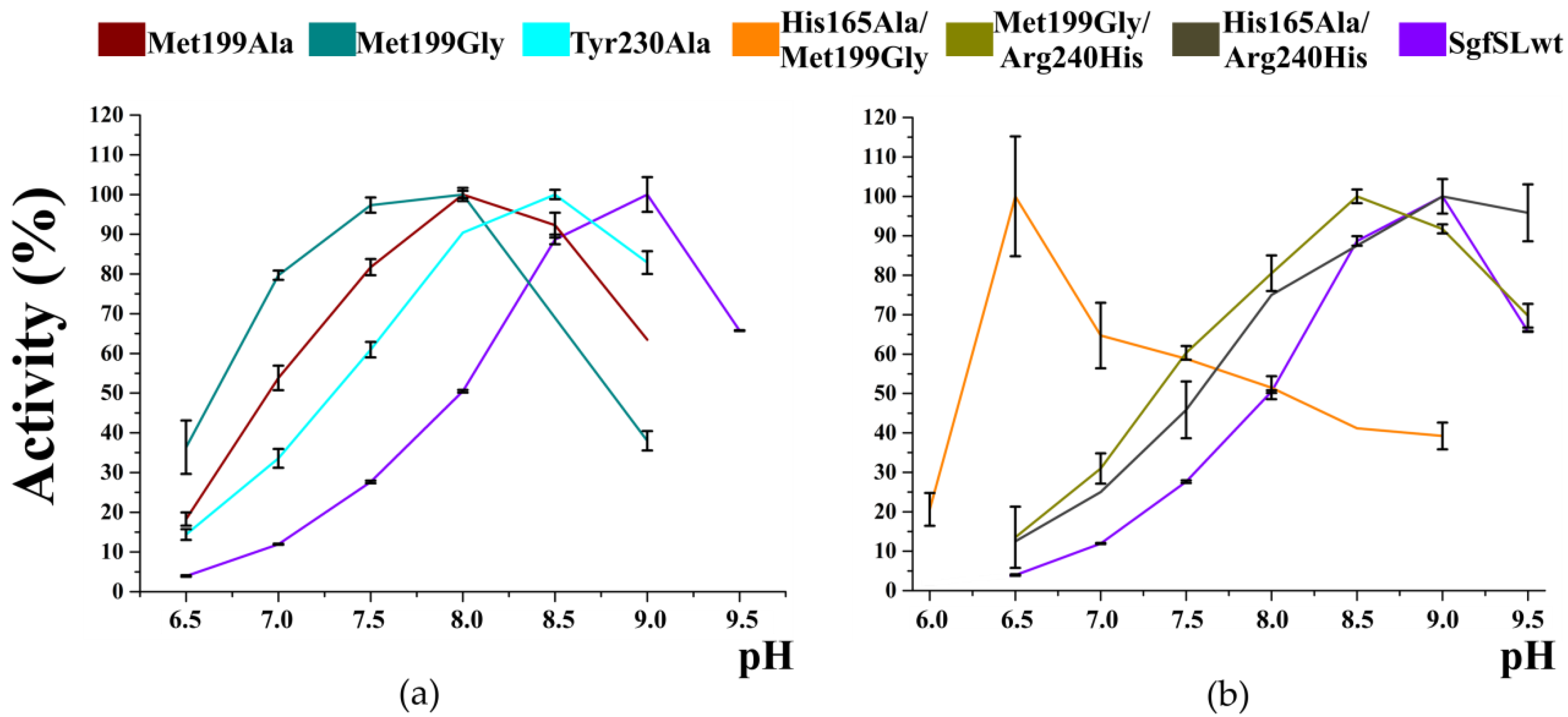
3.4. The Combination of the Mutation in SBP and T2 Tunnels Increased the SgfSL Activity at Alkaline pH
4. Materials and Methods
4.1. Plasmid Construction
- Met199Ala_For:5′-CACACCATCGTGTTCAATGACGCGACAATCAACAACAGGCCCGCC-3′
- Met199Ala_Rev:5′-GGCGGGCCTGTTGTTGATTGTCGCGTCATTGAACACGATGGTGTG-3′
- Met199Gly_For:5′-ACCCACACCATTGTGTTCAACGACGGTACGATCAACAACAGGCCC-3′
- Met199Gly_Rev:5′-GGGCCTGTTGTTGATCGTACCGTCGTTGAACACAATGGTGTGGGT-3′
- Tyr230Ala_For:5′-TCATGATCACGCACGGCGAGGCCTACCACACCTTCCACAT-3′
- Tyr230Ala_Rev:5′-ATGTGGAAGGTGTGGTAGGCCTCGCCGTGCGTGATCATGA-3′
- For:5′-AGATCTGGGTACCATCGAGGGTCGTGCAGGAGCAGCGCCCGCCGGGGGA-3′
- Rev:5′-GGCTGCAAGCTTTCAGTGAGCGTGCTCCTGCGGGT-3′
4.2. Purification of SgfSLwt and Mutants
4.3. Activity Assays
4.3.1. Kinetic Parameters of SgfSLwt and Mutants
4.3.2. Inhibition by Sodium Azide
4.3.3. Optimal Temperature for ABTS Oxidation, Thermal Stability, and Absorbance Change Determination after Heat Treatment of SgfSLwt and Mutants
4.3.4. Decolorization of Dyes
4.4. Crystallization and Crystallography
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TNC | TriNuclear Center |
| 3D | Three-domain |
| 2D | Two-domain |
| SgfSL | Streptomyces griseoflavus Small Laccase |
References
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Thurston, C.F. The structure and function of fungal laccases. Microbiology 1994, 140, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Zerva, A.; Simić, S.; Topakas, E.; Nikodinovic-Runic, J. Applications of microbial laccases: Patent review of the past decade (2009–2019). Catalysts 2019, 9, 1023. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H. Chemistry of Lacquer (Urushi). J. Chem. Soc. Trans. 1883, 43, 472–486. [Google Scholar] [CrossRef] [Green Version]
- El-Bendary, M.A.; Ezzat, S.M.; Ewais, E.A.; Al-Zalama, M.A. Optimization of spore laccase production by Bacillus amyloliquefaciens isolated from wastewater and its potential in green biodecolorization of synthetic textile dyes. Prep. Biochem. Biotechnol. 2021, 51, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhao, M.; Wang, T.N.; Zhao, L.Y.; Du, M.H.; Li, T.L.; Li, D. Bin Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresour. Technol. 2012, 115, 35–40. [Google Scholar] [CrossRef]
- Claus, H. Laccases: Structure, reactions, distribution. Micron 2004, 35, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Matera, I.; Gullotto, A.; Tilli, S.; Ferraroni, M.; Scozzafava, A.; Briganti, F. Crystal structure of the blue multicopper oxidase from the white-rot fungus Trametes trogii complexed with p-toluate. Inorg. Chim. Acta 2008, 361, 4129–4137. [Google Scholar] [CrossRef]
