Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum
Abstract
1. Introduction
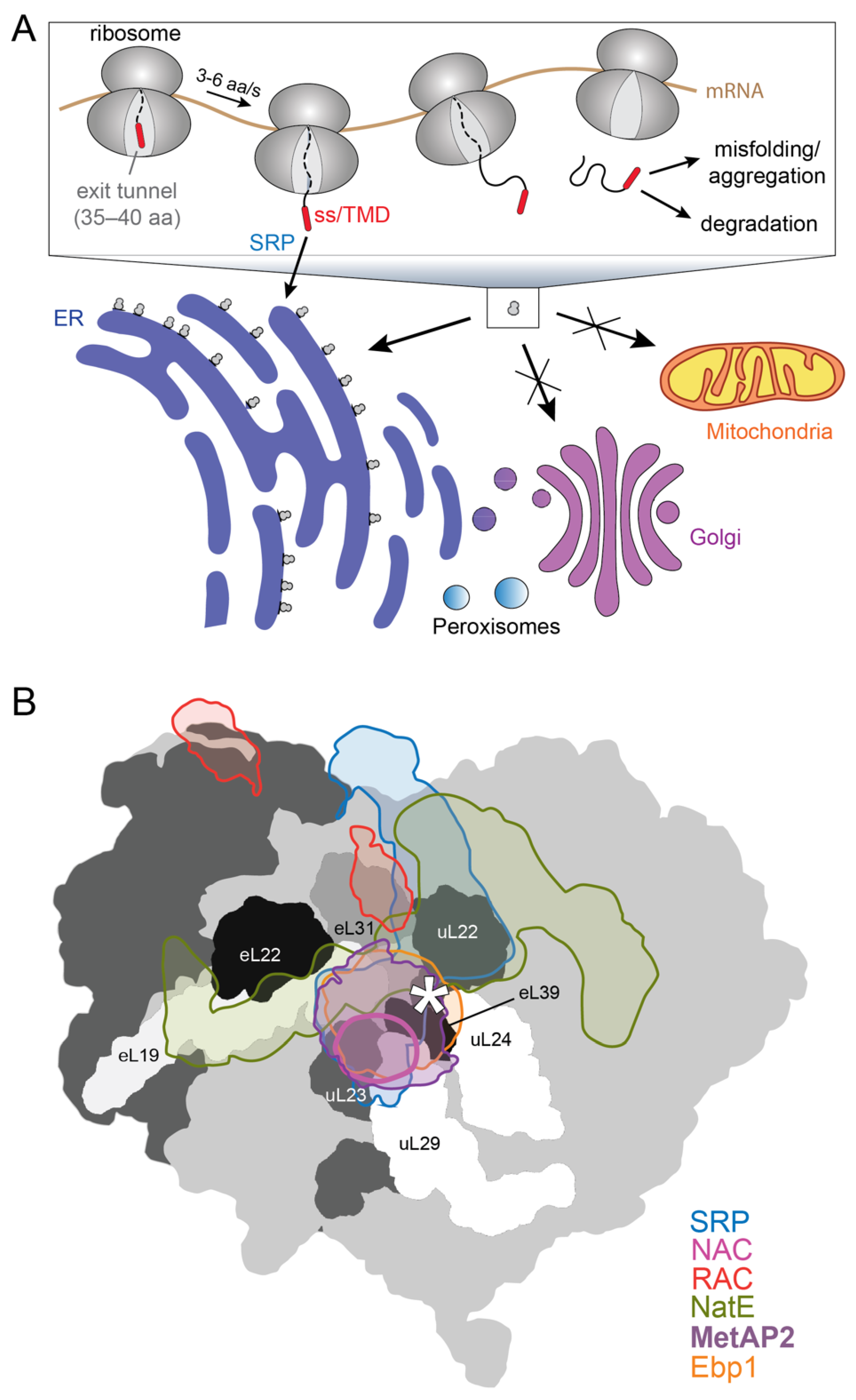
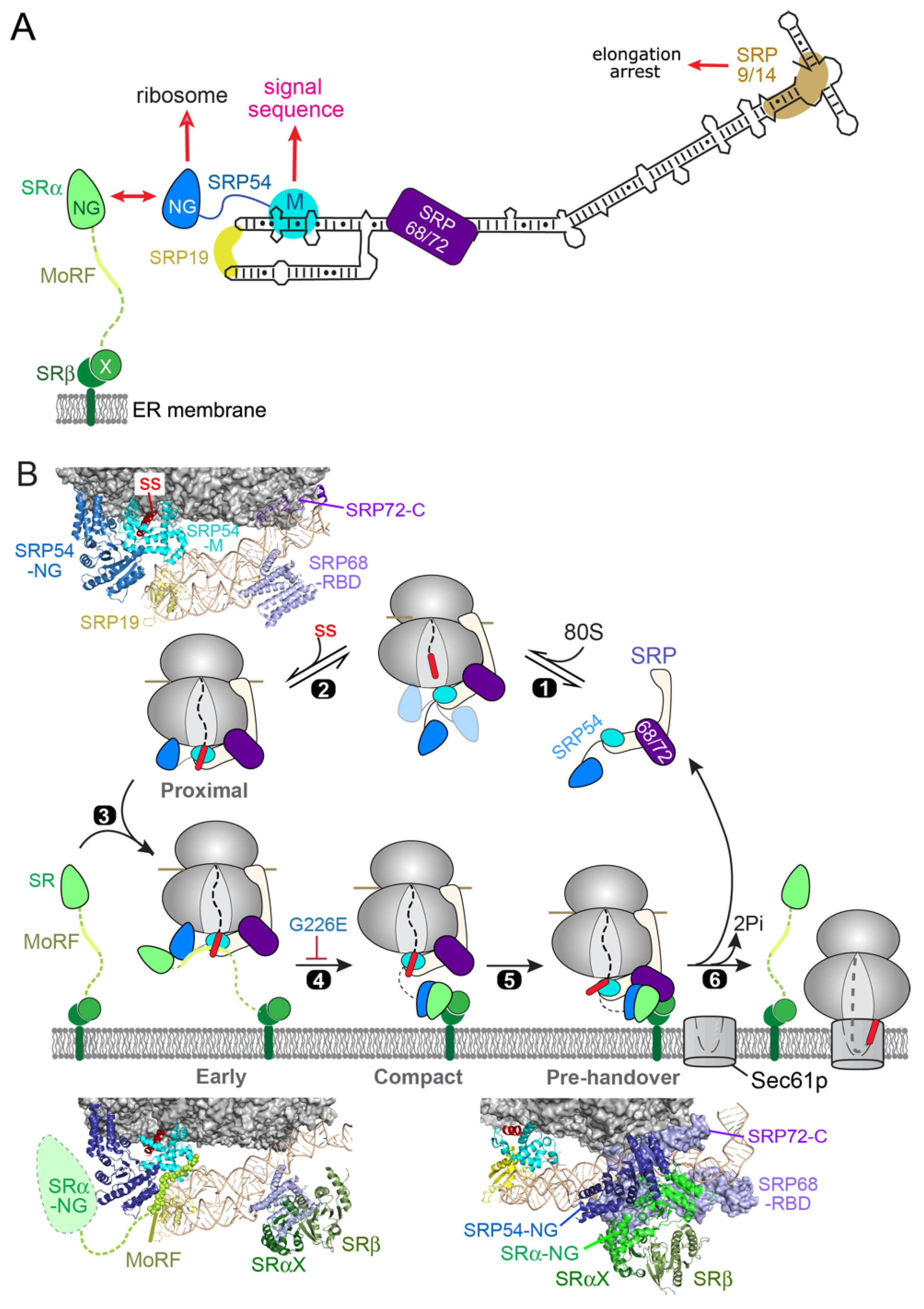
2. SRP-Dependent Cotranslational Protein Targeting

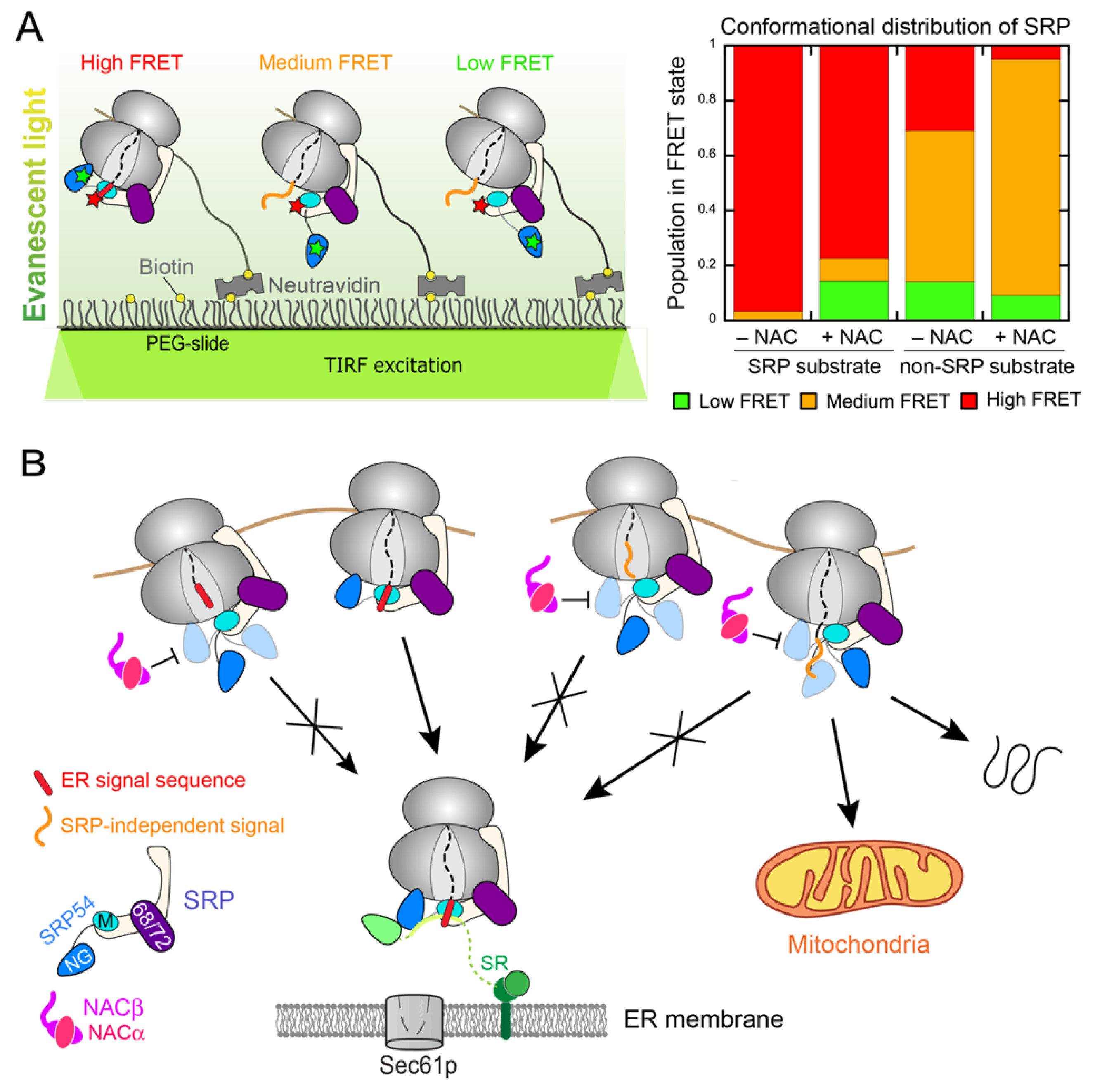
3. NAC: A Triage Factor during Cotranslational Protein Targeting
4. Perspectives and Open Questions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Walter, P.; Johnson, A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Ann. Rev. Cell Biol. 1994, 10, 87–119. [Google Scholar] [CrossRef]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S. Signal recognition particle: An essential protein-targeting machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Keenan, R.J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2021, 1–18. [Google Scholar] [CrossRef]
- Spiess, M.; Junne, T.; Janoschke, M. Membrane Protein Integration and Topogenesis at the ER. Protein J. 2019, 38, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. Signal sequences: The limits of variation. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- Gierasch, L.M. Signal sequences. Biochemistry 1989, 28, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.J.; Chen, J.C.; Miao, Y.; Shao, Y.; Lin, J.; Bock, P.E.; Johnson, A.E. Signal recognition particle binds to ribo-some-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003, 278, 18628–18637. [Google Scholar] [CrossRef] [PubMed]
- Siegel, V.; Walter, P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988, 7, 1769–1775. [Google Scholar] [CrossRef]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.J.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Deuerling, E.; Gamerdinger, M.; Kreft, S.G. Chaperone Interactions at the Ribosome. Cold Spring Harb. Perspect. Biol. 2019, 11, a033977. [Google Scholar] [CrossRef]
- Cassaignau, A.M.E.; Cabrita, L.D.; Christodoulou, J. How Does the Ribosome Fold the Proteome? Annu. Rev. Biochem. 2020, 89, 389–415. [Google Scholar] [CrossRef] [PubMed]
- Kramer, G.; Shiber, A.; Bukau, B. Mechanisms of Cotranslational Maturation of Newly Synthesized Proteins. Annu. Rev. Biochem. 2019, 88, 337–364. [Google Scholar] [CrossRef] [PubMed]
- Batey, R.T.; Rambo, R.P.; Lucast, L.; Rha, B.; Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 2000, 287, 1232–1239. [Google Scholar] [CrossRef]
- Batey, R.T.; Sagar, M.B.; Doudna, J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001, 307, 229–246. [Google Scholar] [CrossRef]
