Concomitant Activation of OSM and LIF Receptor by a Dual-Specific hlOSM Variant Confers Cardioprotection after Myocardial Infarction in Mice
Abstract
1. Introduction
2. Results
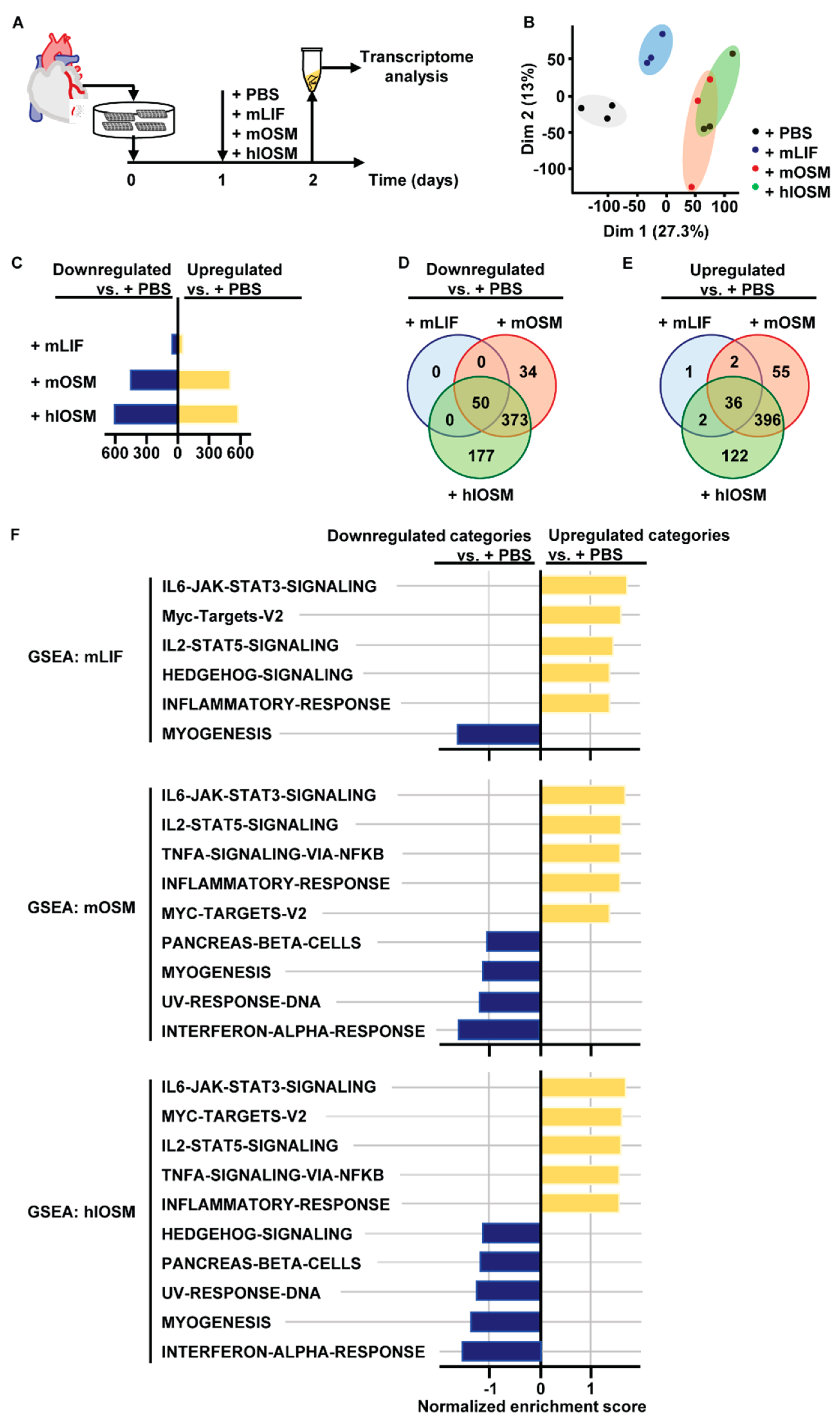
2.1. Transcriptome and Gene Set Enrichment Analysis Characterizes STAT3, STAT5 and c–Myc as Major Common Signaling Molecules Downstream of OSMR and LIFR Activation in Cardiomyocytes
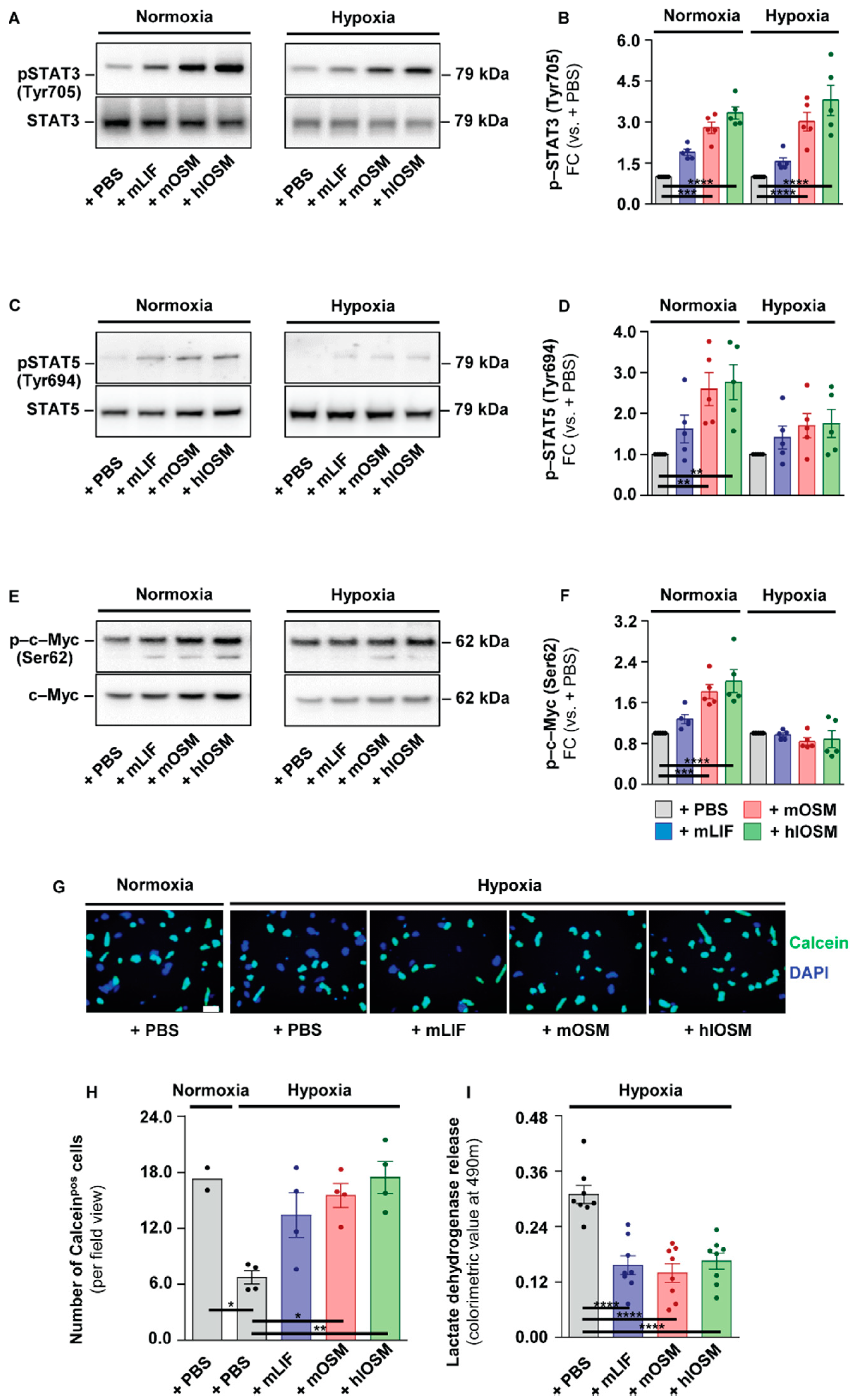
2.2. OSMR– and LIFR–Mediated Activation of STAT3 but Not of STAT5 and c–Myc Is Linked to Increased Survival of Cultured Cardiomyocytes under Hypoxic Conditions
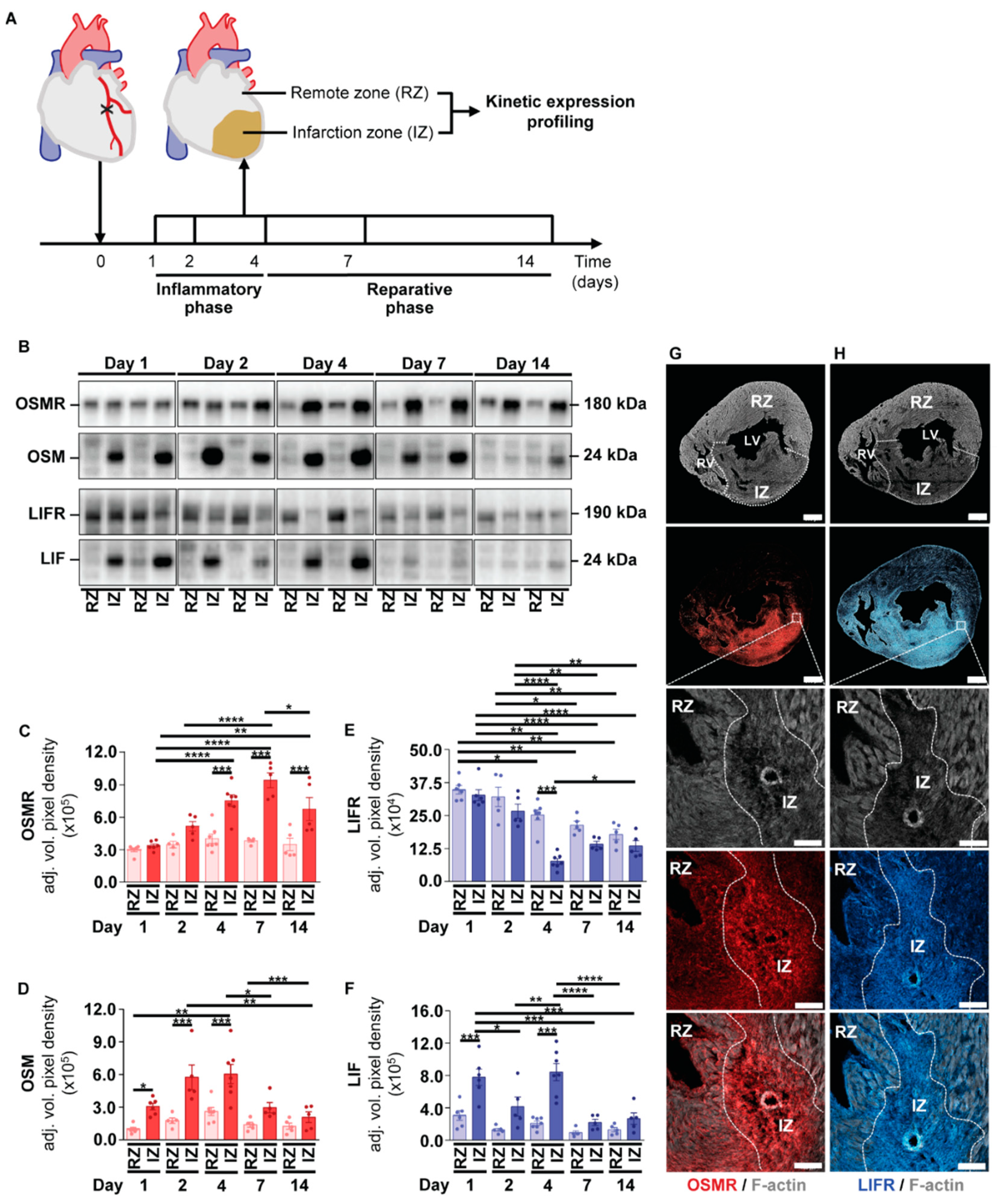
2.3. Kinetic Expression Pattern of OSM, LIF and Their Corresponding Receptors in Cardiac Tissue after the Onset of Myocardial Infarction in Mice
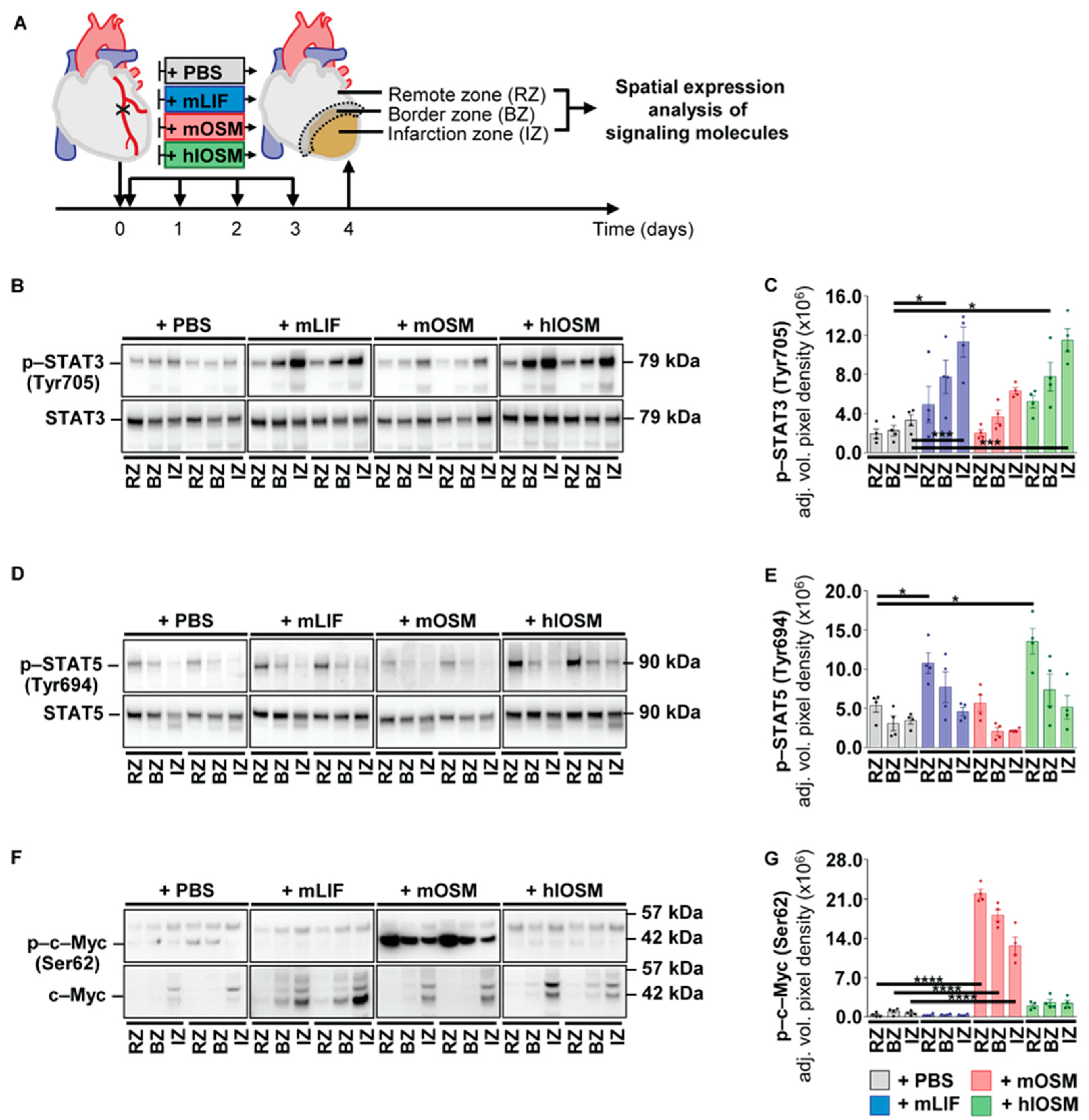
2.4. Post–Infarction Administration of mLIF, mOSM and hlOSM Modulates Activation of STAT3, STAT5 and c–Myc at Distinct Sites within the Myocardium
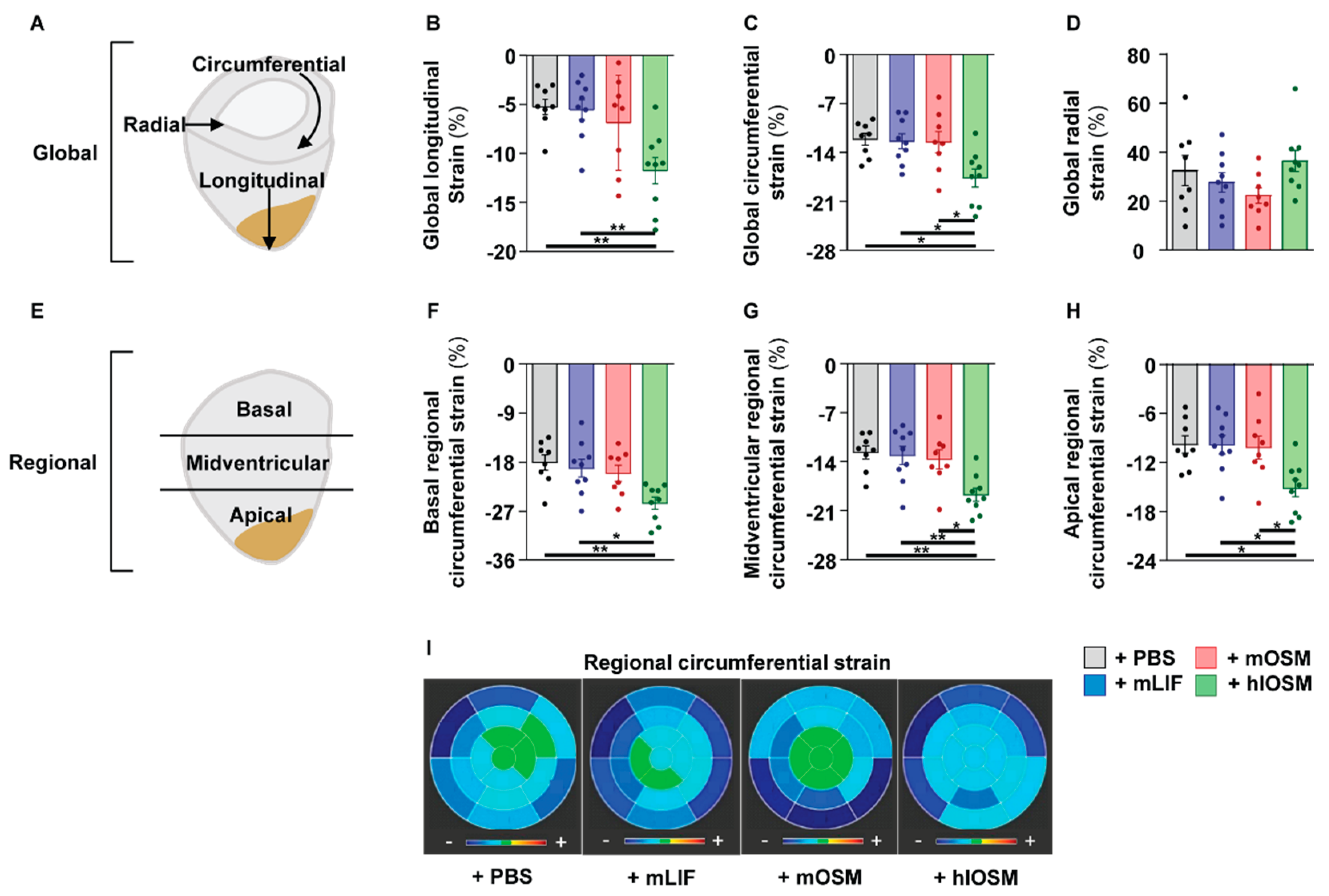
2.5. Simultaneous Activation of the OSMR and LIFR by hlOSM after the Onset of MI Preserves Cardiac Architecture and Contractility
3. Discussion
4. Materials and Methods
4.1. Recombinant Proteins
4.2. Cardiomyocytes Isolation and Cultivation
4.3. Cell Viability Assays
4.4. Myocardial Infarction and Recombinant Protein Administration
4.5. Cardiac Magnetic Resonance Imaging
4.6. RNA Sequence Analysis and Gene Set Enrichment Analysis
4.7. Protein Extraction and Immunoblot Analysis
4.8. Immunofluorescence
4.9. Statistical Analysis and Structural Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frangogiannis, N.G. Regulation of the inflammatory response in cardiac repair. Circ. Res. 2012, 110, 159–173. [Google Scholar] [CrossRef]
- Hedayat, M.; Mahmoudi, M.J.; Rose, N.R.; Rezaei, N. Proinflammatory cytokines in heart failure: Double-edged swords. Heart Fail. Rev. 2010, 15, 543–562. [Google Scholar] [CrossRef]
