Antibiotic Heteroresistance in Klebsiella pneumoniae
Abstract
:1. Introduction
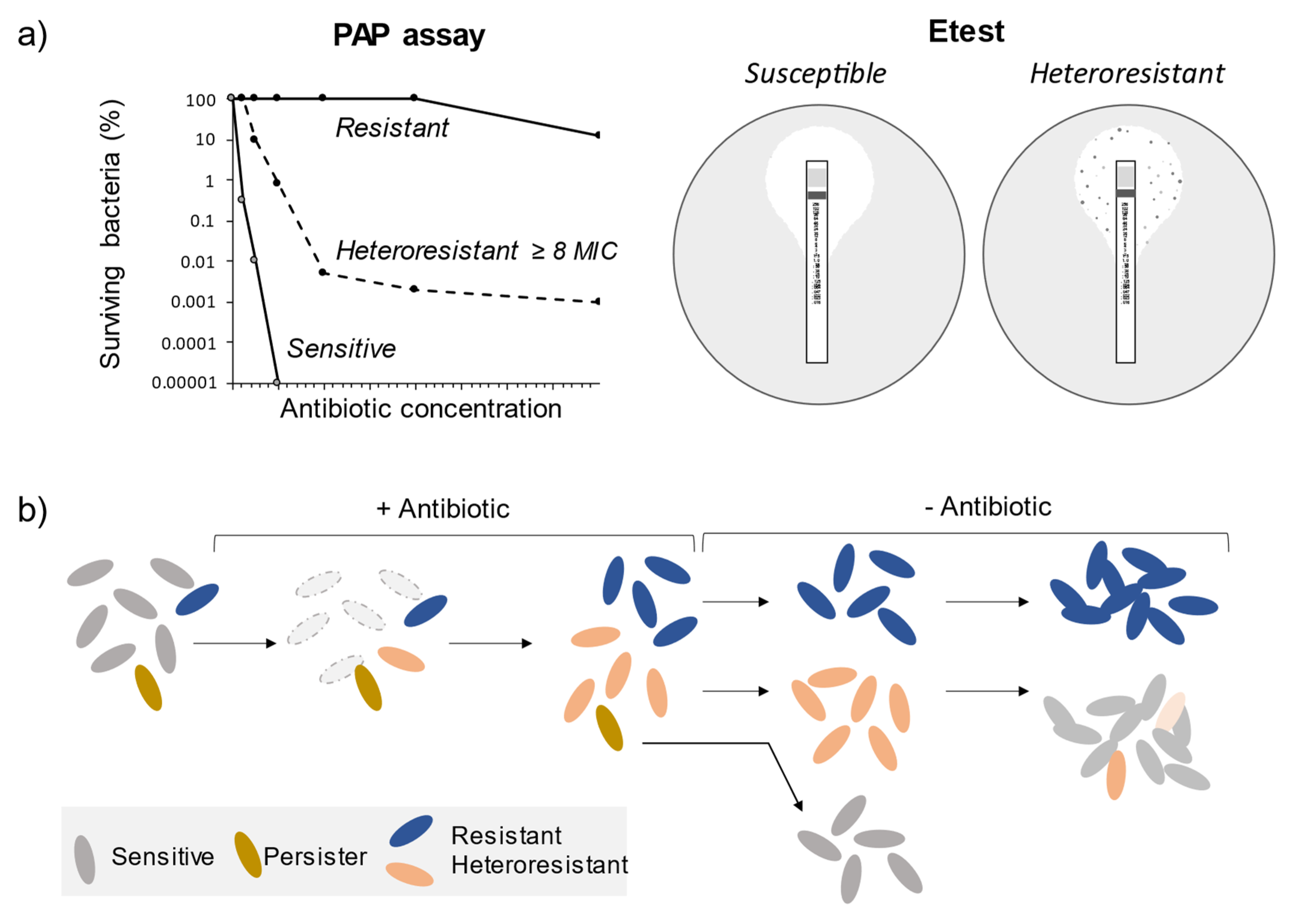
2. Heteroresistance: Definition, Detection Methods, and Underlying Mechanisms
3. Prevalence and Mechanisms of Heteroresistance in K. pneumoniae
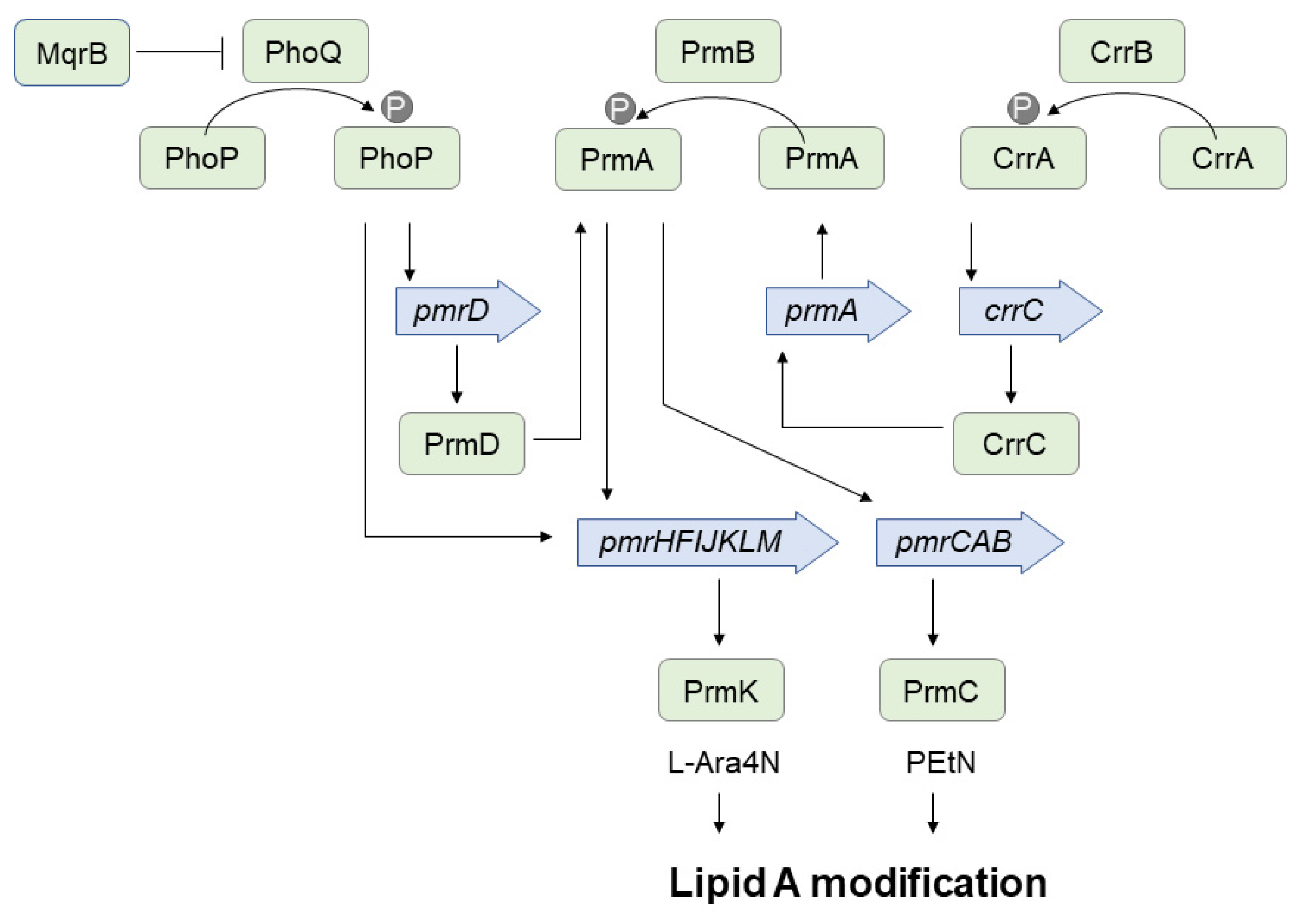
3.1. Heteroresistance to Colistin
3.2. Heteroresistance to Carbapenems
3.3. Heteroresistance to Tetracyclines
3.4. Heteroresistance to Aminoglycosides
4. Therapeutic Strategies against Heteroresistant K. pneumoniae
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMEs | Aminoglycoside modifying enzymes |
| cKP | Classical Klebsiella pneumoniae |
| ESBL | Extended-spectrum beta-lactamases |
| hvKP | Hypervirulent Klebsiella pneumoniae |
| IM | Inner membrane |
| L-Ara4N | 4-amino-4-deoxy-L-arabinose |
| MDR | Multidrug-resistant |
| MIC | Minimum inhibitory concentration |
| OM | Outer membrane |
| PAP | Population analysis profile |
| PEtN | Phosphoethanolamine |
| PDR | Pandrug-resistant |
| SNVs | Single nucleotide variations |
| XDR | Extensively drug-resistant |
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Piperaki, E.T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Bachman, M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, Z.; Ge, Y.; He, F. Rapid emergence of a pandrug-resistant Klebsiella pneumoniae ST11 isolate in an inpatient in a teaching hospital in China after treatment with multiple broad-spectrum antibiotics. Infect. Drug Resist. 2020, 13, 799–804. [Google Scholar] [CrossRef] [Green Version]
- Zowawi, H.M.; Forde, B.M.; Alfaresi, M.; Alzarouni, A.; Farahat, Y.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Li, J.; Schembri, M.A.; et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci. Rep. 2015, 5, 15082. [Google Scholar] [CrossRef]
- Prokesch, B.C.; TeKippe, M.; Kim, J.; Raj, P.; TeKippe, E.M.E.; Greenberg, D.E. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 2016, 16, e190–e195. [Google Scholar] [CrossRef]
- Patel, P.K.; Russo, T.A.; Karchmer, A.W. Hypervirulent Klebsiella pneumoniae. Open Forum Infect. Dis. 2014, 1, ofu028. [Google Scholar] [CrossRef] [Green Version]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 2015, 6, e00630-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zeng, J.; Liu, W.; Zhao, F.; Hu, Z.; Zhao, C.; Wang, Q.; Wang, X.; Chen, H.; Li, H.; et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 2015, 71, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liang, Q.; Liu, W.; Zheng, B.; Liu, L.; Wang, W.; Xu, Z.; Huang, M.; Feng, Y. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: An epidemiological, genomic and functional study. Virulence 2021, 12, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Band, V.I.; Weiss, D.S. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLoS Pathog. 2019, 15, e1007726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef]