- Hainzl, T.; Huang, S.; Merilainen, G.; Brannstrom, K.; Sauer-Eriksson, A.E. Structural basis of signal sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 2011, 18, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Janda, C.Y.; Li, J.; Oubridge, C.; Hernandez, H.; Robinson, C.V.; Nagai, K. Recognition of a signal peptide by the signal recognition particle. Nature 2010, 465, 507–510. [Google Scholar] [CrossRef]
- Krieg, U.C.; Walter, P.; Johnson, A.E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA 1986, 83, 8604–8608. [Google Scholar] [CrossRef]
- Kurzchalia, T.V.; Wiedmann, M.; Girshovich, A.S.; Bochkareva, E.S.; Bielka, H.; Rapoport, T.A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature 1986, 320, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Hegde, R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife 2015, 4, e07975. [Google Scholar] [CrossRef]
- Jomaa, A.; Eitzinger, S.; Zhu, Z.; Chandrasekar, S.; Kobayashi, K.; Shan, S.O.; Ban, N. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 2021, 36, 109350. [Google Scholar] [CrossRef]
- Jomaa, A.; Boehringer, D.; Leibundgut, M.; Ban, N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016, 7, 10471. [Google Scholar] [CrossRef] [PubMed]
- Schaffitzel, C.; Oswald, M.; Berger, I.; Ishikawa, T.; Abrahams, J.P.; Koerten, H.K.; Koning, R.I.; Ban, N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 2006, 444, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Peluso, P.; Herschlag, D.; Nock, S.; Freymann, D.M.; Johnson, A.E.; Walter, P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 2000, 288, 1640–1643. [Google Scholar] [CrossRef] [PubMed]
- Peluso, P.; Shan, S.; Nock, S.; Herschlag, D.; Walter, P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 2001, 40, 15224–15233. [Google Scholar] [CrossRef]
- Egea, P.F.; Shan, S.; Napetschnig, J.; Savage, D.F.; Walter, P.; Stroud, R.M. Substrate twinning activates the signal recog-nition particle and its receptor. Nature 2004, 427, 215–221. [Google Scholar] [CrossRef]
- Focia, P.J.; Shepotinovskaya, I.V.; Seidler, J.A.; Freymann, D.M. Heterodimeric GTPase Core of the SRP Targeting Complex. Science 2004, 303, 373–377. [Google Scholar] [CrossRef]
- Wild, K.; Bange, G.; Motiejunas, D.; Kribelbauer, J.; Hendricks, A.; Segnitz, B.; Wade, R.C.; Sinning, I. Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex. J. Mol. Biol. 2016, 428, 2880–2897. [Google Scholar] [CrossRef]
- Shan, S.; Stroud, R.; Walter, P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004, 2, e320. [Google Scholar] [CrossRef]
- Shan, S.O. ATPase and GTPase Tangos Drive Intracellular Protein Transport. Trends Biochem. Sci. 2016, 41, 1050–1060. [Google Scholar] [CrossRef]
- Saraogi, I.; Shan, S. Molecular mechanism of co-translational protein targeting by the Signal Recognition Particle. Traffic 2011, 12, 535–542. [Google Scholar] [CrossRef]
