Abstract
The major biological methyl donor, S-adenosylmethionine (adoMet) synthesis occurs mainly in the liver. Methionine adenosyltransferase 1A (MAT1A) and glycine N-methyltransferase (GNMT) are two key enzymes involved in the functional implications of that variation. We collected 42 RNA-seq data from paired hepatocellular carcinoma (HCC) and its adjacent normal liver tissue from the Cancer Genome Atlas (TCGA). There was no mutation found in MAT1A or GNMT RNA in the 42 HCC patients. The 11,799 genes were annotated in the RNA-Seq data, and their expression levels were used to investigate the phenotypes of low MAT1A and low GNMT by Gene Set Enrichment Analysis (GSEA). The REACTOME_TRANSLATION gene set was enriched and visualized in a heatmap along with corresponding differences in gene expression between low MAT1A versus high MAT1A and low GNMT versus high GNMT. We identified 43 genes of the REACTOME_TRANSLATION gene set that are powerful prognosis factors in HCC. The significantly predicted genes were referred into eukaryotic translation initiation (EIF3B, EIF3K), eukaryotic translation elongation (EEF1D), and ribosomal proteins (RPs). Cell models expressing various MAT1A and GNMT proved that simultaneous restoring the expression of MAT1A and GNMT decreased cell proliferation, invasion, as well as the REACTOME_TRANSLATION gene EEF1D, consistent with a better prognosis in human HCC. We demonstrated new findings that downregulation or defect in MAT1A and GNMT genes can enrich the protein-associated translation process that may account for poor HCC prognosis. This is the first study demonstrated that MAT1A and GNMT, the 2 key enzymes involved in methionine cycle, could attenuate the function of ribosome translation. We propose a potential novel mechanism by which the diminished GNMT and MAT1A expression may confer poor prognosis for HCC.
1. Introduction
Hepatocellular carcinoma (HCC) is the most frequent primary liver cancer, and incidence of about 10.1 cases per 100,000 persons, and HCC is ranked as the sixth most common neoplasm and the third leading cause of cancer death [1]. HCC occurs mainly in the context of cirrhosis, hepatitis B or C virus infection, or nonalcoholic steatohepatitis, and its incidence is expected to increase [2]. Perturbations in folate dependent methylation pathways have been associated with cancer occurrence [3] and many human pathological conditions [4,5]. Genes involved in the folate-mediated one carbon (1C) metabolism have been important therapeutic targets for numerous human diseases [6,7,8,9] including HCC [10,11]. Enzymes of methionine cycle including methionine adenosyltransferases (MATs) and glycine N-methyltransferase (GNMT), are essential for the synthesis and utilization of S-adenosylmethionine (adoMet), the universal methyl donor and precursor for polyamine and glutathione synthesis [12,13,14].
MAT and GNMT genes are commonly diminished in human HCC and hepatoma cell lines [15,16,17,18,19]. MAT1A or GNMT dysregulation contributes to HCC progression; spontaneous HCC has been observed in the mat1a-knockout as well as in the gnmt-knockout mice [20,21,22]. Deletion of gnmt promotes the susceptibility to liver cancer in mice [22]; gnmt knockout mice exhibited elevated hepatic adoMet levels and S-adenosylhomocysteine hydrolase (SAHH) expression in the liver [22]. These studies suggested that defective MAT and/or GNMT proteins could be early markers in human HCC development.
Pleiotropic effect of MATs has been associated with global DNA hypomethylation and liver cancer progression and prognosis [16]. Mat1a knockout mice are predisposed to liver injury and hepatocarcinogenesis that displays increased proliferation [23].
Mammals have three distinct forms of MAT (MATI, MATII and MATIII), encoded by two distinct genes (MAT1A and MAT2A). MATs in the liver and in extrahepatic tissues are products of two genes, MAT1A and MAT2A, respectively. MATII consists of α2 catalytic subunit (encoded by MAT2A) and β regulatory subunit (encoded by MAT2B). We recently discovered that MAT1A and GNMT were mostly expressed in the cytoplasm, whereas MAT2A showed both cytoplasmic and nuclear immunoreactivity, and that a higher cytoplasmic/nuclear (C/N) MAT2A expression ratio is correlated with poor overall survival in breast cancer patients [24].
HCC is characterized by the low expression of the liver-specific MAT1A gene that encodes the MATI/III isozymes; and the high expression of MAT2A that encodes the MATII isozyme, as well as high expression of MAT2B that encodes a β-subunit without catalytic action but can regulate MATII enzymatic activity [23].
We have demonstrated that, in the GNMT diminished HCC cell-line HepG2, restoration of GNMT assisted methylfolate-dependent homocysteine remethylation [25]. In gnmt transgenic and knockout mouse models, we discovered that gnmt expression can improve folate retention and bioavailability in the liver [25], decrease antifolate drug toxicity [25], improve DNA integrity, and reduce uracil misincorporation in the DNA [26,27]. We also demonstrated that GNMT expression enhances homocysteine transsulfuration and remethylation fluxes when methionine is in excess, and GNMT assists homocysteine clearance when needed [28].
Mechanisms of HCC inhibition by GNMT include: the suppression of dep domain-containing mTOR-interacting protein (DEPTOR) to the activation of mTOR targets SK6 and 4E-BP, that further impedes PI3K/AKT signaling pathway, the repression of the proteasomal degradation of phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 (PREX2) by the E3 ubiquitin ligase HectH9, the maintenance of adequate proteins levels that related in the anti-oxidation and detoxification response and 1C metabolism pathways that could impede HCC development [28]. In gnmt-KO mice developed steatosis, fibrosis, and HCC, the methylation of RASSF1 and SOCS2 promoters and H3K27, which may result in epigenetic modulation of critical HCC carcinogenic pathways [21]. The above series of studies showed the essential and complex role of MAT1A and GNMT on maintaining optimal adoMet homeostasis, methylation balance, DNA integrity, and HCC prevention. In the present study we explored novel role of defected MAT1A and GNMT on HCC by curating gene sets from online pathway databases, publications in PubMed, and knowledge of domain experts by gene set enrichment analysis (GSEA) for profiling the effects of MAT1A and GNMT. We included 42 RNA-Seq data of matched HCC and adjacent normal liver tissues from TCGA and searched for MAT1A and GNMT-mediated novel biological processes/metabolic pathways by GSEA, and further investigated whether the newly identified genes could be involved HCC occurrence and development.
2. Results
2.1. HCC Patients with Low MAT1A and Low GNMT Expressions Had Poor Survival
Data from GEPIA demonstrated MAT1A and GNMT expressions were significantly higher in the adjacent normal tissues (n = 160) than those in the HCC tumor tissues (n = 369) (p < 0.001, Figure 1A,B). On the other hand, there was no statistical significance found in MAT2A between the adjacent normal and the HCC tumor tissues (Figure 1C). The gene expression of MAT1A and GNMT was highly correlated (Pearson’s correlation, p < 0.001, R = 0.52; Figure 1D). Kaplan–Meier plot (http://gepia2.cancer-pku.cn/#index accessed on 1 December 2020) showed that HCC patients with low MAT1A and low GNMT expression had poor survival rate (p = 0.0071 and p = 0.013, respectively, Figure 1E,F). In contrast, the survival curves indicated that high expression of MAT2A is associated with poor overall survival (Figure 1G).