- Vistnes, M.; Hoiseth, A.D.; Rosjo, H.; Nygard, S.; Pettersen, E.; Soyseth, V.; Hurlen, P.; Christensen, G.; Omland, T. Lack of pro-inflammatory cytokine mobilization predicts poor prognosis in patients with acute heart failure. Cytokine 2013, 61, 962–969. [Google Scholar] [CrossRef]
- Hartman, M.H.T.; Groot, H.E.; Leach, I.M.; Karper, J.C.; van der Harst, P. Translational overview of cytokine inhibition in acute myocardial infarction and chronic heart failure. Trends Cardiovasc. Med. 2018, 28, 369–379. [Google Scholar] [CrossRef]
- Kubin, T.; Poling, J.; Kostin, S.; Gajawada, P.; Hein, S.; Rees, W.; Wietelmann, A.; Tanaka, M.; Lorchner, H.; Schimanski, S.; et al. Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 2011, 9, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, D.; Wei, L.; Zhao, Z.; Qi, X.; Li, Z.; Sun, D. OSM Enhances Angiogenesis and Improves Cardiac Function after Myocardial Infarction. BioMed Res. Int. 2015, 2015, 317905. [Google Scholar] [CrossRef]
- Zou, Y.; Takano, H.; Mizukami, M.; Akazawa, H.; Qin, Y.; Toko, H.; Sakamoto, M.; Minamino, T.; Nagai, T.; Komuro, I. Leukemia inhibitory factor enhances survival of cardiomyocytes and induces regeneration of myocardium after myocardial infarction. Circulation 2003, 108, 748–753. [Google Scholar] [CrossRef]
- Kanda, M.; Nagai, T.; Takahashi, T.; Liu, M.L.; Kondou, N.; Naito, A.T.; Akazawa, H.; Sashida, G.; Iwama, A.; Komuro, I.; et al. Leukemia Inhibitory Factor Enhances Endogenous Cardiomyocyte Regeneration after Myocardial Infarction. PLoS ONE 2016, 11, e0156562. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.F.; Pirolli, T.J.; Jayasankar, V.; Morine, K.J.; Moise, M.A.; Fisher, O.; Gardner, T.J.; Patterson, P.H.; Woo, Y.J. Targeted overexpression of leukemia inhibitory factor to preserve myocardium in a rat model of postinfarction heart failure. J. Thorac. Cardiovasc. Surg. 2004, 128, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Zgheib, C.; Zouein, F.A.; Kurdi, M.; Booz, G.W. Chronic treatment of mice with leukemia inhibitory factor does not cause adverse cardiac remodeling but improves heart function. Eur. Cytokine Netw. 2012, 23, 191–197. [Google Scholar] [CrossRef]