- Kallio, J.P.; Auer, S.; Jänis, J.; Andberg, M.; Kruus, K.; Rouvinen, J.; Koivula, A.; Hakulinen, N. Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds. J. Mol. Biol. 2009, 392, 895–909. [Google Scholar] [CrossRef]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry 2002, 41, 7325–7333. [Google Scholar] [CrossRef]
- Durão, P.; Bento, I.; Fernandes, A.T.; Melo, E.P.; Lindley, P.F.; Martins, L.O. Perturbations of the T1 copper site in the CotA laccase from Bacillus subtilis: Structural, biochemical, enzymatic and stability studies. J. Biol. Inorg. Chem. 2006, 11, 514–526. [Google Scholar] [CrossRef]
- Osipov, E.; Polyakov, K.; Kittl, R.; Shleev, S.; Dorovatovsky, P.; Tikhonova, T.; Hann, S.; Ludwig, R.; Popov, V. Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2913–2923. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, K.; Kogi, H.; Tsujimura, S.; Sakurai, T. Modifications of laccase activities of copper efflux oxidase, CueO by synergistic mutations in the first and second coordination spheres of the type I copper center. Biochem. Biophys. Res. Commun. 2013, 431, 393–397. [Google Scholar] [CrossRef]
- Gunne, M.; Höppner, A.; Hagedoorn, P.L.; Urlacher, V.B. Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus. FEBS J. 2014, 281, 4307–4318. [Google Scholar] [CrossRef] [PubMed]
- Toscano, M.D.; De Maria, L.; Lobedanz, S.; Østergaard, L.H. Optimization of a Small Laccase by Active-Site Redesign. ChemBioChem 2013, 14, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Olbrich, A.C.; Schild, J.N.; Urlacher, V.B. Correlation between the T1 copper reduction potential and catalytic activity of a small laccase. J. Inorg. Biochem. 2019, 201, 110843. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Solomon, E.I. Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. 2015, 72, 869–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabdulkhakov, A.; Kolyadenko, I.; Kostareva, O.; Mikhaylina, A.; Oliveira, P.; Tamagnini, P.; Lisov, A.; Tishchenko, S. Investigations of accessibility of T2/T3 copper center of two-domain laccase from streptomyces griseoflavus ac-993. Int. J. Mol. Sci. 2019, 20, 3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabdulkhakov, A.; Kolyadenko, I.; Oliveira, P.; Tamagnini, P.; Mikhaylina, A.; Tishchenko, S. The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993. J. Biomol. Struct. Dyn. 2021, 0, 1–8. [Google Scholar] [CrossRef]