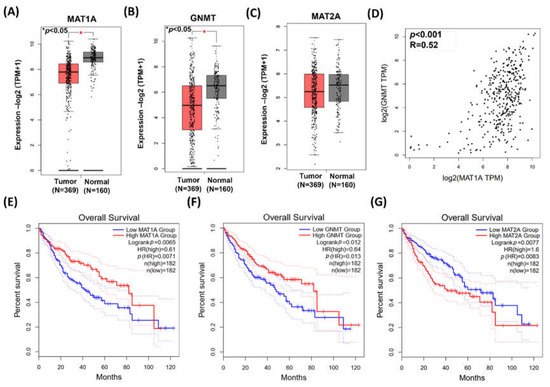
Figure 1.
The expressions of MAT1A, GNMT, and MAT2A in HCC. Gene expression profiling interactive analysis (GEPIA) GEPIA2 database was employed for bioinformatics analysis. (A–C) MAT1A, GNMT, and MAT2A expression in paired tumor and adjacent normal tissue. *, p < 0.05. (D) Pearson’s correlation was used to elucidate MAT1A expression in relation to GNMT expression. (E,F) Kaplan–Meier plots showed that high expression of MAT1A and GNMT (F) are both associated with better overall survival rate of patients with HCC. HR, hazard ratio (G) Kaplan–Meier plot showed that elevated MAT2A expression is associated poor overall survival rate of patients with HCC. HR, hazard ratio.
2.2. HCC RNA-seq Data Collected and Analysis from TCGA Database
Based on the above data, we then explored the data from 42 paired HCC tumor and adjacent normal tissues collected from the Cancer RNA-Seq Nexus [29], and identified the enriched pathways associated with defected MAT1A and GNMT expression. Low expression of MAT1A and GNMT1 were confirmed in the tumor tissues as compared to those in the adjacent normal tissues (for MAT1A and GNMT, p < 0.0001 and p = 0.012, respectively (Figure 2A), consistent with the results obtained from the web server. Pearson’s correlation also revealed a statistically significant correlation between MAT1A and GNMT expression (R= 0.489, p = 0.002, Figure 2B).
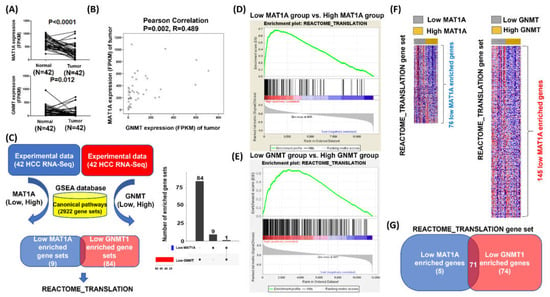
Figure 2.
Searching for the potential biological processes associated with low expression of MAT1A and GNMT. (A) MAT1A and GNMT expressions in paired tumor and adjacent normal tissues in the HCC cases (n = 42). (B) Pearson’s correlation for MAT1A and GNMT expression in the 42 HCC cases. (C) Flow chart for searching the candidate biological processes that could be driven by low expression of MAT1A and GNMT; GSEA was used to process the 42 HCC RNA-seq data. REACTOME_TRANSLATION gene set was found to be significantly enriched in HCC with low MAT1A. The median of mRNA expression level was used as the cutoff point for the dichotomization of MAT1A and GNMT. A score greater than median was defined as “high” expression, whereas a score of less than or equal to median was defined as “low” expression, and the categories further were mapped into GSEA databases. (D) and in HCC with low GNMT (E). (F) Heatmaps of the REACTOME_ TRANSLATION enrichment genes in HCC with high and low MAT1A and GNMT categories (G) A total of 71 overlapped genes of the REACTOME_ TRANSLATION gene set that are enriched in both low MAT1A and low GNMT HCC samples.
Gene set enrichment analysis (GSEA) was then performed to explore novel function of MAT1A and GNMT, in which the gene expression level greater than the median was defined as “high”, whereas less than the median was defined as “low” (Figure 2C). The low MAT1A category has 10 gene sets enriched as compared with the high MAT1A category; the low GNMT category has 85 gene sets enriched compared with the high GNMT category from GSEA (Figure 2C). The Venn plots revealed that the REACTOME_TRANSLATION gene set was enriched in both categories (Figure 2C, left panel is Venn diagram, and right is Venn bar). The statistical significance (nominal p value) of the ES for the two categories was calculated using an empirical phenotype-based permutation test procedure that preserves the complex correlation structure of the gene expression data for MAT1A (Figure 2D) and GNMT (Figure 2E). Heatmaps of the REACTOME_TRANSLATION genes were made in low and high expressions of MAT1A and GNMT (Figure 2F). Further overlapping analyses of low MAT1A (versus high MAT1A) expression and low GNMT (versus high GNMT) expression revealed 71 genes involved the false discovery rate (FDR) against a chance finding at the typical threshold of 0.05 (Figure 2G). The Venn diagram in Figure 2G summarizes the intersection of 71 genes identified from low MAT1A (versus high MAT1A) expression and low GNMT (versus high GNMT) expression.
2.3. Forty-Three REACTOME_TRANSLATION Genes Selected from Low Expressions of GNMT and MAT1A Are Associated with Poor HCC Prognosis
Among the 71 common genes in the REACTOME_ TRANSLATION gene set, further prognostic analysis demonstrated that 43 out of the 71 were statistically associated with poor overall survival of HCC (Table 1). The 43 Kaplan–Meier survival plots were performed after the samples were classified into high- and low-expression groups according to the median scores (Figure 3).

Table 1.
List of 71 overlapped genes in the REACTOME_TRANSLATION gene set that are enriched in both low MAT1A and low GNMT HCC samples.
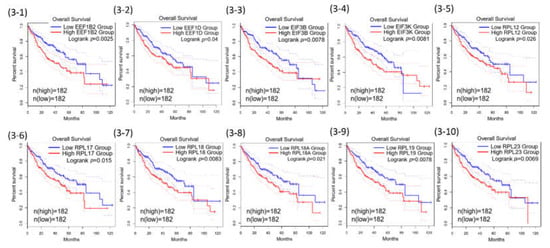
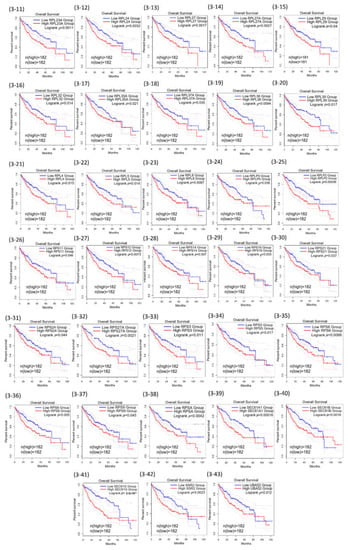
Figure 3.
