Novel Tetrazole-Based Antimicrobial Agents Targeting Clinical Bacteria Strains: Exploring the Inhibition of Staphylococcus aureus DNA Topoisomerase IV and Gyrase
Abstract
1. Introduction
2. Results and Discussion
2.1. In Silico Structure-Based Pharmacological Prediction
2.1.1. Antibacterial Activity
- 3 (confidence value 0.1880), 9 (confidence value 0.2211), and 12 (confidence value 0.1718) for activity against Staphylococcus aureus subsp. aureus MW2 resistant strain.
- 2 (confidence value 0.1431) and 7 (confidence value 0.2540) for activity against Bacillus subtilis subsp. subtilis str. 168 strain.
- 2 (confidence value 0.1158), 7 (confidence value 0.1011), and 9 (confidence value 0.1511) for activity against Mycobacterium smegmatis strain.
2.1.2. Toxicological Parameters
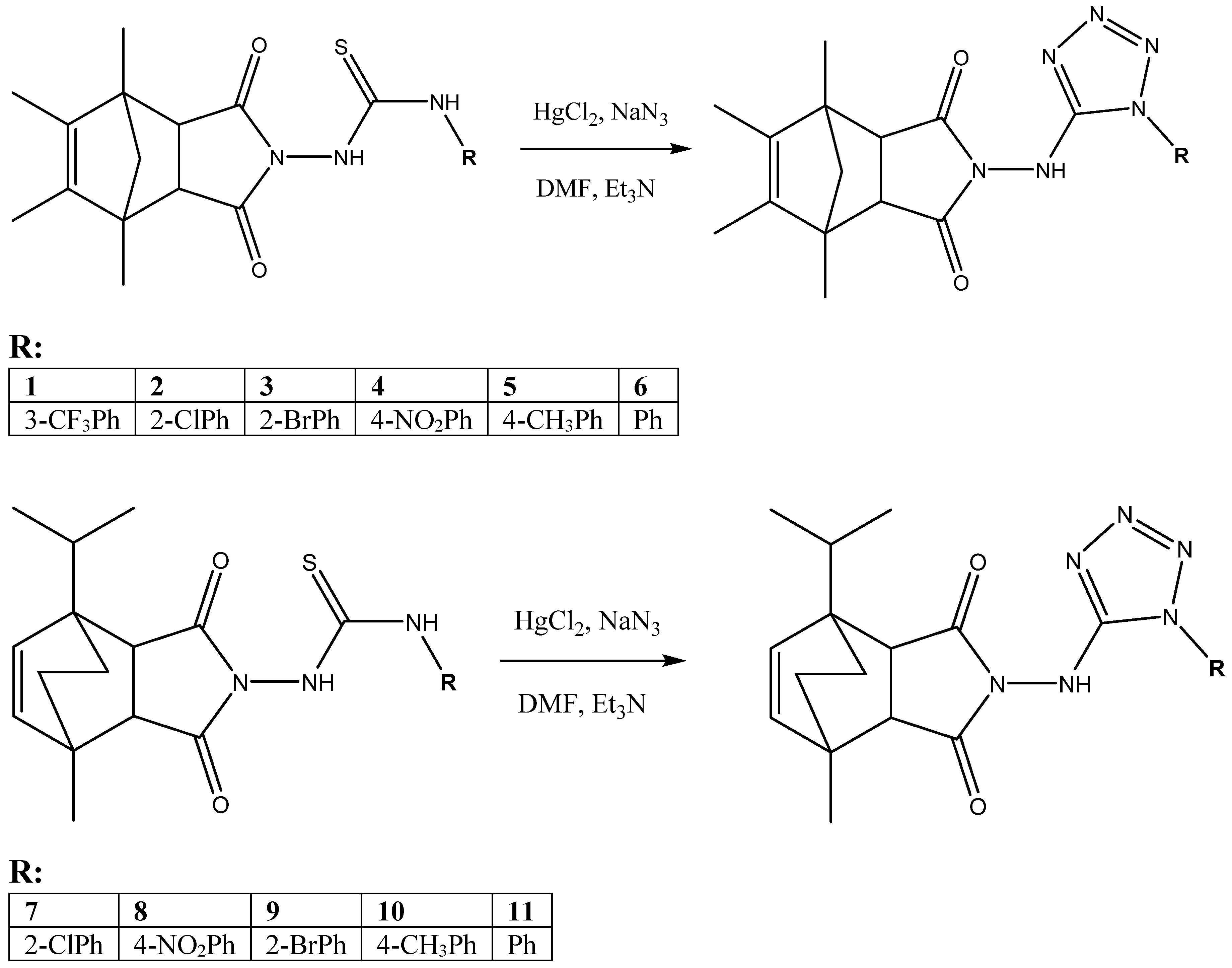
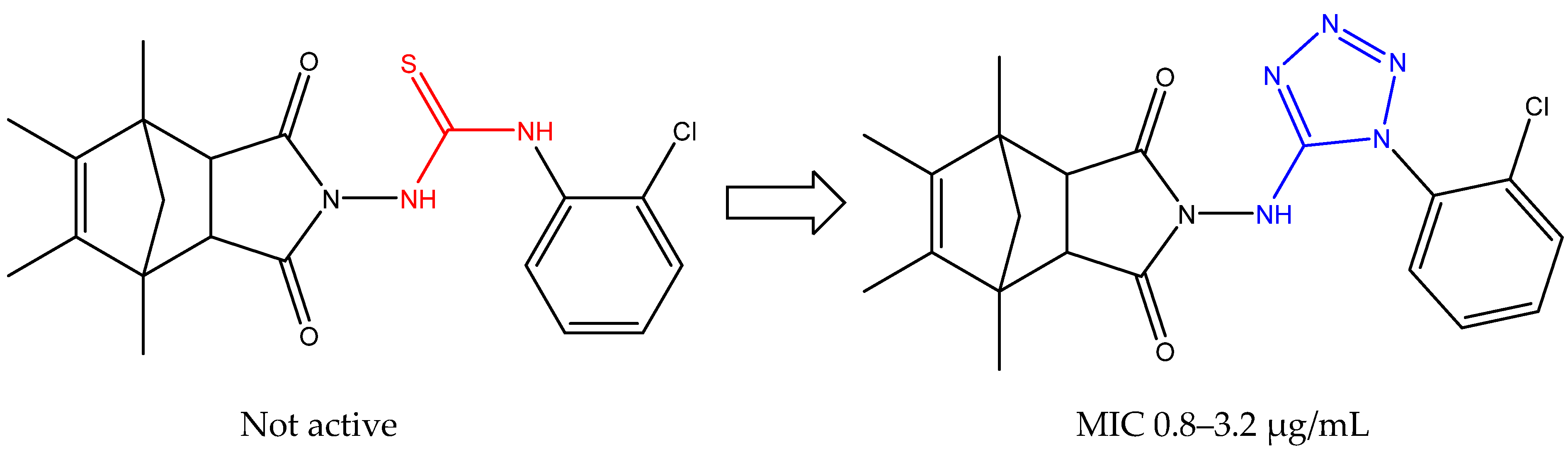
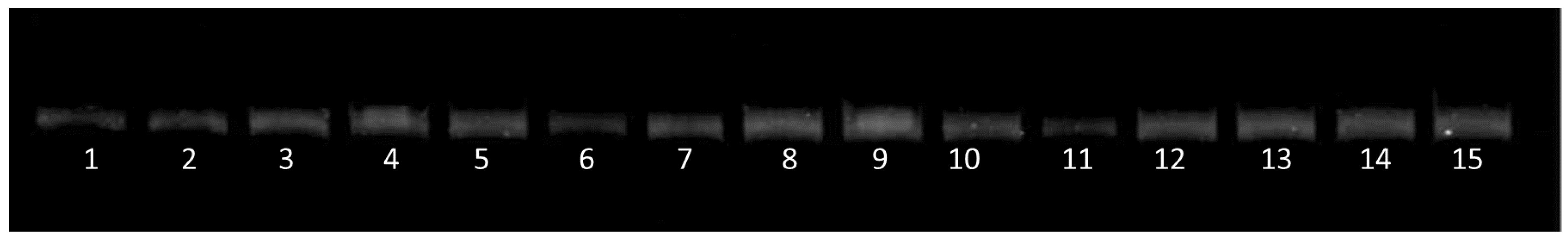
2.2. Chemistry
2.3. Biological Studies
2.3.1. In Vitro Antibacterial Activity Studies
2.3.2. Inhibition of Catalytic Activities of S. aureus Topoisomerases
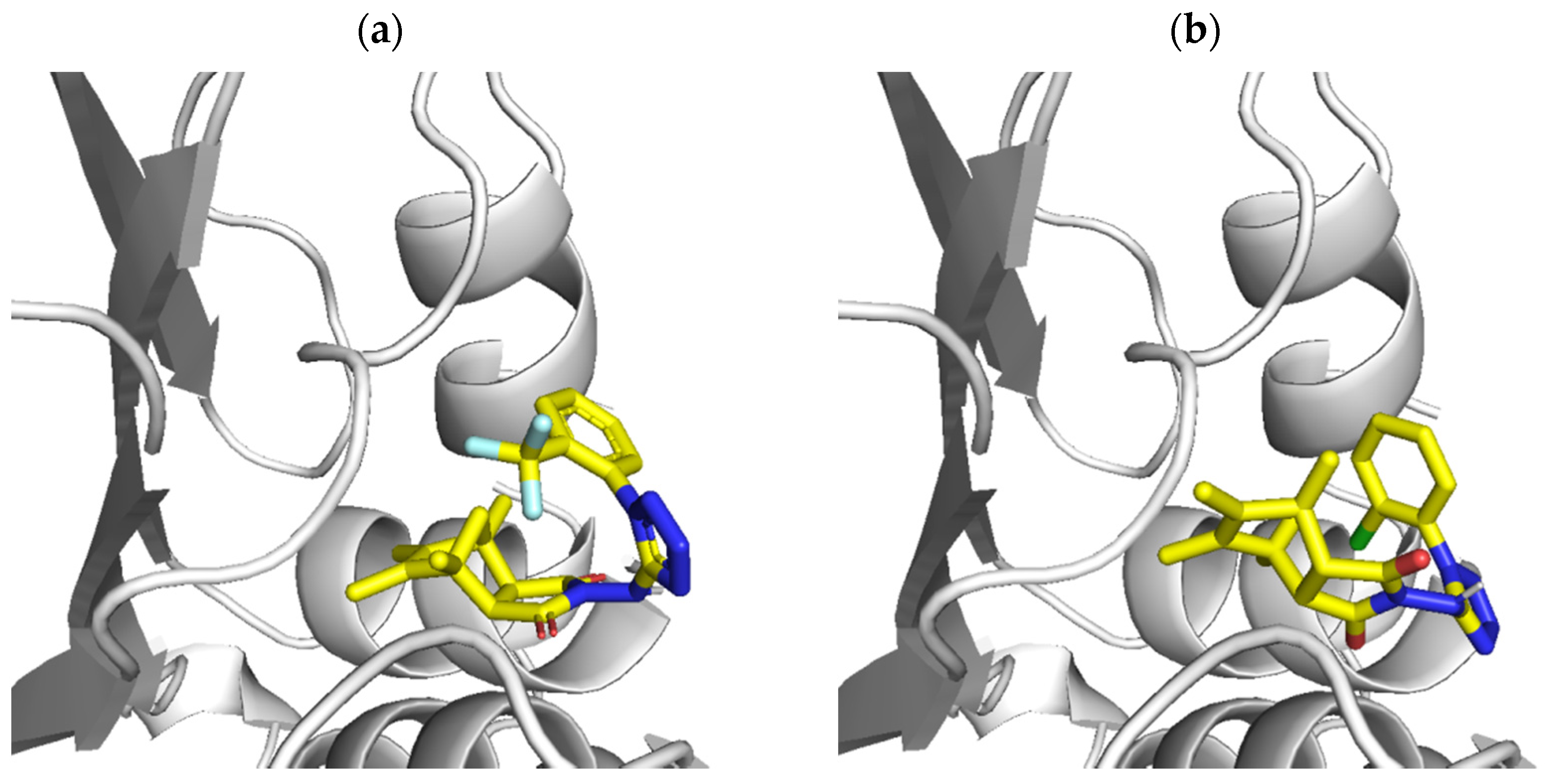
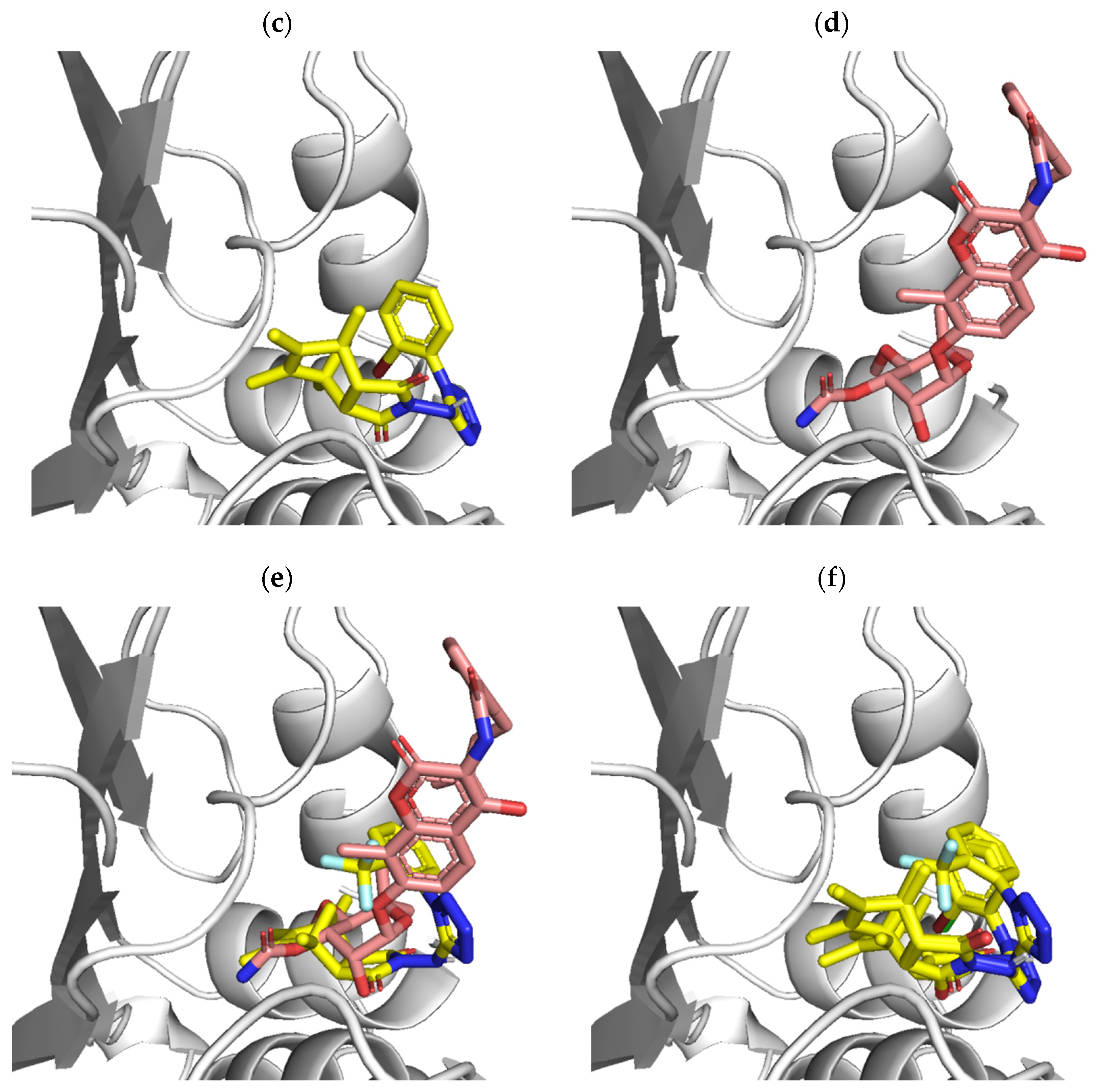
2.4. Molecular Docking
3. Materials and Methods
3.1. Apparatus, Materials, and Analysis
3.2. Imide-Tetrazole Hybrids Preparation: Derivatives of 3a,4,7,7a-Tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione and 4-Isopropyl-7-methyl-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione
3.2.1. 4,5,6,7-Tetramethyl-2-((1-(3-(trifluoromethyl)phenyl)-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (1)
3.2.2. 2-((1-(2-Chlorophenyl)-1H-tetrazol-5-yl)amino)-4,5,6,7-tetramethyl-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (2)
3.2.3. 2-((1-(2-Bromophenyl)-1H-tetrazol-5-yl)amino)-4,5,6,7-tetramethyl-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (3)
3.2.4. 4,5,6,7-Tetramethyl-2-((1-(4-nitrophenyl)-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (4)