- Giovannini, M.; Djabali, M.; McElligott, D.; Selleri, L.; Evans, G.A. Tandem linkage of genes coding for leukemia inhibitory factor (LIF) and oncostatin M (OSM) on human chromosome 22. Cytogenet. Genome Res. 1993, 64, 240–244. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Muller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334 Pt 2, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Chollangi, S.; Mather, T.; Rodgers, K.K.; Ash, J.D. A unique loop structure in oncostatin M determines binding affinity toward oncostatin M receptor and leukemia inhibitory factor receptor. J. Biol. Chem. 2012, 287, 32848–32859. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, M.; Hara, T.; Kim, H.; Murate, T.; Miyajima, A. Oncostatin M and leukemia inhibitory factor do not use the same functional receptor in mice. Blood 1997, 90, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Han, S.S.; Yun, H.; Son, D.J.; Tompkins, V.S.; Peng, L.; Chung, S.T.; Kim, J.S.; Park, E.S.; Janz, S. NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol. Cancer 2010, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Barre, B.; Vigneron, A.; Coqueret, O. The STAT3 transcription factor is a target for the Myc and riboblastoma proteins on the Cdc25A promoter. J. Biol. Chem. 2005, 280, 15673–15681. [Google Scholar] [CrossRef]
- Adrian-Segarra, J.M.; Sreenivasan, K.; Gajawada, P.; Lorchner, H.; Braun, T.; Poling, J. The AB loop of oncostatin M (OSM) determines species-specific signaling in humans and mice. J. Biol. Chem. 2018, 293, 20181–20199. [Google Scholar] [CrossRef]
- Hilfiker-Kleiner, D.; Hilfiker, A.; Fuchs, M.; Kaminski, K.; Schaefer, A.; Schieffer, B.; Hillmer, A.; Schmiedl, A.; Ding, Z.; Podewski, E.; et al. Signal transducer and activator of transcription 3 is required for myocardial capillary growth, control of interstitial matrix deposition, and heart protection from ischemic injury. Circ. Res. 2004, 95, 187–195. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Rajasingh, J.; Lambers, E.; Qin, G.; Losordo, D.W.; Kishore, R. IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR. Circ. Res. 2009, 104, e9–e18. [Google Scholar] [CrossRef]