Kaplan–Meier curves for the 43 of REACTOME_TRANSLATION genes.
Among these targets, eukaryotic translation elongation factor 1 delta (EEF1D) has been reported to modulate proliferation and epithelial-mesenchymal transition in oral squamous cell carcinoma [30] and promote glioma proliferation, migration, and invasion through epithelial-mesenchymal transition and PI3K/Akt pathway [31]. For eukaryotic translation initiation factor 3, subunit B (EIF3B) expression was found to be upregulated in gastric cancer tissues; it is strongly associated with proliferating cell nuclear antigen (PCNA) expression and is associated with poor outcomes in gastric cancer patients [32]. These studies suggest that EEF1D and EIF3B may play an oncogenic role in human cancer progression and whether they can serve as independent prognostic factors for HCC patients was further investigated in our cell models.
2.4. Expression of MAT1A and GNMT Decreased Cell Proliferation, Invasion
To investigate the impacts of MAT1A and GNMT expression on human hepatoma cells, we utilized cell models that expressed various levels of these 2 genes [33]. The cell numbers of wildtype HepG2 (WT) and HepG2 derived MAT1A and GNMT-expressing cell-lines (MAT1A+GNMT+) are shown in Figure 4A. Wild-type HepG2 grew faster than the cell-line stably transfected with MAT1A and GNMT (MAT1A+GNMT+); MAT1A+GNMT+ cells exhibited decreased cell growth (Figure 4A). The results of cell proliferation inspired us to further investigate the impacts of MAT1A and GNMT expression on cell invasion ability. Boyden chamber assay revealed that expressing MAT1A and GNMT in cells diminished with both genes significantly reduced the invasion ability of HepG2 cells (Figure 4B).
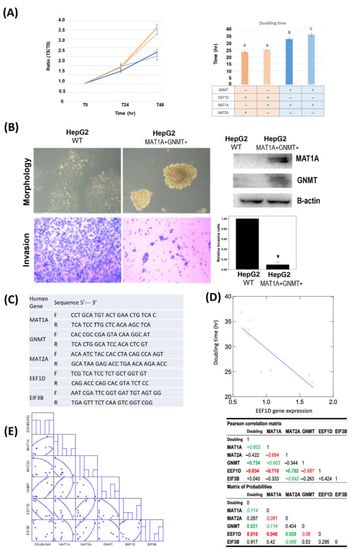
Figure 4.
MAT1A and GNMT expression decreased cell proliferation and invasion in HepG2 cells. MAT1A and GNMT expression also decreased reactome protein EEF1D, and EIF3B mRNA levels. (A) Doubling time of 4 HepG2 derived stable cell lines with various MAT1A and GNMT-expression levels were determined by cell counting. Points, mean; bars, SE. A: Control cells. B: Clone #1, MAT1A and GNMT-overexpressing cells. C: Clone #2, MAT1A and GNMT-overexpressing cells. (B) Overexpression of MAT1A and GNMT decreases invasive ability of HCC cells. MAT1A, GNMT, and β-actin (loading control) protein expression levels were evaluated by immunoblotting in HepG2 and HepG2/MAT1A/GNMT. *, p < 0.05. (C) List of quantitative PCR primers. (D) Pearson’s correlation matrix of GNMT, MAT1A, MAT2A, EEF1D, and EIF3B mRNA expression. (E) Linear plot of correlation estimated from doubling time and EEF1D mRNA expression.
Since the expression of MAT1A and GNMT in HepG2 cells appeared to impede HCC cell proliferation, we further investigated the correlations between the cell doubling time and the expression levels of MAT1A, MAT2A, GNMT, as well as selected REACTOME gene EEF1D and EIF3B by quantitative real-time PCR using designated primers (Figure 4C). MAT1A and GNMT expression levels both positively correlated with cell doubling time (Pearson’s correlation for MAT1A: R= 0.603, p = 0.114; GNMT: R= 0.754, p = 0.031, Figure 4D) whereas no correlation was found between doubling time and MAT2A. EEF1D inversely correlated with MAT1A (R= −0.710, p = 0.048) and GNMT (R= −0.687, p = 0.06). In contrast to MAT1A and GNMT that were inversely correlated with EEF1D, MAT2A, a gene frequently highly expressed in HCC, was positively correlated with EEF1D (R= 0.762, p = 0.028) (Figure 4E). EEF1D expression was found to have a strongly inverse correlation with cell doubling time (R= −0.834, p = 0.010) (Figure 4E). These results indicated that EEF1D may play a significant role in HCC progression independent of protein translation, and MAT1A, GNMT and MAT2A may modulate HCC proliferation/and or progression through the expression of REATOME gene EEF1D.
2.5. MAT1A and GNMT Are Negatively Correlatived with EEF1D and EIF3B in
Using GSEA to process the 42 HCC RNA-seq data, we identified overlapped genes of the REACTOME_ TRANSLATION gene set that are enriched in both low MAT1A and low GNMT HCC samples. In our cell models, MAT1A and GNMT expression significantly decreased cell proliferation and invasion in HepG2 cells. In particular, EEF1D expression was inversely correlated with MAT1A and GNMT expression, as well as with cell doubling time. HCC RNA-seq data were then collected from The Cancer Genome Atlas (TCGA) datasets and analyzed by Gene Expression Profiling Interactive Analysis (GEPIA) [34]. Correlation of EEF1D, EIF3B, MAT1A, and GNMT mRNA expression were performed using GEPIA plotters (Figure 5). These results indicated that our approaches searching for the potential biological processes associated with low expression of MAT1A and GNMT were affective.
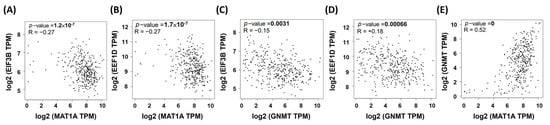
Figure 5.
The correlations between MAT1A and GNMT with reactome genes in liver hepatocellular carcinoma patients using the Cancer Genome Atlas (TCGA) web server program. Pearson’s correlations showed that MAT1A mRNA levels inversely correlated with (A) EIF3B and (B) EEF1D; GNMT mRNA levels inversely correlated with EIF3B (C) and EEF1D (D); MAT1A (E) and GNMT expressions are positively correlated.
3. Discussion
In the present study, bioinformatic analysis based on TCGA HCC database revealed the significant associations between the expression level of REACTOME_TRANSLATION genes and poor prognosis of HCC. These results raised the possibility that low expression of MAT1A and GNMT confers poor prognosis of HCC patients via REACTOME_ TRANSLATION biological process.