- Estrozi, L.F.; Boehringer, D.; Shan, S.; Ban, N.; Schaffitzel, C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat. Struct. Mol. Biol. 2011, 18, 88–90. [Google Scholar] [CrossRef]
- Zhang, X.; Schaffitzel, C.; Ban, N.; Shan, S. Multiple conformational changes in a GTPase complex regulate protein targeting. Proc. Natl. Acad. Sci. USA 2009, 106, 1754–1759. [Google Scholar] [CrossRef]
- Zhang, X.; Kung, S.; Shan, S. Demonstration of a two-step mechanism for assembly of the SRP–SRP receptor complex: Implications for the catalytic role of SRP RNA. J. Mol. Biol. 2008, 381, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Ataide, S.F.; Schmitz, N.; Shenk, K.; Ke, A.; Shan, S.; Doudna, A.; Ban, N. The crystal structure of the Signal Recognition Particle in complex with its receptor. Science 2011, 381, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Arslan, S.; Akopian, D.; Ha, T.; Shan, S. Activated GTPase movement on an RNA scaffold drives cotranslational protein targeting. Nature 2012, 492, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shan, S.O. Fidelity of cotranslational protein targeting by the signal recognition particle. Annu. Rev. Biophys. 2014, 43, 381–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rashid, R.; Wang, K.; Shan, S. Sequential checkpoints govern fidelity during co-translational protein targeting. Science 2010, 328, 757–760. [Google Scholar] [CrossRef]
- Akopian, D.; Dalai, K.; Shen, K.; Duong, F.; Shan, S. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 2013, 200, 397–405. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Disassembly and reconstitution of signal recognition particle. Cell 1983, 34, 525–533. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endo-plasmic reticulum. Nature 1982, 299, 691–698. [Google Scholar] [CrossRef]
- Bahari, L.; Parlitz, R.; Eitan, A.; Stjepanovic, G.; Bochkareva, E.S.; Sinning, I.; Bibi, E. Membrane targeting of ribosomes and their release require distinct and separable functions of FtsY. J. Biol. Chem. 2007, 282, 32168–32175. [Google Scholar] [CrossRef]
- Parlitz, R.; Eitan, A.; Stjepanovic, G.; Bahari, L.; Bange, G.; Bibi, E.; Sinning, I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J. Biol. Chem. 2007, 282, 32176–32184. [Google Scholar] [CrossRef]
- De Leeuw, E.; Poland, D.; Mol, O.; Sinning, I.; ten Hagen-Jongman, C.M.; Oudega, B.; Luirink, J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997, 416, 225–229. [Google Scholar] [CrossRef]