- Obana, M.; Maeda, M.; Takeda, K.; Hayama, A.; Mohri, T.; Yamashita, T.; Nakaoka, Y.; Komuro, I.; Takeda, K.; Matsumiya, G.; et al. Therapeutic activation of signal transducer and activator of transcription 3 by interleukin-11 ameliorates cardiac fibrosis after myocardial infarction. Circulation 2010, 121, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, D.; Obana, M.; Miyawaki, A.; Maeda, M.; Nakayama, H.; Fujio, Y. Cardiac-specific ablation of the STAT3 gene in the subacute phase of myocardial infarction exacerbated cardiac remodeling. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H471–H480. [Google Scholar] [CrossRef]
- Heusch, G.; Musiolik, J.; Kottenberg, E.; Peters, J.; Jakob, H.; Thielmann, M. STAT5 activation and cardioprotection by remote ischemic preconditioning in humans: Short communication. Circ. Res. 2012, 110, 111–115. [Google Scholar] [CrossRef]
- Soond, S.M.; Latchman, D.S.; Stephanou, A. STAT signalling in the heart and cardioprotection. Expert Rev. Mol. Med. 2006, 8, 1–16. [Google Scholar] [CrossRef]
- Poling, J.; Gajawada, P.; Lorchner, H.; Polyakova, V.; Szibor, M.; Bottger, T.; Warnecke, H.; Kubin, T.; Braun, T. The Janus face of OSM-mediated cardiomyocyte dedifferentiation during cardiac repair and disease. Cell Cycle 2012, 11, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Hadebe, N.; Cour, M.; Lecour, S. The SAFE pathway for cardioprotection: Is this a promising target? Basic Res. Cardiol. 2018, 113, 9. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef]
- Lafontant, P.J.; Burns, A.R.; Donnachie, E.; Haudek, S.B.; Smith, C.W.; Entman, M.L. Oncostatin M differentially regulates CXC chemokines in mouse cardiac fibroblasts. Am. J. Physiol. Cell Physiol. 2006, 291, C18–C26. [Google Scholar] [CrossRef]