3.2.5. 4,5,6,7-Tetramethyl-2-((1-(p-tolyl)-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (5)
3.2.6. 4,5,6,7-Tetramethyl-2-((1-phenyl-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-methanoisoindole-1,3(2H)-dione (6)
3.2.7. 2-((1-(2-Chlorophenyl)-1H-tetrazol-5-yl)amino)-4-isopropyl-7-methyl-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione (7)
3.2.8. 4-Isopropyl-7-methyl-2-((1-(4-nitrophenyl)-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione (8)
3.2.9. 2-((1-(2-Bromophenyl)-1H-tetrazol-5-yl)amino)-4-isopropyl-7-methyl-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione (9)
3.2.10. 4-Isopropyl-7-methyl-2-((1-(p-tolyl)-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione (10)
3.2.11. 4-Isopropyl-7-methyl-2-((1-phenyl-1H-tetrazol-5-yl)amino)-3a,4,7,7a-tetrahydro-1H-4,7-ethanoisoindole-1,3(2H)-dione (11)
3.3. Biological Assays
3.4. Molecular Docking Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, A.; Martinez-Martin, N.; Mercadillo, M.; Galan, J.C.; Ghysels, B.; Matthijs, S.; Cornelis, P.; Wiehlmann, L.; Tümmler, B.; Baquero, F.; et al. The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 2008, 3, e1619. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Bismuth, R.; Zilhao, R.; Sakamoto, H.; Guesdon, J.L.; Courvalin, P. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob. Agents Chemother. 1990, 34, 1611–1614. [Google Scholar] [CrossRef]
- van Hoek, A.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.; Aarts, H. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed]
- Dantas, G.; Sommer, M.O. Context matters—The complex interplay between resistome genotypes and resistance phenotypes. Curr. Opin. Microbiol. 2012, 15, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Ecology and evolution of chromosomal gene transfer between environmental microorganisms and pathogens. Microbiol. Spectr. 