- Dewachter, L.; Fauvart, M.; Michiels, J. Bacterial heterogeneity and antibiotic survival: Understanding and combatting persistence and heteroresistance. Mol. Cell 2019, 76, 255–267. [Google Scholar] [CrossRef]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Larsson, J.; Hjort, K.; Elf, J.; Andersson, D.I. The highly dynamic nature of bacterial heteroresistance impairs its clinical detection. Commun. Biol. 2021, 4, 521. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Ozturk, T.; Satola, S.W.; Jacob, J.T.; Weiss, D.S. Widespread cefiderocol heteroresistance in carbapenem-resistant gram-negative pathogens. Lancet Infect. Dis. 2021, 21, 597–598. [Google Scholar] [CrossRef]
- Seo, J.; Wi, Y.M.; Kim, J.M.; Kim, Y.J.; Ko, K.S. Detection of colistin-resistant populations prior to antibiotic exposure in kpc-2-producing Klebsiella pneumoniae clinical isolates. J. Microbiol. 2021, 59, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The High Prevalence of Antibiotic Heteroresistance in Pathogenic Bacteria Is Mainly Caused by Gene Amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef]
- Balaji, V.; Jeremiah, S.S.; Baliga, P.R. Polymyxins: Antimicrobial susceptibility concerns and therapeutic options. Indian J. Med. Microbiol. 2011, 29, 230–242. [Google Scholar] [CrossRef]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [Green Version]
- Bradley, P.; Gordon, N.C.; Walker, T.M.; Dunn, L.; Heys, S.; Huang, B.; Earle, S.; Pankhurst, L.J.; Anson, L.; de Cesare, M.; et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 2015, 6, 10063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Operario, D.J.; Koeppel, A.F.; Turner, S.D.; Bao, Y.; Pholwat, S.; Banu, S.; Foongladda, S.; Mpagama, S.; Gratz, J.; Ogarkov, O.; et al. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLoS ONE 2017, 12, e0176522. [Google Scholar]
- Bauer, D.; Wieland, K.; Qiu, L.; Neumann-Cip, A.C.; Magistro, G.; Stief, C.; Wieser, A.; Haisch, C. Heteroresistant bacteria detected by an extended Raman-based antibiotic susceptibility test. Anal. Chem. 2020, 92, 8722–8731. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Talarico, S.; Yao, L.; He, L.; Self, S.; You, Y.; Zhang, H.; Zhang, Y.; Guo, Y.; Liu, G.; et al. Droplet dDigital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J. Clin. Microbiol. 2018, 56, e00019-18. [Google Scholar] [CrossRef] [Green Version]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial persisters and infection: Past, present, and progressing. Ann. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjort, K.; Nicoloff, H.; Andersson, D.I. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol. Microbiol. 2016, 102, 274–289. [Google Scholar] [CrossRef]
- Anderson, S. E Aminoglycoside heteroresistance in Acinetobacter baumannii AB5075. mSphere 2018, 3, e00271-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girgis, H.S.; Dupai, C.D.; Lund, J.; Reeder, J.; Guillory, J.; Durinck, S.; Liang, Y.; Kaminker, J.; Smith, P.A.; Skippington, E. Single-molecule nanopore sequencing reveals extremetarget copy number heterogeneity inarylomycin-resistant mutants. Proc. Natl. Acad. Sci. USA 2021, 118, e2021958118. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, C.K.; Lee, H.; Jeong, S.H.; Yong, D.; Lee, K. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the BlaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 361–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Hu, D.; Zhang, Q.; Liao, X.P.; Liu, Y.H.; Sun, J. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar typhimurium. Front. Cell. Infect. Microbiol. 2017, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Saridakis, I. Colistin heteroresistance in Acinetobacter spp.: Systematic review and meta-analysis of the prevalence and discussion of the mechanisms and potential therapeutic implications. Int. J. Antimicrob. Agents 2020, 56, 106065. [Google Scholar] [CrossRef]
- Kang, K.N.; Klein, D.R.; Kazi, M.I.; Guérin, F.; Cattoir, V.; Brodbelt, J.S.; Boll, J.M. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol. Microbiol. 2019, 111, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Ikonomidis, A.; Tsakris, A.; Kantzanou, M.; Spanakis, N.; Maniatis, A.N.; Pournaras, S. Efflux system overexpression and decreased OprD contribute to the carbapenem heterogeneity in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2008, 279, 36–39. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Lin, Z.; Sun, X.; Lin, W.; Chen, Z.; Wu, Y.; Qi, G.; Deng, Q.; Qu, D.; Yu, Z. Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mmatli, M.; Mbelle, N.M.; Maningi, N.E.; Osei Sekyere, J. Emerging transcriptional and genomic mechanisms mediating carbapenem and polymyxin resistance in Enterobacteriaceae: A systematic review of current reports. mSystems 2020, 5, e00783-20. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, X.; Zeng, W.; Chen, T.; Liao, W.; Qian, J.; Lin, J.; Zhou, C.; Tian, X.; Cao, J.; et al. Mechanisms of heteroresistance and resistance to imipenem in Pseudomonas aeruginosa. Infect. Drug Resist. 2020, 13, 1419–1428. [Google Scholar] [CrossRef]
- Vogwill, T.; Maclean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef]
- Band, V.I.; Satola, S.W.; Smith, R.D.; Hufnagel, D.A.; Bower, C.; Conley, A.B.; Rishishwar, L.; Dale, S.E.; Hardy, D.J.; Vargas, R.L.; et al. Colistin heteroresistance is largely undetected among carbapenem-resistant Enterobacterales in the United States. mBio 2021, 12, e02881-20. [Google Scholar] [CrossRef]
- Band, V.I.; Satola, S.W.; Burd, E.M.; Farley, M.M.; Jacob, J.T.; Weiss, D.S. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 2018, 9, e02448-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Band, V.I.; Weiss, D.S. Heteroresistance to beta-lactam antibiotics may often be a stage in the progression to antibiotic resistance. PLoS Biol. 2021, 19, e3001346. [Google Scholar] [CrossRef] [PubMed]