To further explore the biological relevance of this differential expression pattern, gene-set enrichment analyses (GSEA) were performed in the RNA-seq data from 42 paired HCC patients. The heatmaps and the Venn diagram revealed that 43 of the 71 REACTOME_TRANSLATION genes (60.1%) overlapping with low expressions of MAT1A and GNMT are associated with poor prognosis in HCC, indicating that REACTOME_TRANSLATION could potentially participate in human HCC tumorigenesis.
Transcriptional dysregulation has been recognized as a hallmark in cancer development, but relatively less is known about the dysregulation of gene expression at the translational level [35]. Translation and translational control are critical in stress adaptation of cancer cells to overcome challenges from the tumor microenvironment, immune recognition, their own continuous replication, and therapeutic modalities. Therefore, changes in the translational machinery can mediate the oncogenic signaling [35].
3.1. Ribosomal Proteins
Our approach in combination with Kaplan–Meier survival plot revealed numerous ribosomal protein (RP) encoding genes that were statistically associated with poor overall survival of HCC. Ribosome is the essential cellular organelle for protein synthesis that consists of ribosomal RNAs (rRNAs) and RPs. Human ribosomes are made up of four rRNAs species and about 80 different RPs. RP genes encode for ribosomal proteins that make up the ribosomal subunits involved in the cellular process of translation. Many RPs also play various roles independent of protein biosynthesis.
Some extra-ribosomal functions of ribosomal proteins have been reported in human colorectal cancers previously. Among the RPs that were found to be associated with poor overall HCC survival in our present study, RPL18 (Figure 3-7) and RPS27 (Figure 3-13) have been reported in cell growth or proliferation regulation; RPL23 (Figure 3-10) and RPL5 (Figure 3-22) have been reported in tumor suppressor gene regulation; RPL35A (Figure 3-17) has been reported in cell apoptosis regulation; RPLP0 (Figure 3-24) was reported to be involved in DNA repair; RPS14 (Figure 3-29) and RPS8 (Figure 3-36) have been reported in self-translation regulation in many human cancers, HCC included [36,37,38]. Other RPs including RPL19, RPL4, RPLP2, RPS10, RPS12, RPS3, RPS5, RPS6, RPS9 have been reported to participate in self-translation regulation, RNA splicing and modification, transcription regulation, DNA repair, and developmental regulation in in other species [36,37,38]. The expression of RPL19 is associated with poor overall survival of HCC in the present study (Figure 3-9). The xenografted tumors with knocked-down RPL19 were significantly smaller than those in the control tumors, and siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer [39,40,41]. These findings and the extra-ribosomal regulatory functions of RPL19 beyond protein synthesis provide a potential target for controlling the human HCC cellular phenotype.
3.2. ER Transmembrane Proteins and Ubiquitin Fusion Protein
Our study also revealed numerous genes encoding for the protein transport protein Sec61 subunits were statistically associated with poor overall survival of HCC, including SEC61A1, SEC61B, and SEC61G. The Sec61 complex is the central component of the protein translocation apparatus of the endoplasmic reticulum (ER) membrane. The Sec61 complex forms a transmembrane channel where proteins are translocated across and integrated into the ER membrane. Targeting human epidermal growth factor receptor 3 (HER3) by interfering with its Sec61-mediated co-translational insertion into the endoplasmic reticulum has been identified as a novel strategy to eliminate HER3 function [42]. Higher expression of SEC61G has been reported in HCC tissues than adjacent tissues. knockdown of SEC61G inhibited cell proliferation and induced cell apoptosis in vitro. SEC61G was required for cell migration and invasion, conferring a potential role for SEC61G in tumor transfer [43].
Ubiquitin A-52 residue ribosomal protein fusion product 1 (UBA52) gene that encodes for the 60S ribosomal protein L40 (RPL40) was statistically associated with poor overall survival of HCC in the present study. Ubiquitin fusion proteins have been overexpressed in colon cancer [44]. Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination promotes cell cycle progression and proliferation of non-small cell lung cancer cells [45].
3.3. Eukaryotic Translation Initiation and Elongation Factors
Our study revealed numerous genes encoding for the eukaryotic translation initiation (EIF3B, EIF3K) and elongation factors (EEF1B2, EEF1D) were statistically associated with poor overall survival of HCC. Eukaryotic translation initiation factor 3 (eIF3), the largest translation initiation factor composed of 13 non-identical polypeptides, plays an important role in protein synthesis that bridges the 43S preinitiation complex and eIF4F-bound mRNA [46]. The aberrant expression of eIF3 subunits was detected in various human cancers, and it was proposed to be a novel target in drug development [47]. Overexpression of the translation elongation factor 1 complex (eEF1) subunits was observed in 72% human cardioesophageal carcinoma [48]. Translation elongation factors have been proposed to play a role in tumorigenesis and affect survival in cancer specific manner [49]. EIF3B expression was upregulated in gastric cancer tissues and is associated with poor outcomes in gastric cancer patients [47]. EIF3B promoted gastric cancer cell proliferation, enhanced tumor cell migration and invasion through epithelial-mesenchymal transition (EMT) and the Stat3 signaling pathway in numerous human cancers [47]. Downregulation of EIF3B inhibited proliferation and metastasis of gastric cancer [50]. Knockdown of EIF3B in gastric cancer cells suppressed the growth of xenograft tumors and lung metastatic colonization in vivo [51]. The role of EIF3B in HCC remained to be determined.
In the present study, we demonstrated that important 1C metabolic gene MAT1A and GNMT are both strong prognostic indicators for HCC. Data from HCC patients with both genes diminished revealed that the REATOME pathway could participate in HCC tumorigenesis. Using cell models with various level of MAT1A and GNMT, we further identified a REATOME gene EEF1D; its expression level is not only inversely related to both genes but also is closed related to cell proliferation.
MAT1A and GNMT are both critical regulators for S-adenosylmethionine homeostasis of (adoMet) and methylation status [27,28]. Perturbations in folate dependent methylation pathways have been associated with increased human cancer risk. Lower serum folate has been shown to be associated poor survival of gastric cancer patients [52]. GNMT is a folate binding protein that can promote methylene-folate dependent pyrimidine and formyl-folate dependent purine synthesis in HCC [27]. We have demonstrated that GNMT facilitates the conservation of methyl groups by limiting homocysteine remethylation fluxes, controls transmethylation kinetics and S-adenosylmethionine (adoMet) homeostasis [28]. Restoring GNMT assists methylfolate-dependent reactions and ameliorates the consequences of folate depletion. GNMT expression in vivo improves folate retention and bioavailability in the liver [25]. Loss of GNMT impairs nucleotide biosynthesis; restoring GNMT expression enhances nucleotide biosynthesis and improves DNA integrity by reducing uracil misincorporation in DNA both in vitro and in vivo [26]. GNMT therefore has a protective role in cellular defense against DNA damage and human cancer [26].