- de Leeuw, E.; te Kaat, K.; Moser, C.; Memestrina, G.; Demel, R.; de Kruijff, B.; Oudegam, B.; Luirink, J.; Sinning, I. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 2000, 19, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Hwang Fu, Y.H.; Huang, W.Y.C.; Shen, K.; Groves, J.T.; Miller, T.; Shan, S.O. Two-step membrane binding by the bacterial SRP receptor enable efficient and accurate Co-translational protein targeting. eLife 2017, 6, e25885. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, B.; Wild, K.; Pool, M.R.; Sinning, I. Structure and Switch Cycle of SRbeta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes. Structure 2015, 23, 1838–1847. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schlenker, O.; Hendricks, A.; Sinning, I.; Wild, K. The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains. J. Biol. Chem. 2006, 281, 8898–8906. [Google Scholar] [CrossRef]
- Hwang Fu, Y.H.; Chandrasekar, S.; Lee, J.H.; Shan, S.O. A molecular recognition feature mediates ribosome-induced SRP-receptor assembly during protein targeting. J. Cell Biol. 2019, 218, 3307–3319. [Google Scholar] [CrossRef]
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I. Pool MR. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 10133. [Google Scholar] [CrossRef]
- Lee, J.H.; Chandrasekar, S.; Chung, S.; Hwang Fu, Y.H.; Liu, D.; Weiss, S.; Shan, S. Sequential activation of human signal recognition particle by the ribosome and signal sequence drives efficient protein targeting. Proc. Natl. Acad. Sci. USA 2018, 115, E5487–E5496. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jomaa, A.; Chung, S.; Hwang Fu, Y.H.; Qian, R.; Sun, X.; Hsieh, H.H.; Chandrasekar, S.; Bi, X.; Mattei, S.; et al. Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Sci. Adv. 2021, 7, eabg0942. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Jomaa, A.; Lee, J.H.; Chandrasekar, S.; Boehringer, D.; Shan, S.O.; Ban, N. Structure of a prehandover mammalian ribo-somal SRP.SRP receptor targeting complex. Science 2018, 360, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Schurch, C.; Schaefer, T.; Muller, J.S.; Hanns, P.; Arnone, M.; Dumlin, A.; Schärer, J.; Sinning, I.; Wild, K.; Skokowa, J.; et al. SRP54 mutations induce congenital neutropenia via dominant-negative effects on XBP1 splicing. Blood J. Am. Soc. Hematol. 2021, 137, 1340–1352. [Google Scholar] [CrossRef] [PubMed]