- Napier, B.A.; Band, V.; Burd, E.M.; Weiss, D.S. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob. Agents Chemother. 2014, 58, 5594–5597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Band, V.I.; Crispell, E.K.; Napier, B.A.; Herrera, C.M.; Tharp, G.K.; Vavikolanu, K.; Pohl, J.; Read, T.D.; Bosinger, S.E.; Trent, M.S.; et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 2016, 1, 16053. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, A.; Howden, B.P.; Bell, J.M.; Gao, W.; Owen, R.J.; Turnidge, J.D.; Nation, R.L.; Li, J. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008, 62, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G.; Tzampaz, E.; Sianou, E.; Tzavaras, I.; Sofianou, D. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2011, 66, 946–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayol, A.; Nordmann, P.; Brink, A.; Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015, 59, 2780–2784. [Google Scholar] [CrossRef] [Green Version]
- Halaby, T.; Kucukkose, E.; Janssen, A.B.; Rogers, M.R.C.; Doorduijn, D.J.; van der Zanden, A.G.M.; al Naiemi, N.; Vandenbroucke-Grauls, C.M.J.E.; van Schaik, W. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 2016, 60, 6837–6843. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.; Sousa, A.M.; Alves, D.; Lourenço, A.; Pereira, M.O. Heteroresistance to colistin in Klebsiella pneumoniae is triggered by small colony variants sub-populations within biofilms. Pathog. Dis. 2016, 74, ftw036. [Google Scholar] [CrossRef] [Green Version]
- Bardet, L.; Baron, S.; Leangapichart, T.; Okdah, L.; Diene, S.M.; Rolain, J.-M. Deciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, France. Antimicrob. Agents Chemother. 2017, 61, e00356-17. [Google Scholar] [CrossRef] [Green Version]
- Juhász, E.; Iván, M.; Pintér, E.; Pongrácz, J.; Kristóf, K. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J. Glob. Antimicrob. Resist. 2017, 11, 167–170. [Google Scholar] [CrossRef]
- Cain, A.K.; Boinett, C.J.; Barquist, L.; Dordel, J.; Fookes, M.; Mayho, M.; Ellington, M.J.; Goulding, D.; Pickard, D.; Wick, R.R.; et al. Morphological, genomic and transcriptomic responses of Klebsiella pneumoniae to the last-line antibiotic colistin. Sci. Rep. 2018, 8, 9868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, H.S.; Kim, S.Y.; Wi, Y.M.; Peck, K.R.; Ko, K.S. Colistin heteroresistance in Klebsiella pneumoniae isolates and diverse mutations of PmrAB and PhoPQ in resistant subpopulations. J. Clin. Med. 2019, 8, 1444. [Google Scholar] [CrossRef] [Green Version]
- Morales-León, F.; Lima, C.A.; González-Rocha, G.; Opazo-Capurro, A.; Bello-Toledo, H. Colistin Heteroresistance among extended spectrum β-lactamases-producing Klebsiella pneumoniae. Microorganisms 2020, 8, 1279. [Google Scholar] [CrossRef] [PubMed]
- Adams-Sapper, S.; Nolen, S.; Donzelli, G.F.; Lal, M.; Chen, K.; da Silva, L.H.J.; Moreira, B.M.; Riley, L.W. Rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2015, 59, 3281–3289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tato, M.; Morosini, M.; García, L.; Albertí, S.; Coque, M.T.; Cantón, R. Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: Consequences for routine susceptibility testing. J. Clin. Microb. 2010, 48, 4089–4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Li, Q.; Bai, J.; Ding, M.; Yan, X.; Wang, G.; Zhu, B.; Zhou, Y. Heteroresistance to amikacin in carbapenem-resistant Klebsiella pneumoniae Strains. Front. Microbiol. 2021, 12, 3502. [Google Scholar] [CrossRef]