The present study indicated that restoring MAT1A and GNMT expression may suppress EEF1D expression that is potentially oncogenic in human cancer progression. Dynamic alteration of the epigenome are important regulatory processes in a biological function. Cancer is a multifactorial disease characterized by aberrant epigenetic controls. The significant correlations among EEF1D and MAT1A, GNMT, and MAT2A in our cell models raise the possibility that its expression might be controlled by 1C metabolism mediated epigenetic modification. DNA methylation levels of EEF1D’s first CpG island have been reported to be negatively correlated with its gene expression levels in cattle [53], supporting our postulation that one carbon metabolism may control gene expression of EEF1D via modulation of methylation. We examined promoter and gene body methylation status of EEF1D using the methylation bank (MethBank http://bigd.big.ac.cn/methbank/ accessed on 1 October 2021). MethBank is a comprehensive methylation database that features consensus reference methylomes (CRMs), single-base resolution methylomes (SRMs), single-cell methylation maps and open platform for epigenome-wide association studies and integrates DNA & RNA methylation tools. Complete mCG methylation was shown in the gene body of EEF1D in normal healthy liver (Figure 6).
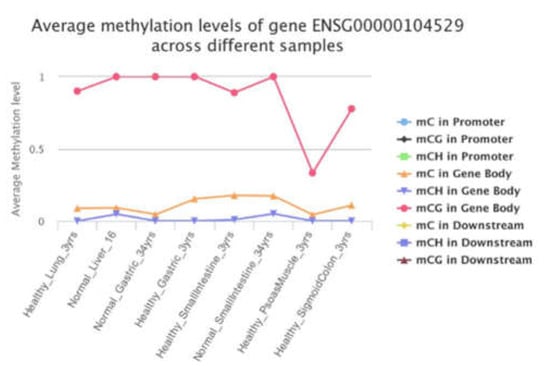
Figure 6.
Promoter and gene body methylation status of EEF1D.
Whether EEF1D methylation status is altered in human HCC, and whether MAT1A and GNMT functions affect EEF1D expression or other Reactome pathway genes, remains to be determined. Methylation profiling in cells with various MAT1A and GNMT expression are underway.
4. Materials and Methods
4.1. Data Collection
We collected a total of 42 of paired tumor and tumor-adjacent normal HCC tissue RNA-Seq datasets from the Cancer RNA-Seq Nexus (CRN, http://syslab4.nchu.edu.tw/CRN accessed on 1 October 2019) that has a user-friendly web interface designed to facilitate cancer research and personalized medicine [29,54].
4.2. Gene Set Enrichment Analysis
The median of mRNA expression was used as the cutoff point for the dichotomization of MAT1A and GNMT. A score greater than median was defined as “high” expression, whereas a score of less than or equal to median was defined as “low” expression, and the categories further were mapped into GSEA databases. This was performed using java GSEA Desktop Application from the Broad Institute at MIT. The MSigDB gene sets are divided into eight major collections: hallmark gene sets are coherently expressed signatures derived by aggregating many MSigDB gene sets to represent well-defined biological states or processes (H), positional gene sets for each human chromosome and cytogenetic band (C1), curated gene sets from online pathway databases, publications in PubMed, and knowledge of domain experts (C2), regulatory target gene sets based on gene target predictions for microRNA seed sequences and predicted transcription factor binding sites.(C3), computational gene sets defined by mining large collections of cancer-oriented microarray data (C4), GO gene sets consist of genes annotated by the same GO terms (C5), oncogenic gene sets defined directly from microarray gene expression data from cancer gene perturbations (C6), and immunologic gene sets defined directly from microarray gene expression data from immunologic studies (C7). Our data set had 11,799 genes, and the collections cp (canonical pathways: Biocarta, KEGG, and Reactome, total 2922 gene sets) was used for mapping by running GSEA (https://www.gsea-msigdb.org/gsea/index.jsp, accessed on 1 December 2020).
4.3. Survival Analysis 25
GEPIA2 performs survival analyses based on gene or isoform expression levels [34]. Given a list of custom cancer types, GEPIA2 would provide a heat map and show the survival analysis based on multiple cancer types [34]. The red blocks denote the higher and blue ones the lower risk, with an increase in the gene or isoform expression. The blocks with darkened frames indicate statistical significance in prognostic analyses. Since gene survival analyses can be context-dependent, this function allows users to screen for the prognostic impact of a gene or an isoform across different cancer types (http://gepia2.cancer-pku.cn/#index, accessed on 1 October 2021).
4.4. Cell Models with and without GNMT and MAT1A Expression
To elucidate how MAT1A and GNMT may affect cell proliferation, invasion, and Reactome pathway genes, stable cell-lines established from the human hepatoma cell-line HepG2 were used. One stable cell-line was established by cotransfecting pGNMT and pTK-Hyg plasmid (Clontech, Takara Bio USA, Inc., San Jose, CA, USA) DNAs that represented a model with normal GNMT function (GNMT+). The negative control cell-line, GNMT- was established by stable transfection with pFLAG-CMV-5. Establishment of stable cell-lines was described in detail previously and GNMT expression was confirmed by Western blot analyses [25]. Human MAT1A cDNA clone pCMVSPORT6 was obtained from National Yang-Ming University VYM Genome research center (Taipei, Taiwan, ROC). The full-length cDNA clone of human MAT1A was cloned into the mammalian expression vector pcDNA3. Human hepatoma cell line HepG2 cells expressing GNMT were further transfected with pcDNA3-MAT1a and underwent G418 (Sigma-Aldrich, Inc., St. Louis, MO, USA) selection. Multiple clones were selected, and MAT1A and GNMT expression was analyzed by real-time PCR (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Detailed procedures on the establishment of cell models and the characteristics of these cell-lines will be submitted elsewhere (Ko et al. manuscript in preparation).
4.5. Cell Proliferation in HCC Cell-Lines Expressing Various MAT1a and GNMT
All chemicals were purchased from Sigma-Aldrich, Inc. via the local distributor in Taiwan unless otherwise specified. Wildtype HepG2 and HepG2 derived cell-lines with various MAT1A and GNMT expressions were cultured in αminimum essential medium α (αMEM) with 10% fetal bovine serum (FBS), 1% Penicillin-Streptomycin-Amphotericin B Solution (PSA), and cultured at 37 °C in a 5% CO2 incubator. The cell growth and doubling time were compared between wildtype HepG2 and other cell-lines expressing MAT1A and GNMT.
4.6. Correlations between GNMT, MAT1A, and MAT2A and REACTOME Genes in Human HCC Cell-Lines
Total RNA was isolated by trizol and RNA integrity was checked by electrophoresis. RNA was converted to cDNA. Gene expression was determined by quantitative real-time PCR (ABI7000, Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). using Sybr green. Primers used for target genes are shown in Figure 4B. The expression of each gene was calculated by normalizing the threshold cycle value of target gene to that of the control housekeeping gene 18sRNA [55].