- Juaire, K.D.; Lapouge, K.; Becker, M.M.M.; Kotova, I.; Michelhans, M.; Carapito, R.; Wild, K.; Bahram, S.; Sinning, I. Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia. Structure 2021, 29, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.H.; Lee, J.H.; Chandrasekar, S.; Shan, S.O. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Zhang, X.; Shan, S. Synergiestic action between the SRP RNA and translating ribosome allows efficient delivery of correct cargos during co-translational protein targeting. RNA 2011, 17, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Raue, U.; Oellerer, S.; Rospert, S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 2007, 282, 7809–7816. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, W.; Wang, L.; Zhang, X.C.; Li, X.; Rao, Z. Crystal structures of NAC domains of human nascent polypep-tide-associated complex (NAC) and its alphaNAC subunit. Protein. Cell 2010, 1, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, R.D.; Hofmann, D.; Merz, F.; Nikolay, R.; Rauch, T.; Graf, C.; Deuerling, E. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 2006, 281, 2847–2857. [Google Scholar] [CrossRef]
- Pech, M.; Spreter, T.; Beckmann, R.; Beatrix, B. Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J. Biol. Chem. 2010, 285, 19679–19687. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Kobayashi, K.; Wallisch, A.; Kreft, S.G.; Sailer, C.; Schlomer, R.; Sachs, N.; Jomaa, A.; Stengel, F.; Ban, N.; et al. Early Scanning of Nascent Polypeptides inside the Ribosomal Tunnel by NAC. Mol. Cell 2019, 75, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, R.D.; Deuerling, E. Molecular guardians for newborn proteins: Ribosome-associated chaperones and their role in protein folding. Cell. Mol. Life Sci. 2005, 62, 2727–2738. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In Vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Arsenovic, P.T.; Maldonado, A.T.; Colleluori, V.D.; Bloss, T.A. Depletion of the C. elegans NAC engages the unfolded protein response, resulting in increased chaperone expression and apoptosis. PLoS ONE 2012, 7, e44038. [Google Scholar] [CrossRef]
- Deng, J.M.; Behringer, R.R. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res. 1995, 4, 264–269. [Google Scholar] [CrossRef]
- Bloss, T.A.; Witze, E.S.; Rothman, J.H. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caeno-rhabditis elegans. Nature 2003, 424, 1066–1071. [Google Scholar] [CrossRef]
- Markesich, D.C.; Gajewski, K.M.; Nazimiec, M.E.; Beckingham, K. bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery*. Development 2000, 127, 559–572. [Google Scholar] [CrossRef]
- Koplin, A.; Preissler, S.; Ilina, Y.; Koch, M.; Scior, A.; Erhardt, M.; Deuerling, E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010, 189, 57–68. [Google Scholar] [CrossRef]
- Ott, A.K.; Locher, L.; Koch, M.; Deuerling, E. Functional Dissection of the Nascent Polypeptide-Associated Complex in Sac-charomyces cerevisiae. PLoS ONE 2015, 10, e0143457. [Google Scholar] [CrossRef]