2018, 6, 1–16. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel, F.A. Staphylococcus aureus (including Staphylococcal Toxic Shock). In Principles and Practice of Infectious Diseases; Mandell, G.L., Bennett, J.E., Dolin, R., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2000; pp. 2069–2092. [Google Scholar]
- Mylotte, J.M.; McDermott, C.; Spooner, J.A. Prospective study of 114 consecutive episodes of Staphylococcus aureus bacteremia. Rev. Infect. Dis. 1987, 9, 891–907. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Dis. Mon. 2020, 66, 100971. [Google Scholar] [CrossRef]
- Blair, J.; Webber, M.; Baylay, A.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Bbosa, G.; Mwebaza, N.; Odda, J.; Kyegombe, D.; Ntale, M. Antibiotics/antibacterial drug use, their marketing and promotion during the post-antibiotic golden age and their role in emergence of bacterial resistance. Health 2014, 6, 410–425. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Poole, K. Overcoming antimicrobial resistance by targeting resistance mechanisms. J. Pharm. Pharmacol. 2001, 53, 283–294. [Google Scholar] [CrossRef]
- Ruppé, É.; Woerther, P.L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care 2015, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.R.; Holubar, M.; David, M.Z. Antimicrobial resistance in methicillin-resistant Staphylococcus aureus to newer antimicrobial agents. Antimicrob. Agents Chemother. 2019, 63, e01216–e01219. [Google Scholar] [CrossRef] [PubMed]
- Bielenica, A.; Szulczyk, D.; Olejarz, W.; Madeddu, S.; Giliberti, G.; Materek, I.B.; Koziol, A.E.; Struga, M. 1H-Tetrazol-5-Amine and 1,3-Thiazolidin-4-One Derivatives Containing 3-(Trifluoromethyl)Phenyl Scaffold: Synthesis, Cytotoxic and Anti-HIV Studies. Biomed. Pharmacother. 