- Pournaras, S.; Kristo, I.; Vrioni, G.; Ikonomidis, A.; Poulou, A.; Petropoulou, D.; Tsakris, A. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J. Clin. Microb. 2010, 48, 2601–2604. [Google Scholar] [CrossRef] [Green Version]
- López-Camacho, E.; Paño-Pardo, J.R.; Sotillo, A.; Elías-López, C.; Martínez-Martínez, L.; Gómez-Gil, R.; Mingorance, J. Meropenem heteroresistance in clinical isolates of OXA-48–producing Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2019, 93, 162–166. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Lin, L.; Pan, Y.; Chen, J. Characterization of tigecycline-heteroresistant Klebsiella pneumoniae clinical isolates from a Chinese tertiary care teaching hospital. Front. Microbiol. 2021, 12, 2178. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, Q.; Wen, L.; Chen, J. Combined effect of polymyxin B and tigecycline to overcome heteroresistance in carbapenem-resistant Klebsiella pneumoniae. Microbiol. Spectr. 2021, 9, e0015221. [Google Scholar] [CrossRef]
- Naparstek, L.; Carmeli, Y.; Chmelnitsky, I.; Banin, E.; Navon-Venezia, S. Reduced susceptibility to chlorhexidine among extremely-drug-resistant strains of Klebsiella pneumoniae. J. Hosp. Infect. 2012, 81, 15–19. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of polymyxin resistance. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2019; Volume 1145, pp. 55–71. [Google Scholar]
- Sabnis, A.; Hagart, K.L.H.; Klöckner, A.; Becce, M.; Evans, L.E.; Furniss, R.C.D.; Mavridou, D.A.I.; Murphy, R.; Stevens, M.M.; Davies, J.C.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife 2021, 10, e65836. [Google Scholar] [CrossRef]
- Elias, R.; Duarte, A.; Perdigão, J. A molecular perspective on colistin and Klebsiella pneumoniae: Mode of action, resistance genetics, and phenotypic susceptibility. Diagnostics 2021, 11, 1165. [Google Scholar] [CrossRef] [PubMed]
- McConville, T.H.; Annavajhala, M.K.; Giddins, M.J.; Macesic, N.; Herrera, C.M.; Rozenberg, F.D.; Bhushan, G.L.; Ahn, D.; Mancia, F.; Trent, M.S.; et al. CrrB positively regulates high-level polymyxin resistance and virulence in Klebsiella pneumoniae. Cell Rep. 2020, 3, 108313. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Herold, M.; Vidaillac, C.; Duval, R.E.; Dague, E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 2015, 70, 2261–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berglund, B. Acquired resistance to colistin via chromosomal and plasmid-mediated mechanisms in Klebsiella pneumoniae. Infect. Microb. Dis. 2019, 1, 10–19. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Lopes, S.P.; Jorge, P.; Sousa, A.M.; Pereira, M.O. Discerning the role of polymicrobial biofilms in the ascent, prevalence, and extent of heteroresistance in clinical practice. Crit. Rev. Microbiol. 2021, 47, 162–191. [Google Scholar] [CrossRef]
- Van Acker, H.; van Dijck, P.; Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar] [CrossRef]