- Shen, K.; Gamerdinger, M.; Chan, R.; Gense, K.; Martin, E.M.; Sachs, N.; Knight, P.D.; Schlömer, R.; Calabrese, A.N.; Stewart, K.L.; et al. Dual Role of Ribosome-Binding Domain of NAC as a Potent Suppressor of Protein Aggregation and Aging-Related Proteinopathies. Mol. Cell 2019, 74, 729–741. [Google Scholar] [CrossRef]
- Martin, E.M.; Jackson, M.P.; Gamerdinger, M.; Gense, K.; Karamonos, T.K.; Humes, J.R.; Deuerling, E.; Ashcroft, A.E.; Radford, S.E. Conformational flexibility within the nascent polypeptide-associated complex enables its interactions with structurally diverse client proteins. J. Biol. Chem. 2018, 293, 8554–8568. [Google Scholar] [CrossRef] [PubMed]
- Kirstein-Miles, J.; Scior, A.; Deuerling, E.; Morimoto, R.I. The nascent polypeptide-associated complex is a key regulator of pro-teostasis. EMBO J. 2013, 32, 1451–1468. [Google Scholar] [CrossRef] [PubMed]
- Funfschilling, U.; Rospert, S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 1999, 10, 3289–3299. [Google Scholar] [CrossRef] [PubMed]
- Beddoe, T.; Lithgow, T. Delivery of nascent polypeptides to the mitochondrial surface. Biochim. Biophys. Acta. 2002, 1592, 35–49. [Google Scholar] [CrossRef]
- Yogev, O.; Karniely, S.; Pines, O. Translation-coupled translocation of yeast fumarase into mitochondria in Vivo. J. Biol. Chem. 2007, 282, 29222–29229. [Google Scholar] [CrossRef]
- Ponce-Rojas, J.C.; Avendano-Monsalve, M.C.; Yanez-Falcon, A.R.; Jaimes-Miranda, F.; Garay, E.; Torres-Quiroz, F.; DeLuna, A.; Funes, S. αβ′-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017, 284, 814–830. [Google Scholar] [CrossRef]
- Yotov, W.V.; Moreau, A.; St-Arnaud, R. The alpha chain of the nascent polypeptide-associated complex functions as a tran-scriptional coactivator. Mol. Cell. Biol. 1998, 18, 1303–1311. [Google Scholar] [CrossRef]
- Quelo, I.; Hurtubise, M.; St-Arnaud, R. alphaNAC requires an interaction with c-Jun to exert its transcriptional coactivation. Gene Expr. 2002, 10, 255–262. [Google Scholar] [CrossRef]
- Wiedmann, B.; Sakai, H.; Davis, T.A.; Wiedmann, M. A protein complex required for signal-sequence-specific sorting and translocation. Nature 1994, 370, 434–440. [Google Scholar] [CrossRef]
- Lauring, B.; Sakai, H.; Kreibich, G.; Wiedmann, M. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1995, 92, 5411–5415. [Google Scholar] [CrossRef]
- Powers, T.; Walter, P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 1996, 6, 331–338. [Google Scholar] [CrossRef]
- Wiedmann, B.; Prehn, S. The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett. 1999, 458, 51–54. [Google Scholar] [CrossRef]