2017, 94, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Szulczyk, D.; Bielenica, A.; Głogowska, A.; Augustynowicz-Kopeć, E.; Dobrowolski, M.; Roszkowski, P.; Stępień, K.; Chrzanowska, A.; Struga, M. Development of (4-Methoxyphenyl)-1H-Tetrazol-5-Amine Regioisomers as a New Class of Selective Antitubercular Agents. Eur. J. Med. Chem. 2020, 186, 111882. [Google Scholar] [CrossRef]
- Szulczyk, D.; Bielenica, A.; Roszkowski, P.; Dobrowolski, M.A.; Olejarz, W.; Napiórkowska, M.; Struga, M. Cytotoxicity Evaluation of Novel Bis(2-aminoethyl)amine Derivatives. Molecules 2020, 25, 2816. [Google Scholar] [CrossRef] [PubMed]
- Szulczyk, D.; Bielenica, A.; Roszkowski, P.; Dobrowolski, M.A.; Olejarz, W.; Kmiecik, S.; Podsiad, M.; Struga, M. Synthetic Transition from Thiourea-Based Compounds to Tetrazole Derivatives: Structure and Biological Evaluation of Synthesized New N-(Furan-2-ylmethyl)-1H-tetrazol-5-amine Derivatives. Molecules 2021, 26, 323. [Google Scholar] [CrossRef] [PubMed]
- Szulczyk, D.; Bielenica, A.; Dobrowolski, M.A.; Dobrzycki, L.; Krawiecka, M.; Kuran, B.; Struga, M. Synthesis and structure evaluation of new complex butylarylpiperazin-1-yl derivatives. Med. Chem. Res. 2014, 23, 1519–1536. [Google Scholar] [CrossRef]
- Dobrowolski, M.A.; Roszkowski, P.; Struga, M.; Szulczyk, D. The Unexpected Product of Diels-Alder Reaction between “Indanocyclon” and Maleimide. J. Mol. Struct. 2017, 1130, 573–578. [Google Scholar] [CrossRef]
- Szulczyk, D.; Struga, M. 4-Hydroxy-1-methyl-7-(propan-2-yl)-4-azatricyclo[5.2.2.02,6]undec-8-ene-3,5-dione. Molbank 2012, 2012, M767. [Google Scholar] [CrossRef]
- Bielenica, A.; Stefańska, J.; Kozioł, A.E.; Iuliano, F.; Collu, D.; Sanna, G.; Jóźwiak, M.; Struga, M. Thiourea Derivatives of 4-Azatricyclo[5.2.2.0(2.6)]Undec-8-Ene-3,5-Dione—Synthesis and Biological Activity. Acta Pol. Pharm. 2016, 73, 693–703. [Google Scholar] [PubMed]