- Mateljak, I.; Monza, E.; Lucas, M.F.; Guallar, V.; Aleksejeva, O.; Ludwig, R.; Leech, D.; Shleev, S.; Alcalde, M. Increasing Redox Potential, Redox Mediator Activity, and Stability in a Fungal Laccase by Computer-Guided Mutagenesis and Directed Evolution. ACS Catal. 2019, 9, 4561–4572. [Google Scholar] [CrossRef] [Green Version]
- Yin, Q.; Zhou, G.; Peng, C.; Zhang, Y.; Kües, U.; Liu, J.; Xiao, Y.; Fang, Z. The first fungal laccase with an alkaline pH optimum obtained by directed evolution and its application in indigo dye decolorization. AMB Express 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Zhao, N.; Yin, Q.; Zhang, T.; Xu, X.; Fang, W.; Hong, Y.; Fang, Z.; Xiao, Y. Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour. Technol. 2014, 162, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Machczynski, M.C.; Vijgenboom, E.; Samyn, B.; Canters, G.W. Characterization of SLAC: A small laccase from Streptomyces coelicolor with unprecedented activity. Protein Sci. 2004, 13, 2388–2397. [Google Scholar] [CrossRef] [Green Version]
- Molina-Guijarro, J.M.; Pérez Torres, J.; Muñoz-Dorado, J.; Guillén Carretero, F.; Moya Lobo, R.; Herńndez Cutuli, M.; Arias Fernández, M.E. Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea. Int. Microbiol. 2009, 12, 13–21. [Google Scholar] [CrossRef]
- Gunne, M.; Urlacher, V.B. Characterization of the Alkaline Laccase Ssl1 from Streptomyces sviceus with Unusual Properties Discovered by Genome Mining. PLoS ONE 2012, 7, e52360. [Google Scholar] [CrossRef] [PubMed]
- Trubitsina, L.I.; Tishchenko, S.V.; Gabdulkhakov, A.G.; Lisov, A.V.; Zakharova, M.V.; Leontievsky, A.A. Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes. Biochimie 2015, 112, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Durão, P.; Silva, C.S.; Pereira, M.M.; Todorovic, S.; Hildebrandt, P.; Bento, I.; Lindley, P.F.; Martins, L.O. The role of Glu498 in the dioxygen reactivity of CotA-laccase from Bacillus subtilis. Dalton Trans. 2010, 39, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Damas, J.M.; Chen, Z.; Brissos, V.; Martins, L.O.; Soares, C.M.; Lindley, P.F.; Bento, I. The role of Asp116 in the reductive cleavage of dioxygen to water in CotA laccase: Assistance during the proton-transfer mechanism. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 186–193. [Google Scholar] [CrossRef]
- Heinzkill, M.; Bech, L.; Halkier, T.; Schneider, P.; Anke, T. Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl. Environ. Microbiol. 1998, 64, 1601–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabdulkhakov, A.G.; Kostareva, O.S.; Kolyadenko, I.A.; Mikhaylina, A.O.; Trubitsina, L.I.; Tishchenko, S.V. Incorporation of Copper Ions into T2/T3 Centers of Two-Domain Laccases. Mol. Biol. 2018, 52, 23–29. [Google Scholar] [CrossRef]