- Raden, D.; Gilmore, R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol. Biol. Cell 1998, 9, 117–130. [Google Scholar] [CrossRef]
- Neuhof, A.; Rolls, M.M.; Jungnickel, B.; Kalies, K.U.; Rapoport, T.A. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell 1998, 9, 103–115. [Google Scholar] [CrossRef][Green Version]
- Lauring, B.; Kreibich, G.; Wiedmann, M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc. Natl. Acad. Sci. USA 1995, 92, 9435–9439. [Google Scholar] [CrossRef]
- Zhang, Y.; Berndt, U.; Golz, H.; Tais, A.; Oellerer, S.; Wolfle, T.; Fitzke, E.; Rospert, S. NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol. Biol. Cell 2012, 16, 3027–3040. [Google Scholar] [CrossRef]
- Del Alamo, M.; Hogan, D.J.; Pechmann, S.; Albanese, V.; Brown, P.O.; Frydman, J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011, 9, e1001100. [Google Scholar]
- Wang, S.; Sakai, H.; Wiedmann, M. NAC covers ribosome-associated nascent chains thereby forming a protective envi-ronment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 1995, 130, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ariosa, A.; Lee, J.H.; Wang, S.; Saraogi, I.; Shan, S.O. Regulation by a chaperone improves substrate selectivity during cotrans-lational protein targeting. Proc. Natl. Acad. Sci. USA 2015, 112, E3169–E3178. [Google Scholar] [CrossRef]
- Yang, C.I.; Hsieh, H.H.; Shan, S.O. Timing and specificity of cotranslational nascent protein modification in bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 23050–23060. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 2008, 134, 634–645. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981, 91, 557–561. [Google Scholar] [CrossRef]
- Siegel, V.; Walter, P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J. Cell Biol. 1985, 100, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Wolin, S.L.; Walter, P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989, 109, 2617–2622. [Google Scholar] [CrossRef] [PubMed]
- Halic, M.; Becker, T.; Pool, M.R.; Spahn, C.M.T.; Grassucci, R.A.; Frank, J.; Beckmann, R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004, 427, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Mary, C.; Scherrer, A.; Huck, L.; Lakkaraju, A.K.; Thomas, Y.; Johnson, A.E.; Strub, K. Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein. RNA 2010, 16, 969–979. [Google Scholar] [CrossRef]
- Lakkaraju, A.K.K.; Mary, C.; Scherrer, A.; Johnson, A.E.; Strub, K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 2008, 133, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Pechmann, S.; Chartron, J.W.; Frydman, J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP In Vivo. Nat. Struct. Mol. Biol. 2014, 21, 1100–1105. [Google Scholar] [CrossRef]