- Struga, M.; Kossakowski, J.; Kedzierska, E.; Fidecka, S.; Stefańska, J. Synthesis and pharmacological activity of urea and thiourea derivatives of 4-azatricyclo[5.2.2.02,6]undec-8-ene-3,5-dione. Chem. Pharm. Bull. 2007, 55, 796–799. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pogodin, P.V.; Lagunin, A.A.; Rudik, A.V.; Druzhilovskiy, D.S.; Filimonov, D.A.; Poroikov, V.V. AntiBac-Pred: A Web Application for Predicting Antibacterial Activity of Chemical Compounds. J. Chem. Inf. Mod. 2019, 59, 4513–4518. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Approved Standard M7-a7. Clinical and Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; CLSI: Malvern, PA, USA, 2006. [Google Scholar]
- Szulczyk, D.; Dobrowolski, M.A.; Roszkowski, P.; Bielenica, A.; Stefańska, J.; Koliński, M.; Kmiecik, S.; Jóźwiak, M.; Wrzosek, M.; Olejarz, W.; et al. Design and Synthesis of Novel 1H-Tetrazol-5-Amine Based Potent Antimicrobial Agents: DNA Topoisomerase IV and Gyrase Affinity Evaluation Supported by Molecular Docking Studies. Eur. J. Med. Chem. 2018, 156, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated force field Topology Builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comp. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Resistance Gene(s) | Gene Product(s) | Mechanism(s) of Resistance | Location(s) |
|---|---|---|---|---|
| β-Lactams | (1) blaZ (2) mecA | (1) β-Lactamase (2) PBP2a | (1) Enzymatic hydrolysis of β-lactam nucleus (2) Reduced affinity for PBP | (1) Pl:Tn (2) C:SCCmec |
| Glycopeptides | (1) Unknown (VISA) (2) VanHA | (1) Altered peptidoglycan (2) D-Ala-D-Lac | (1) Trapping of vancomycin in the cell wall (2) Synthesis of dipeptide with reduced affinity for vancomycin | (1) C (2) Pl:Tn |
| Quinolones | (1) parC (2) gyrA or gyrB | (1) ParC (or GrlA) component of topoisomerase IV (2) GyrA or GyrB components of gyrase | (1, 2) Mutations in the QRDR region, reducing affinity of enzyme-DNA complex for quinolones | (1) C (2) C |
