Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors
Abstract
:1. Introduction
2. Results
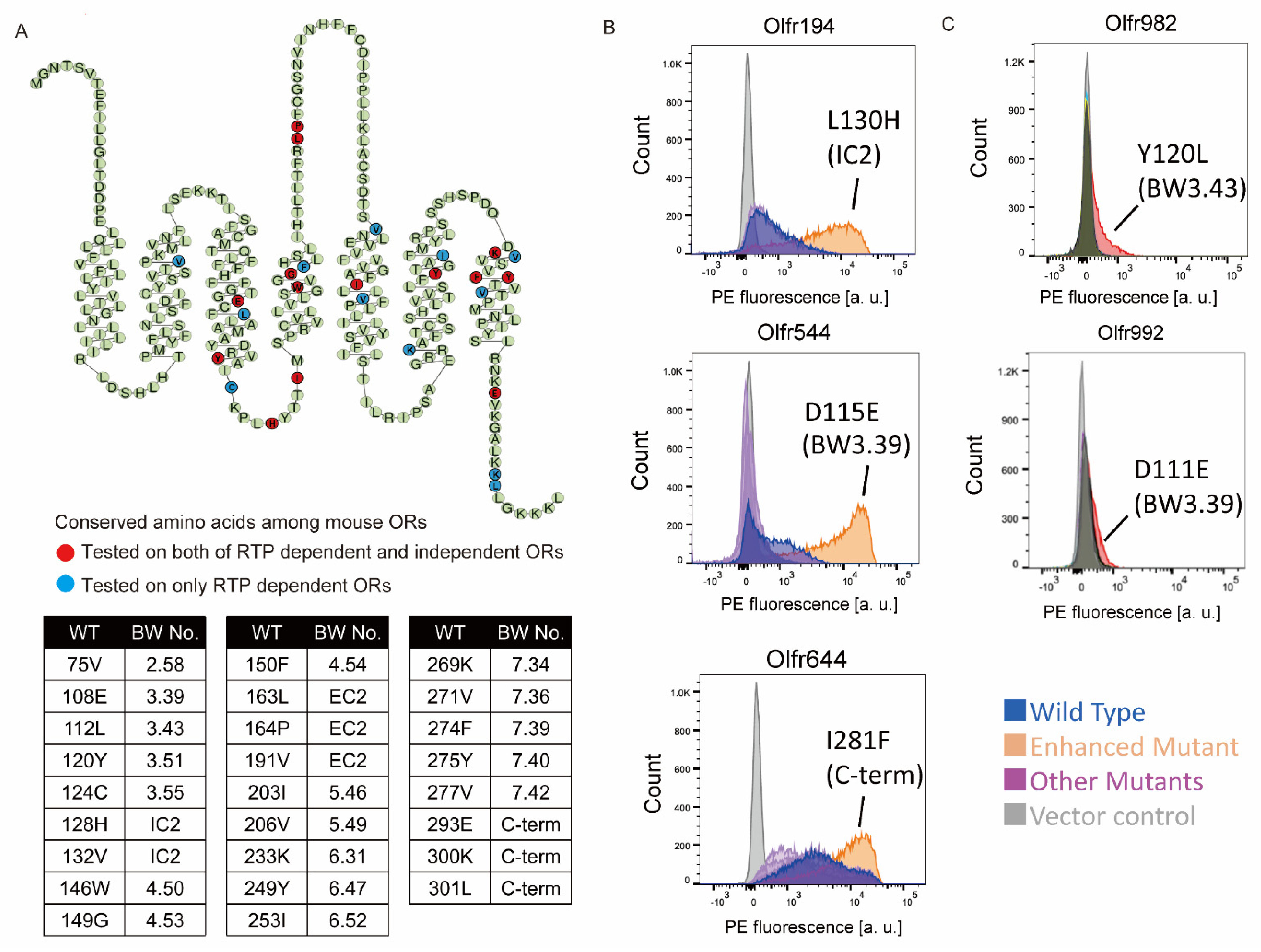