4.7. Transwell Invasion Assay
Cell invasion was investigated using Matrigel invasion chambers with a pore size of 8 μm (Costar; Corning Life Sciences, Cambridge, MA, USA) as described [56]. Briefly, wildtype HepG2 and stable cell-lines expressing various levels of MAT1A and GNMT (4 × 104 cells per chamber) in serum-free medium were seeded in the upper chamber, and 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used as a chemoattractant in the bottom well. After incubation for 24 h at 37 °C, the non-invasive cells on the upper surface of the membrane were removed with a cotton swab, and the invasive cells on the bottom side were fixed in 100% methanol at room temperature for 5 min, stained with 1% crystal violet at room temperature for 10 min and counted using a microscope (Nikon Eclipse 80i; Nikon Corporation Tokyo, Japan) under ×200 magnification with three replicate wells of view per cells.
4.8. Western Blot Analysis
The cells were harvested using a curet and centrifuged at 1000× g for 10 min at 4 °C and then lysed in ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Catalog number: 89900, Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 100 µL protease inhibitor cocktail (Roche, San Francisco, CA, USA). Equal amounts of protein (30 µg) were separated by SDS-PAGE (10% gel) and subsequently transferred to a polyvinylidene difluoride membrane. Subsequent to blocking with 5% skimmed milk at room temper-ature for 1 h, the membranes were incubated at 4 °C overnight with primary antibodies, including anti-GNMT (1:1000; GTX64826; GeneTex, Irvine, CA, USA), anti-MAT1A (1:5000; GTX132095; GeneTex), anti-β-actin (1:1000; GTX629630; GeneTex), followed by incubation at room temperature for 2 h with HRP-conjugated polyclonal secondary antibody (1:5000; GTX213110-01/GTX213111-01; GeneTex) [11]. All Western blots were visualized using the enhanced plus chemiluminescence assay kit (EMD Millipore, Billerica, MA, USA), according to the manufacturer’s protocol. Protein expression levels in cells were quantified by ImageJ software (Analytik Jena US LLC, Upland, CA, USA, https://imagej.nih.gov/ij/, accessed on 11 November 2021).
4.9. Statistical Analysis
Pearson’s correlation was conducted using SYSTAT (SYSTAT Software Inc., Chicago, IL, USA) and SPSS (PASW Statistics 18.0, SPSS Inc., Chicago, IL, USA). Independent-sample t-test was respectively used for binary variables and continuous variables to compare liver tumor and their adjacent liver tissues. The p-value of the test was 2-tailed with a level of significance (α) = 0.05. A p-value of less than 0.05 indicated statistical significance.
5. Conclusions
Dynamic alteration of the epigenome are important regulatory mechanisms in biological function. Our study indicated that restoring MAT1A and GNMT expression may suppress EEF1D expression that is potentially oncogenic in human cancer progression. This is the first study demonstrated that MAT1A and GNMT, the 2 key enzymes involved in methionine cycle, could potentially attenuate cancer progression via suppression of ribosome translation. More studies on methylation profiling in cells with various MAT1A and GNMT expression are warranted.
Author Contributions
Conceptualization, P.-M.C. and E.-P.I.C.; methodology, P.-M.C., C.-H.T. and H.-A.K.; validation, P.-M.C., C.-H.T. and E.-P.I.C.; formal analysis, P.-M.C.; investigation, E.-P.I.C.; resources, J.-R.L. and C.-C.L.; data curation, J.-R.L. and C.-C.L.; writing-original draft preparation, P.-M.C. and E.-P.I.C.; writing-review and editing, C.-C.H., H.-H.H. and E.-P.I.C.; visualization, P.-M.C. and C.-H.T.; supervision, E.-P.I.C.; project administration, E.-P.I.C.; funding acquisition, C.-C.H., H.-H.H. and E.-P.I.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants 110-2320-B-005-008-MY3 (C.E.P.*), M110-2320-B-005-003-MY3(C.E.P.*), 110-2321-B-005-008 (C.E.P.; H.H.H; C.C.H.*), 109-2321-B-005-025 (C.E.P.;C.C.H.;H.H.H*), 108-2321-B-005 -004 (C.E.P.;C.C.H.;H.H.H*), 107-2321-B-005 -009 (C.E.P.;C.C.H.;H.H.H*), 109-2811-B-005-528 (C.P.M), 108-2321-B005-004 (C.E.P.*), 107-2321-B005-009 (C.E.P.*), 107-2621-M005-008-MY3 (C.E.P.*), 107-2320-B005-003-MY3 (C.E.P.*), 107-2811-B-005 -515 (C.P.M), 108-2811-B-005-521 (C.P.M), and 109-2811-B-005 -528 (C.P.M) from Ministry of Science and Technology (MOST). It is also funded in part by the Ministry of Education Taiwan under the Higher Education Sprout Project (NCHU-IDCSA) (C.E.P.*) and in part by the TCVGH-NCHU1107602 and by TCVGH-NCHU1117603.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
There is no conflict of interest in all authors.
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Martínez-Chantar, M.L.; Avila, M.A.; Lu, S.C. Hepatocellular Carcinoma: Updates in Pathogenesis, Detection and Treatment. Cancers 2020, 12, 2729. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Yang, T.P.; Berry, R.; Bailey, L.B. Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Adv. Nutr. Int. Rev. J. 2012, 3, 21–38. [Google Scholar] [CrossRef]
- Chiang, E.-P.; Bagley, P.J.; Selhub, J.; Nadeau, M.; Roubenoff, R. Abnormal vitamin B6 status is associated with severity of symptoms in patients with rheumatoid arthritis. Am. J. Med. 2003, 114, 283–287. [Google Scholar] [CrossRef]
- Chiang, E.-P.; Selhub, J.; Bagley, P.J.; Dallal, G.; Roubenoff, R. Pyridoxine supplementation corrects vitamin B6 deficiency but does not improve inflammation in patients with rheumatoid arthritis. Arthritis Res. 2005, 7, R1404–R1411. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.-P.; E Smith, D.; Selhub, J.; Dallal, G.; Wang, Y.-C.; Roubenoff, R. Inflammation causes tissue-specific depletion of vitamin B6. Arthritis Res. 2005, 7, R1254–R1262. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-Y.; Tzen, J.T.C.; Lin, S.-J.; Wu, Y.-T.; Chiang, E.-P.I. Long-Term Prednisolone Treatments Increase Bioactive Vitamin B6Synthesis In Vivo. J. Pharmacol. Exp. Ther. 2010, 337, 102–109. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Wu, M.-T.; Tang, F.-Y.; Chen, D.-Y.; Ko, H.-A.; Shane, B.; Huang, W.-N.; Chiang, E.-P.I. MTHFRC677T polymorphism increases MTX sensitivity via the inhibition ofS-adenosylmethionine andde novopurine synthesis. Clin. Sci. 2019, 133, 253–267. [Google Scholar] [CrossRef]