- Amzulescu, M.S.; de Craene, M.; Langet, H.; Pasquet, A.; Vancraeynest, D.; Pouleur, A.C.; Vanoverschelde, J.L.; Gerber, B.L. Myocardial strain imaging: Review of general principles, validation, and sources of discrepancies. Eur. Heart J. Cardiovasc. Imaging 2019, 20, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Mangion, K.; McComb, C.; Auger, D.A.; Epstein, F.H.; Berry, C. Magnetic Resonance Imaging of Myocardial Strain After Acute ST-Segment-Elevation Myocardial Infarction: A Systematic Review. Circ. Cardiovasc. Imaging 2017, 10, e006498. [Google Scholar] [CrossRef]
- Sengupta, P.P.; Narula, J. Cardiac strain as a universal biomarker: Interpreting the sounds of uneasy heart muscle cells. JACC Cardiovasc. Imaging 2014, 7, 534–536. [Google Scholar] [CrossRef][Green Version]
- Hutchins, G.M.; Bulkley, B.H. Infarct expansion versus extension: Two different complications of acute myocardial infarction. Am. J. Cardiol. 1978, 41, 1127–1132. [Google Scholar] [CrossRef]
- Richardson, W.J.; Holmes, J.W. Why Is Infarct Expansion Such an Elusive Therapeutic Target? J. Cardiovasc. Transl. Res. 2015, 8, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Fukuda, K.; Pan, J.; Makino, S.; Baba, A.; Hori, S.; Ogawa, S. a potent cardiac hypertrophic cytokine, activates the JAK/STAT pathway in rat cardiomyocytes. Circ. Res. 1997, 81, 656–663. [Google Scholar] [CrossRef]
- Sano, M.; Fukuda, K.; Kodama, H.; Pan, J.; Saito, M.; Matsuzaki, J.; Takahashi, T.; Makino, S.; Kato, T.; Ogawa, S. Interleukin-6 family of cytokines mediate angiotensin II-induced cardiac hypertrophy in rodent cardiomyocytes. J. Biol. Chem. 2000, 275, 29717–29723. [Google Scholar] [CrossRef]
- Schuster, E.H.; Bulkley, B.H. Expansion of transmural myocardial infarction: A pathophysiologic factor in cardiac rupture. Circulation 1979, 60, 1532–1538. [Google Scholar] [CrossRef]