| Aminoglycosides (e.g., gentamicin) Trimethoprim-sulfamethoxazole (TMP-SMZ) | Aminoglycoside-modifying enzymes (e.g., aac, aph) (1) Sulfonamide: sulA (2) TMP: dfrB | Acetyltransferase, phosphotransferase (1) Dihydropteroate synthase (2) Dihydrofolate reductase (DHFR) | Acetylating and/or phosphorylating enzymes modify aminoglycosides (1) Overproduction of p-aminobenzoic acid by enzyme (2) Reduced affinity for DHFR | Pl, Pl:Tn (1) C (2) C |
| Oxazolidinones | rrn | 23S rRNA | Mutations in domain V of 23S rRNA component of the 50S ribosome. Interferes with ribosomal binding | C |
| Quinupristin-dalfopristin (Q-D) | (1) Q: ermA, ermB, ermC (2) D: vat, vatB | (1) Ribosomal methylases (2) Acetyltransferases | (1) Reduce binding to the 23S ribosomal subunit (2) Enzymatic modification of dalfopristin | (1) Pl, C (2) Pl |
| Toxicological Test | Unit | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMES toxicity | (Yes/No) | No | No | No | Yes | No | No | No | Yes | No | No | No |
| Max. tolerated dose (human) | (log mg/kg/day) | 0.309 | 0.361 | 0.355 | −0.002 | 0.402 | 0.402 | 0.267 | −0.055 | 0.272 | −0.685 | 0.297 |
| hERG I inhibitor | (Yes/No) | No | No | No | No | No | No | No | No | No | No | No |
| hERG II inhibitor | (Yes/No) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Oral Rat Acute Toxicity (LD50) | (mol/kg) | 2.749 | 2.589 | 2598 | 2.483 | 2.417 | 2.47 | 2.636 | 2.542 | 2.558 | 1.834 | 2.517 |
| Oral Rat Chronic Toxicity (LOAEL) | (log mg/kg_bw/day) | 1.025 | 1.218 | 1.207 | 1.345 | 1.487 | 1.42 | 1.213 | 1.337 | 1.245 | 1.944 | 1.416 |
| Hepatotoxicity | (Yes/No) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Skin Sensitization | (Yes/No) | No | No | No | No | No | No | No | No | No | No | No |
| T. Pyriformis toxicity | (log ug/L) | 0.294 | 0.297 | 0.297 | 0.287 | 0.301 | 0.3 | 0.29 | 0.286 | 0.291 | 0.29 | 0.291 |