- Sou, N.-L.; Huang, Y.-H.; Chen, D.-Y.; Chen, Y.-M.; Tang, F.-Y.; Ko, H.-A.; Fan, Y.-H.; Lin, Y.-Y.; Wang, Y.-C.; Chih, H.-M.; et al. Folinate Supplementation Ameliorates Methotrexate Induced Mitochondrial Formate Depletion In Vitro and In Vivo. Int. J. Mol. Sci. 2021, 22, 1350. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-H.; Quan, Z.-Y.; Piao, J.-M.; Zhang, T.-T.; Jiang, M.-H.; Shin, M.-H.; Choi, J.-S. Plasma Folate and Vitamin B12 Levels in Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 1032. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-T.; Ye, W.-T.; Wang, Y.-C.; Chen, P.-M.; Liu, J.-Y.; Tai, C.-K.; Tang, F.-Y.; Li, J.-R.; Liu, C.-C.; Chiang, E.-P.I. MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA. Int. J. Mol. Sci. 2021, 22, 9392. [Google Scholar] [CrossRef] [PubMed]
- Martínez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Más, D.; Valdivié, M.; Hu, C.-A.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Froese, D.S.; Fowler, B.; Baumgartner, M.R. Vitamin B 12, folate, and the methionine remethylation cycle—Biochemistry, pathways, and regulation. J. Inherit. Metab. Dis. 2019, 42, 673–685. [Google Scholar] [CrossRef]
- Geltink, R.I.K.; Pearce, E.L. The importance of methionine metabolism. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18, e13034. [Google Scholar] [CrossRef]
- Frau, M.; Feo, F.; Pascale, R.M. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J. Hepatol. 2013, 59, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Mato, J. S-adenosylmethionine in Liver Health, Injury, and Cancer. Physiol. Rev. 2012, 92, 1515–1542. [Google Scholar] [CrossRef] [PubMed]
- Frau, M.; Tomasi, M.L.; Simile, M.M.; Demartis, M.I.; Salis, F.; Latte, G.; Calvisi, D.F.; Seddaiu, M.A.; Daino, L.M.E.; Feo, C.F.; et al. Role of transcriptional and posttranscriptional regulation of methionine adenosyltransferases in liver cancer progression. Hepatology 2012, 56, 165–175. [Google Scholar] [CrossRef]
- Ramani, K.; Lu, S.C. Methionine adenosyltransferases in liver health and diseases. Liver Res. 2017, 1, 103–111. [Google Scholar] [CrossRef]
- Lu, S.C.; Alvarez, L.; Huang, Z.-Z.; Chen, L.; An, W.; Corrales, F.J.; Avila, M.A.; Kanel, G.; Mato, J.M. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 5560–5565. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Chantar, M.L.; Vázquez-Chantada, M.; Ariz, U.; Martínez, N.; Varela, M.; Luka, Z.; Capdevila, A.; Rodríguez, J.; Aransay, A.M.; Matthiesen, R.; et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology 2007, 47, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-P.; Li, Y.-S.; Chen, Y.-J.; Chiang, E.-P.; Li, A.F.-Y.; Lee, Y.-H.; Tsai, T.-F.; Hsiao, M.; Hwang, S.-F.; Chen, Y.-M.A. Glycine N-methyltransferase−/−mice develop chronic hepatitis and glycogen storage disease in the liver. Hepatology 2007, 46, 1413–1425. [Google Scholar] [CrossRef] [PubMed]
- Simile, M.M.; Latte, G.; Feo, C.F.; Feo, F.; Calvisi, D.F.; Pascale, R.M. Alterations of methionine metabolism in hepatocarcinogenesis: The emergent role of glycine N-methyltransferase in liver injury. Ann. Gastroenterol. 2018, 31, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.-Y.; Wu, H.-J.; Wang, S.-M.; Chen, P.-M.; Tang, F.-Y.; Chiang, E.-P. MAT2A Localization and Its Independently Prognostic Relevance in Breast Cancer Patients. Int. J. Mol. Sci. 2021, 22, 5382. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Chen, Y.-M.; Lin, Y.-J.; Liu, S.-P.; Chiang, E.-P.I. GNMT Expression Increases Hepatic Folate Contents and Folate-Dependent Methionine Synthase-Mediated Homocysteine Remethylation. Mol. Med. 2011, 17, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Lin, W.-L.; Lin, Y.-J.; Tang, F.-Y.; Chen, Y.-M.; Chiang, E.-P.I. A novel role of the tumor suppressor GNMT in cellular defense against DNA damage. Int. J. Cancer 2014, 134, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Wu, M.-T.; Lin, Y.-J.; Tang, F.-Y.; Ko, H.-A.; Chiang, E.-P. Regulation of Folate-Mediated One-Carbon Metabolism by Glycine N-Methyltransferase (GNMT) and Methylenetetrahydrofolate Reductase (MTHFR). J. Nutr. Sci. Vitaminol. 2015, 61, S148–S150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Tang, F.-Y.; Chen, S.-Y.; Chen, Y.-M.; Chiang, E.-P.I. Glycine-N Methyltransferase Expression in HepG2 Cells Is Involved in Methyl Group Homeostasis by Regulating Transmethylation Kinetics and DNA Methylation. J. Nutr. 2011, 141, 777–782. [Google Scholar] [CrossRef]
- Li, J.-R.; Sun, C.-H.; Li, W.; Chao, R.-F.; Huang, C.-C.; Zhou, X.J.; Liu, C.-C. Cancer RNA-Seq Nexus: A database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016, 44, D944–D951. [Google Scholar] [CrossRef]
- Flores, I.L.; Kawahara, R.; Miguel, M.C.; Granato, D.C.; Domingues, R.R.; Macedo, C.C.S.; Carnielli, C.M.; Yokoo, S.; Rodrigues, P.C.; Monteiro, B.; et al. EEF1D modulates proliferation and epithelial–mesenchymal transition in oral squamous cell carcinoma. Clin. Sci. 2016, 130, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Zhou, M.; Lin, J.; Wu, Z.; Ding, S.; Luo, J.; Zhan, Z.; Cai, Y.; Xue, S.; Song, Y. EEF1D Promotes Glioma Proliferation, Migration, and Invasion through EMT and PI3K/Akt Pathway. BioMed Res. Int. 2020, 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, X.; Luan, F.; Fu, T.; Gao, C.; Du, H.; Guo, T.; Han, J.; Huangfu, L.; Cheng, X.; et al. EIF3B is associated with poor outcomes in gastric cancer patients and promotes cancer progression via the PI3K/AKT/mTOR signaling pathway. Cancer Manag. Res. 2019, 11, 7877–7891. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.-L.; Sou, N.-L.; Tang, F.-Y.; Ko, H.-A.; Yeh, W.-T.; Peng, J.-H.; Chiang, E.-P.I. Tracing Metabolic Fate of Mitochondrial Glycine Cleavage System Derived Formate In Vitro and In Vivo. Int. J. Mol. Sci. 2020, 21, 8808. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Vaklavas, C.; Blume, S.W.; Grizzle, W.E. Translational Dysregulation in Cancer: Molecular Insights and Potential Clinical Applications in Biomarker Development. Front. Oncol. 2017, 7, 158. [Google Scholar] [CrossRef]