- Vandeyar, M.A.; Weiner, M.P.; Hutton, C.J.; Batt, C.A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene 1988, 65, 129–133. [Google Scholar] [CrossRef]
- Heinfling, A.; Martínez, M.J.; Martínez, A.T.; Bergbauer, M.; Szewzyk, U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol. Lett. 1998, 165, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef]
- Nguyen, H.H.T.; Nakagawa, K.H.; Hedman, B.; Elliott, S.J.; Lidstrom, M.E.; Hodgson, K.O.; Chan, S.I. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu(I) ions and their implications. J. Am. Chem. Soc. 1996, 118, 12766–12776. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; Herrera De Los Santos, M.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Fact. 2019, 18, 1–33. [Google Scholar] [CrossRef]
- D’Arcy, A.; Bergfors, T.; Cowan-Jacob, S.W.; Marsh, M. Microseed matrix screening for optimization in protein crystallization: What have we learned? Acta Crystallogr. Sect. Struct. Biol. Commun. 2014, 70, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Kraft, P.; Bergamaschi, A.; Broennimann, C.; Dinapoli, R.; Eikenberry, E.F.; Henrich, B.; Johnson, I.; Mozzanica, A.; Schlepütz, C.M.; Willmott, P.R.; et al. Performance of single-photon-counting PILATUS detector modules. J. Synchrotron Radiat. 2009, 16, 368–375. [Google Scholar] [CrossRef]
- Mueller, U.; Förster, R.; Hellmig, M.; Huschmann, F.U.; Kastner, A.; Malecki, P.; Pühringer, S.; Röwer, M.; Sparta, K.; Steffien, M.; et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 2015, 130, 1–10. [Google Scholar] [CrossRef]
- Rigaku, O.D. CrysAlis PRO; Rigaku Oxford Diffraction: Yarnton, UK, 2019. [Google Scholar]
- Kabsch, W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System v2.5. Available online: https://pymol.org/2/ (accessed on 18 December 2021).
| Met199Ala | Met199Gly | Tyr230Ala | His165Ala/ Met199Gly | His165Ala/ Arg240His | Met199Gly/ Arg240His | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P 21 | P 21 | P 21 | P 21 | P 21 | P 21 |
| Cell parameters: | ||||||
| a,b,c (Å) | 74.98, 94.12, 119.73 | 75.18, 94.36, 119.79 | 77.26, 94.98, 116.27 | 76.23, 94.59, 120.87 | 74.76, 94.29 120.48 | 74.06, 93.72, 119.23 |
| α = γ = 90, β (°) | 91.16 | 91.14 | 91.67 | 91.39 | 91.27 | 91.30 |
| Collection temperature (K) | 100 | 100 | 100 | 295 | 100 | 100 |
| Resolution (Å) | 50.00–1.75 (1.80–1.75) a | 50.00–1.75 (1.80–1.75) a | 50.00–1.60 (1.64–1.60) a | 25.00–2.20 (2.28–2.20) a | 50.00–1.30 (1.38-1.30) a | 25.0–1.85 (1.92–1.85) a |