- Spagnolo, A.M.; Orlando, P.; Panatto, D.; Perdelli, F.; Cristina, M.L. An overview of carbapenem-resistant Klebsiella pneumoniae: Epidemiology and control measures. Rev. Med. Microbiol. 2014, 25, 7–14. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bonomo, R.A.; Schofield, C.J.; Mulholland, A.J.; Spencer, J. Natural variants modify Klebsiella pneumonia carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance. J. Biol. Chem. 2021, 296, 100126. [Google Scholar] [CrossRef]
- Li, X.; Quan, J.; Ke, H.; Wu, W.; Feng, Y.; Yu, Y.; Jiang, Y. Emergence of a KPC variant conferring resistance to ceftazidime-avibactam in a widespread ST11 carbapenem-resistant Klebsiella pneumoniae clone in China. Front. Microbiol. 2021, 12, 2419. [Google Scholar] [CrossRef]
- Sugawara, E.; Kojima, S.; Nikaido, H. Klebsiella pneumoniae major porins OmpK35 and OmpK36 allow more efficient diffusion of β-lactams than their Escherichia coli homologs OmpF and OmpC. J. Bacteriol. 2016, 198, 3200–3208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Ni, R.T.; Onishi, M.; Mizusawa, M.; Kitagawa, R.; Kishino, T.; Matsubara, F.; Tsuchiya, T.; Kuroda, T.; Ogawa, W. The Role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci. Rep. 2020, 10, 81877. [Google Scholar] [CrossRef] [PubMed]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bialek-Davenet, S.; Lavigne, J.P.; Guyot, K.; Mayer, N.; Tournebize, R.; Brisse, S.; Leflon-Guibout, V.; Nicolas-Chanoine, M.H. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Xing, B.; Yang, X.; Fu, Y.; Feng, Y.; Zhang, Y. Molecular epidemiology of aminoglycosides resistance on Klebsiella pneumonia in a hospital in China. Int. J. Clin. Exp. Med. 2015, 8, 1381–1385. [Google Scholar]
- Foudraine, D.E.; Strepis, N.; Stingl, C.; ten Kate, M.T.; Verbon, A.; Klaassen, C.H.W.; Goessens, W.H.F.; Luider, T.M.; Dekker, L.J.M. Exploring antimicrobial resistance to beta-lactams, aminoglycosides and fluoroquinolones in E. coli and K. pneumoniae using proteogenomics. Sci. Rep. 2021, 11, 12472. [Google Scholar] [CrossRef] [PubMed]
- Band, V.I.; Hufnagel, D.A.; Jaggavarapu, S.; Sherman, E.X.; Wozniak, J.E.; Satola, S.W.; Farley, M.M.; Jacob, J.T.; Burd, E.M.; Weiss, D.S. Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 2019, 4, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; He, Y.; Yu, X.; Cai, Y.; Zeng, J.; Cai, R.; Lu, Y.; Chen, L.; Chen, C.; Huang, B. Ceftazidime/avibactam improves the antibacterial efficacy of polymyxin B against polymyxin B heteroresistant KPC-2-producing Klebsiella pneumoniae and hinders emergence of resistant subpopulation in vitro. Front. Microbiol. 2019, 10, 2029. [Google Scholar] [CrossRef]
- Gaspari, R.; Spinazzola, G.; Teofili, L.; Avolio, A.W.; Fiori, B.; Maresca, G.M.; Spanu, T.; Nicolotti, N.; de Pascale, G.; Antonelli, M. Protective effect of SARS-CoV-2 preventive measures against ESKAPE and Escherichia coli infections. Eur. J. Clin. Investig. 2021, 51, e13687. [Google Scholar] [CrossRef]