- Erlebacher, J.A.; Weiss, J.L.; Weisfeldt, M.L.; Bulkley, B.H. Early dilation of the infarcted segment in acute transmural myocardial infarction: Role of infarct expansion in acute left ventricular enlargement. J. Am. Coll. Cardiol. 1984, 4, 201–208. [Google Scholar] [CrossRef]
- Pirolo, J.S.; Hutchins, G.M.; Moore, G.W. Infarct expansion: Pathologic analysis of 204 patients with a single myocardial infarct. J. Am. Coll. Cardiol. 1986, 7, 349–354. [Google Scholar] [CrossRef]
- Gotthardt, D.; Trifinopoulos, J.; Sexl, V.; Putz, E.M. JAK/STAT Cytokine Signaling at the Crossroad of NK Cell Development and Maturation. Front. Immunol. 2019, 10, 2590. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jing, X.Y.; Shen, Y.J.; Wang, T.L.; Ou, C.; Lu, S.F.; Cai, Y.; Li, Q.; Chen, X.; Ding, Y.J.; et al. Stat5-dependent cardioprotection in late remote ischaemia preconditioning. Cardiovasc. Res. 2018, 114, 679–689. [Google Scholar] [CrossRef]
- Bolli, R.; Stein, A.B.; Guo, Y.; Wang, O.L.; Rokosh, G.; Dawn, B.; Molkentin, J.D.; Sanganalmath, S.K.; Zhu, Y.; Xuan, Y.T. A murine model of inducible, cardiac-specific deletion of STAT3: Its use to determine the role of STAT3 in the upregulation of cardioprotective proteins by ischemic preconditioning. J. Mol. Cell Cardiol. 2011, 50, 589–597. [Google Scholar] [CrossRef]
- Negoro, S.; Kunisada, K.; Fujio, Y.; Funamoto, M.; Darville, M.I.; Eizirik, D.L.; Osugi, T.; Izumi, M.; Oshima, Y.; Nakaoka, Y.; et al. Activation of signal transducer and activator of transcription 3 protects cardiomyocytes from hypoxia/reoxygenation-induced oxidative stress through the upregulation of manganese superoxide dismutase. Circulation 2001, 104, 979–981. [Google Scholar] [CrossRef]
- Boengler, K.; Hilfiker-Kleiner, D.; Heusch, G.; Schulz, R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res. Cardiol. 2010, 105, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Szczepanek, K.; Chen, Q.; Derecka, M.; Salloum, F.N.; Zhang, Q.; Szelag, M.; Cichy, J.; Kukreja, R.C.; Dulak, J.; Lesnefsky, E.J.; et al. Mitochondrial-targeted Signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J. Biol. Chem. 2011, 286, 29610–29620. [Google Scholar] [CrossRef]