- Chartron, J.W.; Hunt, K.C.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–248. [Google Scholar] [CrossRef] [PubMed]
- Jungnickel, B.; Rapoport, T.A. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 1995, 82, 261–270. [Google Scholar] [CrossRef]
- Gorlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified compoennts of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Okreglak, V.; Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 8019–8024. [Google Scholar] [CrossRef]
- Li, L.; Zheng, J.; Wu, X.; Jiang, H. Mitochondrial AAA-ATPase Msp1 detects mislocalized tail-anchored proteins through a du-al-recognition mechanism. EMBO Rep. 2019, 20, e46989. [Google Scholar] [CrossRef]
- Chen, Y.C.; Umanah, G.K.; Dephoure, N.; Andrabi, S.A.; Gygi, S.P.; Dawson, T.M.; Dawson, V.L.; Rutter, J. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014, 33, 1548–1564. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Walter, P. Msp1/ATAD1 in Protein Quality Control and Regulation of Synaptic Activities. Annu. Rev. Cell Dev. Biol. 2020, 36, 141–164. [Google Scholar] [CrossRef]
- Wang, L.; Myasnikov, A.; Pan, X.; Walter, P. Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction. eLife 2020, 9, e54031. [Google Scholar] [CrossRef]
- Matsumoto, S.; Nakatsukasa, K.; Kakuta, C.; Tamura, Y.; Esaki, M.; Endo, T. Msp1 Clears Mistargeted Proteins by Facilitating Their Transfer from Mitochondria to the ER. Mol. Cell 2019, 76, 191–205.e10. [Google Scholar] [CrossRef]
- McKenna, M.J.; Sim, S.I.; Ordureau, A.; Wei, L.; Harper, J.W.; Shao, S.; Park, E. The endoplasmic reticulum P5A-ATPase is a trans-membrane helix dislocase. Science 2020, 369. [Google Scholar] [CrossRef] [PubMed]
- Dederer, V.; Lemberg, M.K. Transmembrane dislocases: A second chance for protein targeting. Trends Cell Biol. 2021, 31, 898–911. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, H.-H.; Shan, S.-o. Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2022, 23, 281. https://doi.org/10.3390/ijms23010281
Hsieh H-H, Shan S-o. Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum. International Journal of Molecular Sciences. 2022; 23(1):281. https://doi.org/10.3390/ijms23010281
Chicago/Turabian StyleHsieh, Hao-Hsuan, and Shu-ou Shan. 2022. "Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum" International Journal of Molecular Sciences 23, no. 1: 281. https://doi.org/10.3390/ijms23010281
APA StyleHsieh, H.-H., & Shan, S.-o. (2022). Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum. International Journal of Molecular Sciences, 23(1), 281. https://doi.org/10.3390/ijms23010281