- Micozzi, A.; Assanto, G.M.; Cesini, L.; Minotti, C.; Cartoni, C.; Capria, S.; Ciotti, G.; Fegatelli, D.A.; Donzelli, L.; Martelli, M.; et al. Reduced transmission of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae (KPC-KP) in patients with haematological malignancies hospitalized in an Italian hospital during the COVID-19 pandemic. JAC-Antimicrob. Resist. 2021, 3, dlab167. [Google Scholar] [CrossRef] [PubMed]
- Lobie, T.A.; Roba, A.A.; Booth, J.A.; Kristiansen, K.I.; Aseffa, A.; Skarstad, K.; Bjørås, M. Antimicrobial resistance: A challenge awaiting the post-COVID-19 era. Int. J. Infect. Dis. 2021, 111, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef]
- Mędrzycka-Dabrowska, W.; Lange, S.; Zorena, K.; Dabrowski, S.; Ozga, D.; Tomaszek, L. Carbapenem-resistant Klebsiella pneumoniae infections in ICU COVID-19 Patients—A scoping review. J. Clin. Med. 2021, 10, 2067. [Google Scholar] [CrossRef] [PubMed]
- Montrucchio, G.; Corcione, S.; Sales, G.; Curtoni, A.; de Rosa, F.G.; Brazzi, L. Carbapenem-resistant Klebsiella pneumoniae in ICU-admitted COVID-19 patients: Keep an eye on the ball. J. Glob. Antimicrob. Resis. 2020, 23, 398–400. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Sample | Prevalence of Heteroresistance | Mechanisms of Heteroresistance | Stability of Resistant Phenotypes | References | |
|---|---|---|---|---|---|---|
| Colistin | Multidrug-resistant clinical isolates (worldwide) | 15/21 | ND | ND | [48] | |
| Carbapenemase-producing clinical strains (Greece) | 12/20 | ND | Stable (11) Unstable (1) | [49] | ||
| Multidrug-resistant clinical isolate (South Africa) | 1/1 | Single amino acid change in PhoP, activation of the pmrHFIJKLM operon | Unstable | [50] | ||
| ESBL-producing clinical isolates | 5/13 | Mutations in the lpxM, mgrB, phoQ and yciM genes | ND | [51] | ||
| Urine clinical isolate (Portugal) | 1/1 | Biofilm, small colony variant phenotype | ND | [52] | ||
| Stool clinical isolate (France) | 1/1 | Single nucleotide insertion in the mreB gene | Unstable | [53] | ||
| Clinical isolates (Hungary) | 68/140 | ND | ND | [54] | ||
| Colistin susceptible K. pneu-moniae Ecl8 | 1/1 | Mutations in the crrB gene | Unstable | [55] | ||
| Multidrug-resistant urine isolates (USA) | 2/2 | Lower expression of mgrB, higher expression of phoP | Stable | [44] | ||
| Clinical blood isolates (South Korea) | 3/252 | Different amino acid substitutions in PmrAB and PhoPQ | ND | [56] | ||
| ESBL-producing clinical isolates (Chile) | 8/60 | Diverse mutations in PmrAB and PhoPQ, disruption of the mgrB gene | Stable | [57] | ||
| Clinical carbapenem-resistant isolates (South Korea) | 12/12 | Different amino acid substitutions in PmrAB and PhoPQ | Stable | [21] | ||
| Carbapenems | Imipenem | Clinical isolates (Brazil, USA) | 8/15 | Carriage of the blaKPC gene, decreased expression of OmpK36 | Stable (6) Unstable (2) | [58] |
| VIM-1-producing isolates (Spain) | 3/18 | ND | ND | [59] | ||
| Clinical isolates (China) | 75/155 | ND | ND | [60] | ||
| Meropenem | Clinical isolates (Greece) | 6/6 | Overexpression of the blaKPC-2 gene | ND | [61] | |