| Minnow toxicity | (log mM) | −0.453 | −0.621 | −0.767 | −1.597 | −0.913 | −0.306 | −0.95 | −1.914 | −1.606 | −2.571 | −0.635 |
| Strain | Compound (µg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Ciprofloxacin (µg/mL) | |
| S. aureus NCTC 4163 | 0.2 | 0.4 | 0.1 | 16 | >256 | 2 | 32 | 8 | 64 | 4 | 128 | 0.125 |
| S. aureus ATCC 25923 | 0.2 | 0.4 | 0.1 | 8 | >256 | 2 | 32 | 8 | 64 | 4 | 128 | 0.5 |
| S. aureus ATCC 6538 | 0.2 | 0.4 | 0.2 | 32 | >256 | 2 | 32 | 8 | 64 | 4 | 128 | 0.25 |
| S. aureus ATCC 29213 | 0.2 | 0.8 | 0.2 | 16 | >256 | 4 | 64 | 8 | 64 | 8 | 128 | 0.25 |
| S. epidermidis ATCC 12228 | 3.2 | 3.2 | 2 | 32 | >256 | 16 | 128 | 16 | 128 | 32 | 256 | 0.25 |
| S. epidermidis ATCC 35984 | 0.2 | 0.4 | 0.2 | 32 | >256 | 4 | 32 | 16 | 64 | 8 | 128 | 0.125 |
| E. coli ATCC 25922 | 0.4 | 1.6 | 0.4 | 256 | >256 | 8 | 128 | >256 | 128 | 16 | >256 | 0.015 |
| P. aeruginosa ATCC 15442 | 12.8 | 25.6 | 16 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | 0.125 |
| Strain | Compound (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 6 | 8 | 9 | 10 | Ciprofloxacin (µg/mL) | |
| S. epidermidis 5253 | 0.8 | 0.8 | 0.8 | 16 | 4 | 16 | 4 | 16 | 0.25 |
| S. epidermidis 4243 | 0.8 | 0.8 | 0.8 | 32 | 4 | 16 | 2 | 8 | 0.13 |
| S. aureus T5592 | 0.8 | 0.8 | 0.8 | 16 | 4 | 16 | 4 | 16 | 0.25 |
| S. aureus T5591 | 0.8 | 0.8 | 0.8 | 32 | 4 | 16 | 4 | 16 | 0.25 |
| E. coli 520 | 3.2 | 3.2 | 1.6 | 256 | 8 | >256 | 8 | 32 | 0.06 |
| E. coli 600 | 3.2 | 1.6 | 1.6 | 256 | 8 | >256 | 8 | 32 | 0.03 |
| K. pneumoniae 510 | 3.2 | 3.2 | 3.2 | 256 | 16 | >256 | 16 | 32 | 4 |
| P. aeruginosa 659 | 1.6 | 1.6 | 1.6 | 256 | 16 | >256 | 16 | 32 | 0.5 |
| Compounds | IC50 ± S.E.M. for Topoisomerase IV (µg/mL) | IC50 ± S.E.M. for DNA Gyrase (µg/mL) |
|---|---|---|
| Ciprofloxacin | 1.70 ± 0.15 | 3.55 ± 0.13 |