- Zouein, F.A.; Kurdi, M.; Booz, G.W. LIF and the heart: Just another brick in the wall? Eur. Cytokine Netw. 2013, 24, 11–19. [Google Scholar] [CrossRef]
- Chueh, F.Y.; Leong, K.F.; Yu, C.L. Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem. Biophys. Res. Commun. 2010, 402, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Heusch, G.; Musiolik, J.; Gedik, N.; Skyschally, A. Mitochondrial STAT3 activation and cardioprotection by ischemic postconditioning in pigs with regional myocardial ischemia/reperfusion. Circ. Res. 2011, 109, 1302–1308. [Google Scholar] [CrossRef]
- Rosano, G.M.; Fini, M.; Caminiti, G.; Barbaro, G. Cardiac metabolism in myocardial ischemia. Curr. Pharm. Des. 2008, 14, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, P.; Zhao, P.; Angelis, E.; Ruan, H.; Korge, P.; Olson, A.; Wang, Y.; Jin, E.S.; Jeffrey, F.M.; Portman, M.; et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Investig. 2010, 120, 1494–1505. [Google Scholar] [CrossRef]
- Fukuda, R.; Marin-Juez, R.; El-Sammak, H.; Beisaw, A.; Ramadass, R.; Kuenne, C.; Guenther, S.; Konzer, A.; Bhagwat, A.M.; Graumann, J.; et al. Stimulation of glycolysis promotes cardiomyocyte proliferation after injury in adult zebrafish. EMBO Rep. 2020, 21, e49752. [Google Scholar] [CrossRef]
- Ecker, A.; Simma, O.; Hoelbl, A.; Kenner, L.; Beug, H.; Moriggl, R.; Sexl, V. The dark and the bright side of Stat3: Proto-oncogene and tumor-suppressor. Front. Biosci. 2009, 14, 2944–2958. [Google Scholar] [CrossRef]
- O’Connell, T.D.; Rodrigo, M.C.; Simpson, P.C. Isolation and culture of adult mouse cardiac myocytes. Methods Mol. Biol. 2007, 357, 271–296. [Google Scholar] [PubMed]
- Lorchner, H.; Poling, J.; Gajawada, P.; Hou, Y.; Polyakova, V.; Kostin, S.; Adrian-Segarra, J.M.; Boettger, T.; Wietelmann, A.; Warnecke, H.; et al. Myocardial healing requires Reg3beta-dependent accumulation of macrophages in the ischemic heart. Nat. Med. 2015, 21, 353–362. [Google Scholar] [CrossRef]