| OXA-48-producing isolates (Spain) | 24/24 | Decreased OmpK36 expression or activity | Stable | [62] | ||
| Clinical isolates (China) | 38/155 | ND | ND | [60] | ||
| Tetracyclines | Tigecycline | Clinical isolates (China) | 21/334 | Overexpression of the acrAB genes, mutations in the ramR, soxR and rpsJ genes | Stable | [63] |
| Carbapenem-resistant clinical isolates (China) | 49/95 | Upregulated expression of the pmrA, phoP and acrB genes | ND | [64] | ||
| Eravacycline | Clinical isolates (China) | 20/393 | Mutation in the acrR or ramR genes, overexpression of the MacAB efflux pump | ND | [39] | |
| Aminoglycosides | Amikacin | Clinical isolates (China) | 13/155 | Increased expression of aac(6′)-Ib, aph (3′)-Ia, aac (3)-II Mutations in parB, threonine dehydrogenase, Ssb, Sid | Unstable (11) Stable (2) | [60] |
| Amikacin | Clinical isolates (Sweden) | 2/10 | Mutations: ubiJ Δ10 nt, PTS glucose transporter subunit IIA Δ1 nt, cydAΔ57 nt | Unstable | [22] | |
| Gentamycin | 2/10 | aac(3)-IIa amplification | ||||
| Tobramycin | 3/10 | aac(6′)-Ib-ct or aac(3)-IIa amplification | ||||
| Netilmicin | 4/10 | aac(6′)-Ib-ct amplification | ||||
| Chlorhexidine | XDR clinical isolates (Israel) | 113/126 | ND | ND | [65] | |
| Isolates | Drug Combination | References |
|---|---|---|
| MDR, colistin-heteroresistant | Colistin + meropenem | [56] |
| Carbapenem-resistant, heteroresistant to polymyxin B, and resistant, heteroresistant or susceptible to tigecycline (4 isolates) | Polymyxin B + tigecycline | [64] |
| Carbapenem-resistant, heteroresistant to colistin and fosfomycin | Colistin + fosfomycin | [85] |
| Carbapenem-resistant, heteroresistant to fosfomycin and ceftazidime | Fosfomycin + ceftazidime | |
| Pandrug-resistant, heteroresistant to fosfomycin and sulfamethoxazole/trimethoprim | Fosfomycin + sulfamethoxazole/trimethoprim | |
| Pandrug-resistant, heteroresistant to amikacin and piperacillin/tazobactam | Amikacin + piperacillin/tazobactam | |
| Carbapenem-resistant, heteroresistant to polymyxin B (4 isolates) | Polymyxin B + ceftazidime/avibactam | [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stojowska-Swędrzyńska, K.; Łupkowska, A.; Kuczyńska-Wiśnik, D.; Laskowska, E. Antibiotic Heteroresistance in Klebsiella pneumoniae. Int. J. Mol. Sci. 2022, 23, 449. https://doi.org/10.3390/ijms23010449
Stojowska-Swędrzyńska K, Łupkowska A, Kuczyńska-Wiśnik D, Laskowska E. Antibiotic Heteroresistance in Klebsiella pneumoniae. International Journal of Molecular Sciences. 2022; 23(1):449. https://doi.org/10.3390/ijms23010449
Chicago/Turabian StyleStojowska-Swędrzyńska, Karolina, Adrianna Łupkowska, Dorota Kuczyńska-Wiśnik, and Ewa Laskowska. 2022. "Antibiotic Heteroresistance in Klebsiella pneumoniae" International Journal of Molecular Sciences 23, no. 1: 449. https://doi.org/10.3390/ijms23010449
APA StyleStojowska-Swędrzyńska, K., Łupkowska, A., Kuczyńska-Wiśnik, D., & Laskowska, E. (2022). Antibiotic Heteroresistance in Klebsiella pneumoniae. International Journal of Molecular Sciences, 23(1), 449. https://doi.org/10.3390/ijms23010449