- Mao-De, L. Ribosomal Proteins and Colorectal Cancer. Curr. Genom. 2007, 8, 43–49. [Google Scholar] [CrossRef]
- Chen, R.; Dawson, D.W.; Pan, S.; Ottenhof, N.A.; De Wilde, R.F.; Wolfgang, C.L.; May, D.; A Crispin, D.; A Lai, L.; Lay, A.R.; et al. Proteins associated with pancreatic cancer survival in patients with resectable pancreatic ductal adenocarcinoma. Lab. Investig. 2015, 95, 43–55. [Google Scholar] [CrossRef]
- Bi, N.; Sun, Y.; Lei, S.; Zeng, Z.; Zhang, Y.; Sun, C.; Yu, C. Identification of 40S ribosomal protein S8 as a novel biomarker for alcohol-associated hepatocellular carcinoma using weighted gene co-expression network analysis. Oncol. Rep. 2020, 44, 611–627. [Google Scholar] [CrossRef]
- Bee, A.; Ke, Y.; Forootan, S.; Lin, K.; Beesley, C.; Forrest, S.E.; Foster, C.S. Ribosomal Protein L19 Is a Prognostic Marker for Human Prostate Cancer. Clin. Cancer Res. 2006, 12, 2061–2065. [Google Scholar] [CrossRef]
- Huang, C.-J.; Chien, C.-C.; Yang, S.-H.; Chang, C.-C.; Sun, H.-L.; Cheng, Y.-C.; Liu, C.-C.; Lin, S.-C.; Lin, C.-M. Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J. Cell. Mol. Med. 2008, 12, 1936–1943. [Google Scholar] [CrossRef]
- Bee, A.; Brewer, D.; Beesley, C.; Dodson, A.; Forootan, S.; Dickinson, T.; Gerard, P.; Lane, B.; Yao, S.; Cooper, C.S.; et al. siRNA Knockdown of Ribosomal Protein Gene RPL19 Abrogates the Aggressive Phenotype of Human Prostate Cancer. PLoS ONE 2011, 6, e22672. [Google Scholar] [CrossRef]
- Ruiz-Saenz, A.; Sandhu, M.; Carrasco, Y.; Maglathlin, R.L.; Taunton, J.; Moasser, M.M. Targeting HER3 by interfering with its Sec61-mediated cotranslational insertion into the endoplasmic reticulum. Oncogene 2015, 34, 5288–5294. [Google Scholar] [CrossRef]
- Gao, H.; Niu, W.; He, Z.; Gao, C.; Peng, C.; Niu, J. SEC61G plays an oncogenic role in hepatocellular carcinoma cells. Cell Cycle 2020, 19, 3348–3361. [Google Scholar] [CrossRef] [PubMed]
- Barnard, G.F.; Mori, M.; Staniunas, R.J.; Begum, N.A.; Bao, S.; Puder, M.; Cobb, J.; Redman, K.L.; Steele, G.D.; Chen, L.B. Ubiquitin fusion proteins are overexpressed in colon cancer but not in gastric cancer. Biochim. et Biophys. Acta (BBA)Mol. Basis Dis. 1995, 1272, 147–153. [Google Scholar] [CrossRef]
- Wang, F.; Chen, X.; Yu, X.; Lin, Q. Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination promotes cell cycle progression and proliferation of non-small cell lung cancer cells. Am. J. Transl. Re.s 2019, 11, 7166–7185. [Google Scholar]
- Pelletier, J.; Graff, J.; Ruggero, D.; Sonenberg, N. Targeting the eIF4F Translation Initiation Complex: A Critical Nexus for Cancer Development. Cancer Res. 2015, 75, 250–263. [Google Scholar] [CrossRef]
- Yin, Y.; Long, J.; Sun, Y.; Li, H.; Jiang, E.; Zeng, C.; Zhu, W. The function and clinical significance of eIF3 in cancer. Gene 2018, 673, 130–133. [Google Scholar] [CrossRef]
- Veremieva, M.; Khoruzhenko, A.; Zaicev, S.; Negrutskii, B.; El’Skaya, A. Unbalanced expression of the translation complex eEF1 subunits in human cardioesophageal carcinoma. Eur. J. Clin. Investig. 2010, 41, 269–276. [Google Scholar] [CrossRef]
- Hassan, K.; Kumar, D.; Naik, M.; Dixit, M. The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS ONE 2018, 13, e0191377. [Google Scholar] [CrossRef]
- Ma, F.; Li, X.; Ren, J.; Guo, R.; Li, Y.; Liu, J.; Sun, Y.; Liu, Z.; Jia, J.; Li, W. Downregulation of eukaryotic translation initiation factor 3b inhibited proliferation and metastasis of gastric cancer. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Wang, H.; Ru, Y.; Sanchez-Carbayo, M.; Wang, X.; Kieft, J.S.; Theodorescu, D. Translation Initiation Factor eIF3b Expression in Human Cancer and Its Role in Tumor Growth and Lung Colonization. Clin. Cancer Res. 2013, 19, 2850–2860. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-Y.; Chiang, E.-P.; Shih, Y.-T.; Lane, H.-Y.; Lin, J.-T.; Wu, C.-Y. Lower serum folate is associated with development and invasiveness of gastric cancer. World J. Gastroenterol. 2014, 20, 11313–11320. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, J.; Zhang, Q.; Jiang, L. Regulation of DNA methylation on EEF1D and RPL8 expression in cattle. Genet. 2017, 145, 387–395. [Google Scholar] [CrossRef]
- Chen, P.-M.; Li, J.-R.; Liu, C.-C.; Tang, F.-Y.; Chiang, E.-P.I. Metabolic Pathways Enhancement Confers Poor Prognosis in p53 Exon Mutant Hepatocellular Carcinoma. Cancer Inform. 2020, 19, 1176935119899913. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.-P.I.; Wang, Y.-C.; Chen, W.-W.; Tang, F.-Y. Effects of Insulin and Glucose on Cellular Metabolic Fluxes in Homocysteine Transsulfuration, Remethylation, S-Adenosylmethionine Synthesis, and Global Deoxyribonucleic Acid Methylation. J. Clin. Endocrinol. Metab. 2009, 94, 1017–1025. [Google Scholar] [CrossRef][Green Version]
- Cheng, Y.-H.; Chiang, E.-P.I.; Syu, J.-N.; Chao, C.-Y.; Lin, H.-Y.; Lin, C.-C.; Yang, M.-D.; Tsai, S.-Y.; Tang, F.-Y. Treatment of 13-cis retinoic acid and 1,25-dihydroxyvitamin D3 inhibits TNF-alpha-mediated expression of MMP-9 protein and cell invasion through the suppression of JNK pathway and microRNA 221 in human pancreatic adenocarcinoma cancer cells. PLoS ONE 2021, 16, e0247550. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).