- Schneider, J.E.; Wiesmann, F.; Lygate, C.A.; Neubauer, S. How to perform an accurate assessment of cardiac function in mice using high-resolution magnetic resonance imaging. J. Cardiovasc. Magn. Reson. 2006, 8, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lörchner, H.; Adrian-Segarra, J.M.; Waechter, C.; Wagner, R.; Góes, M.E.; Brachmann, N.; Sreenivasan, K.; Wietelmann, A.; Günther, S.; Doll, N.; et al. Concomitant Activation of OSM and LIF Receptor by a Dual-Specific hlOSM Variant Confers Cardioprotection after Myocardial Infarction in Mice. Int. J. Mol. Sci. 2022, 23, 353. https://doi.org/10.3390/ijms23010353
Lörchner H, Adrian-Segarra JM, Waechter C, Wagner R, Góes ME, Brachmann N, Sreenivasan K, Wietelmann A, Günther S, Doll N, et al. Concomitant Activation of OSM and LIF Receptor by a Dual-Specific hlOSM Variant Confers Cardioprotection after Myocardial Infarction in Mice. International Journal of Molecular Sciences. 2022; 23(1):353. https://doi.org/10.3390/ijms23010353
Chicago/Turabian StyleLörchner, Holger, Juan M. Adrian-Segarra, Christian Waechter, Roxanne Wagner, Maria Elisa Góes, Nathalie Brachmann, Krishnamoorthy Sreenivasan, Astrid Wietelmann, Stefan Günther, Nicolas Doll, and et al. 2022. "Concomitant Activation of OSM and LIF Receptor by a Dual-Specific hlOSM Variant Confers Cardioprotection after Myocardial Infarction in Mice" International Journal of Molecular Sciences 23, no. 1: 353. https://doi.org/10.3390/ijms23010353
APA StyleLörchner, H., Adrian-Segarra, J. M., Waechter, C., Wagner, R., Góes, M. E., Brachmann, N., Sreenivasan, K., Wietelmann, A., Günther, S., Doll, N., Braun, T., & Pöling, J. (2022). Concomitant Activation of OSM and LIF Receptor by a Dual-Specific hlOSM Variant Confers Cardioprotection after Myocardial Infarction in Mice. International Journal of Molecular Sciences, 23(1), 353. https://doi.org/10.3390/ijms23010353