| Moxifloxacin | 0.9 ± 0.1 | 12.05 ± 0.50 |
| 1 | 63.2 ± 4.2 | 32.06 ± 4.04 |
| 2 | 58.4 ± 3.5 | 37.52 ± 9.72 |
| 3 | 61.2 ± 2.1 | 45.43 ± 3.54 |
| Ligand (Compound) | Number of Structures in the Largest Cluster | Binding Energy |
|---|---|---|
| 3 | 598 | −5.09 [kcal/mol] |
| 2 | 609 | −4.97 [kcal/mol] |
| 1 | 818 | −5.58 [kcal/mol] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roszkowski, P.; Szymańska-Majchrzak, J.; Koliński, M.; Kmiecik, S.; Wrzosek, M.; Struga, M.; Szulczyk, D. Novel Tetrazole-Based Antimicrobial Agents Targeting Clinical Bacteria Strains: Exploring the Inhibition of Staphylococcus aureus DNA Topoisomerase IV and Gyrase. Int. J. Mol. Sci. 2022, 23, 378. https://doi.org/10.3390/ijms23010378
Roszkowski P, Szymańska-Majchrzak J, Koliński M, Kmiecik S, Wrzosek M, Struga M, Szulczyk D. Novel Tetrazole-Based Antimicrobial Agents Targeting Clinical Bacteria Strains: Exploring the Inhibition of Staphylococcus aureus DNA Topoisomerase IV and Gyrase. International Journal of Molecular Sciences. 2022; 23(1):378. https://doi.org/10.3390/ijms23010378
Chicago/Turabian StyleRoszkowski, Piotr, Jolanta Szymańska-Majchrzak, Michał Koliński, Sebastian Kmiecik, Małgorzata Wrzosek, Marta Struga, and Daniel Szulczyk. 2022. "Novel Tetrazole-Based Antimicrobial Agents Targeting Clinical Bacteria Strains: Exploring the Inhibition of Staphylococcus aureus DNA Topoisomerase IV and Gyrase" International Journal of Molecular Sciences 23, no. 1: 378. https://doi.org/10.3390/ijms23010378
APA StyleRoszkowski, P., Szymańska-Majchrzak, J., Koliński, M., Kmiecik, S., Wrzosek, M., Struga, M., & Szulczyk, D. (2022). Novel Tetrazole-Based Antimicrobial Agents Targeting Clinical Bacteria Strains: Exploring the Inhibition of Staphylococcus aureus DNA Topoisomerase IV and Gyrase. International Journal of Molecular Sciences, 23(1), 378. https://doi.org/10.3390/ijms23010378