| Total No. Of reflections | 1,135,633 (80,057) | 1,148,856 (86,851) | 1,241,259 (93,269) | 317,814 (31,656) | 2,743,665 (406,573) | 484,277 (44,747) |
| No. Of unique reflections | 167,091 (12,290) | 168,209 (12,404) | 220,573 (16,337) | 85,543 (8557) | 405,781 (64,031) | 138,143 (13,817) |
| Rmerge (%) | 6.8 (110.1) | 6.1 (172.2) | 9.2 (103.5) | 16.9 (59.7) | 5.5 (135.8) | 15.1 (63.6) |
| I/σ(I) | 14.65 (1.53) | 14.31 (1.02) | 10.93 (1.55) | 6.15 (1.36) | 18.19 (1.28) | 6.96 (1.06) |
| Completeness (%) | 99.9 (99.6) | 100.0 (100.0) | 99.9 (99.9) | 98.1 (99.3) | 99.1 (97.0) | 99.2 (98.0) |
| CC1/2 | 0.99 (0.64) | 0.99 (0.50) | 0.99 (68.3) | 0.98 (0.52) | 1.00 (0.55) | 0.98 (0.51) |
| Redundancy | 6.79 (6.51) | 6.83 (7.00) | 5.63 (5.71) | 3.7 (3.7) | 6.76 (6.35) | 3.5 (3.2) |
| Refinement | ||||||
| Resolution (Å) | 47.24–1.75 (1.77–1.75) | 47.18–1.75 (1.80–1.75) | 47.10–1.60 (1.62–1.60) | 24.74–2.20 (2.23–2.20) | 47.41-1.30 (1.34-1.30) | 24.68–1.85 (1.87–1.85) |
| No. reflections | 167,072 (5530) | 168,176 (12,756) | 220,558 (7050) | 85,458 (3541) | 405,693 (12,826) | 138,068 (4315) |
| Rwork (%) | 13.18 (21.64) | 16.58 (31.81) | 13.53 (30.44) | 15.73 (23.87) | 13.26 (31.20) | 18.32 (31.76) |
| Rfree (%) | 16.93 (29.38) | 20.13 (31.71) | 16.53 (33.62) | 18.85 (25.93) | 15.69 (34.01) | 22.35 (37.02) |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.006 | 0.007 | 0.006 | 0.008 |
| Bond angles (°) | 0.781 | 0.901 | 0.888 | 0.890 | 0.867 | 0.968 |
| Ramachandran plot (%) | ||||||
| Most favored | 98.37 | 98.00 | 98.55 | 98.18 | 98.79 | 97.82 |
| Additionally allowed | 1.63 | 2.00 | 1.45 | 1.82 | 1.21 | 2.18 |
| Generously allowed | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| PDB ID | 7PFR | 7PES | 7PEN | 7PU0 | 7PUH | 7PTM |
| Substrate | Protein | pHopt | Km(mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| ABTS | SgfSLwt | 4.0 | 0.36 ± 0.07 | 15 ± 1 | 42 |
| Met199Ala | 4.0 | 0.32 ± 0.05 | 33 ± 2 | 102 | |
| Met199Gly | 4.0 | 0.20 ± 0.06 | 45 ± 7 | 226 | |
| Tyr230Ala | 3.5 | 0.17 ± 0.04 | 29 ± 3 | 169 | |
| His165Ala/Met199Gly | 3.5 | 0.09 ± 0.01 | 9 ± 0 | 95 | |
| Met199Gly/Arg240His | 3.5 | 0.10 ± 0.015 | 20 ± 1 | 203 | |
| His165Ala/Arg240His | 3.5 | 0.075 ± 0.03 | 6 ± 0 | 74 | |
| 2.6-DMP | SgfSLwt | 9.0 | 0.32 ± 0.08 | 0.35 ± 0.02 | 1.09 |
| Met199Ala | 8.0 | 0.83 ± 0.08 | 1.30 ± 0.05 | 1.55 | |
| Met199Gly | 8.0 | 0.67 ± 0.12 | 3.39 ± 0.17 | 5.07 | |
| Tyr230Ala | 8.5 | 0.76 ± 0.03 | 1.68 ± 0.07 | 2.22 | |
| His165Ala/Met199Gly | 6.5 | 0.28 ± 0.05 | 0.31 ± 0.01 | 1.11 | |
| Met199Gly/Arg240His | 8.5 | 0.25 ± 0.02 | 4.18 ± 0.09 | 16.51 | |
| His165Ala/Arg240His | 9.0 | 0.15 ± 0.016 | 0.11 ± 0.004 | 0.74 | |
| K4[Fe(CN)6] | SgfSLwt | 4.0 | 0.10 ± 0.02 | 38 ± 2 | 383 |
| Met199Ala | 4.0 | 0.35 ± 0.02 | 54 ± 1 | 155 | |
| Met199Gly | 4.0 | 0.25 ± 0.02 | 66 ± 2 | 261 | |
| Tyr230Ala | 4.0 | 0.31 ± 0.01 | 86 ± 1 | 278 | |
| His165Ala/Met199Gly | 4.0 | 0.24 ± 0.04 | 35 ± 2 | 144 | |
| Met199Gly/Arg240His | 4.0 | 0.17 ± 0.02 | 48 ± 3 | 279 | |
| His165Ala/Arg240His | 4.0 | 0.18 ± 0.02 | 29 ± 1 | 160 |
| Protein | The Residual Activity (%) in the Presence of 10 mM of NaN3 |
|---|---|
| SgfSLwt | 23 ± 1 |
| Met199Ala | 13 ± 2 |
| Met199Gly | 20 ± 1 |
| Tyr230Ala | 20 ± 3 |
| His165Ala/Met199Gly | 116 ± 3 |
| His165Ala/Arg240His | 78 ± 5 |
| Met199Gly/Arg240His | 25 ± 1 |
| Protein | Reaction Rate (%) at Different pH | Vm | ||||||
|---|---|---|---|---|---|---|---|---|
| 6.5 | 7.0 | 7.5 | 8.0 | 8.5 | 9.0 | 9.5 | ||
| SgfSLwt | 3.95 | 11.94 | 27.62 | 50.47 | 88.68 | 100.00 | 65.73 | 1.1 × 10−2 |
| Arg240His | 3.62 | 7.77 | 11.63 | 36.05 | 75.58 | 100.00 | 82.56 | 3.3 × 10−2 |
| His165Ala | 9.44 | 25.42 | 46.25 | 73.37 | 96.97 | 100.00 | 76.39 | 6.0 × 10−3 |
| His165Ala/Arg240His | 12.50 | 25.00 | 45.83 | 75.00 | 87.50 | 100.00 | 95.83 | 2.8 × 10−3 |
| Met199Ala | 18.27 | 53.85 | 81.73 | 100.00 | 92.31 | 63.46 | - | 2.3 × 10−2 |
| Met199Gly | 36.36 | 79.68 | 97.33 | 100.00 | 68.98 | 37.97 | - | 8.3 × 10−2 |
| Tyr230Ala | 14.38 | 33.56 | 60.96 | 90.41 | 100.00 | 82.88 | - | 3.7 × 10−2 |
| His165Ala/Met199Gly | 100.00 | 64.71 | 58.82 | 51.47 | 41.18 | 39.22 | - | 6.9 × 10−3 |
| Met199Gly/Arg240His | 13.53 | 30.96 | 60.32 | 80.50 | 100.00 | 91.74 | 69.72 | 7.7 × 10−2 |

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolyadenko, I.; Scherbakova, A.; Kovalev, K.; Gabdulkhakov, A.; Tishchenko, S. Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993. Int. J. Mol. Sci. 2022, 23, 65. https://doi.org/10.3390/ijms23010065
Kolyadenko I, Scherbakova A, Kovalev K, Gabdulkhakov A, Tishchenko S. Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993. International Journal of Molecular Sciences. 2022; 23(1):65. https://doi.org/10.3390/ijms23010065
Chicago/Turabian StyleKolyadenko, Ilya, Anastasia Scherbakova, Kirill Kovalev, Azat Gabdulkhakov, and Svetlana Tishchenko. 2022. "Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993" International Journal of Molecular Sciences 23, no. 1: 65. https://doi.org/10.3390/ijms23010065
APA StyleKolyadenko, I., Scherbakova, A., Kovalev, K., Gabdulkhakov, A., & Tishchenko, S. (2022). Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993. International Journal of Molecular Sciences, 23(1), 65. https://doi.org/10.3390/ijms23010065