2.1. Screening of the Amino Acid Mutations That Enhance OR Expression
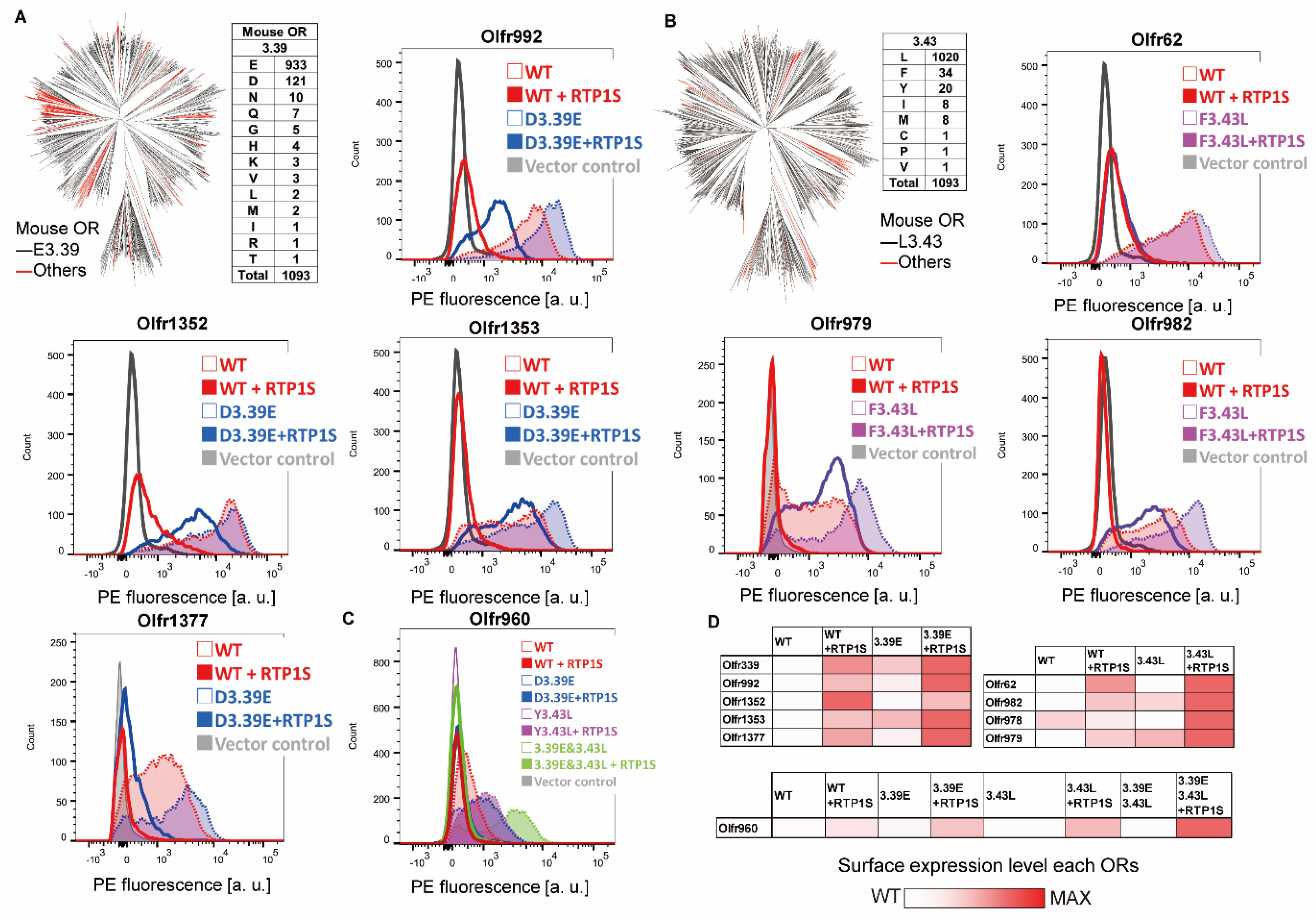
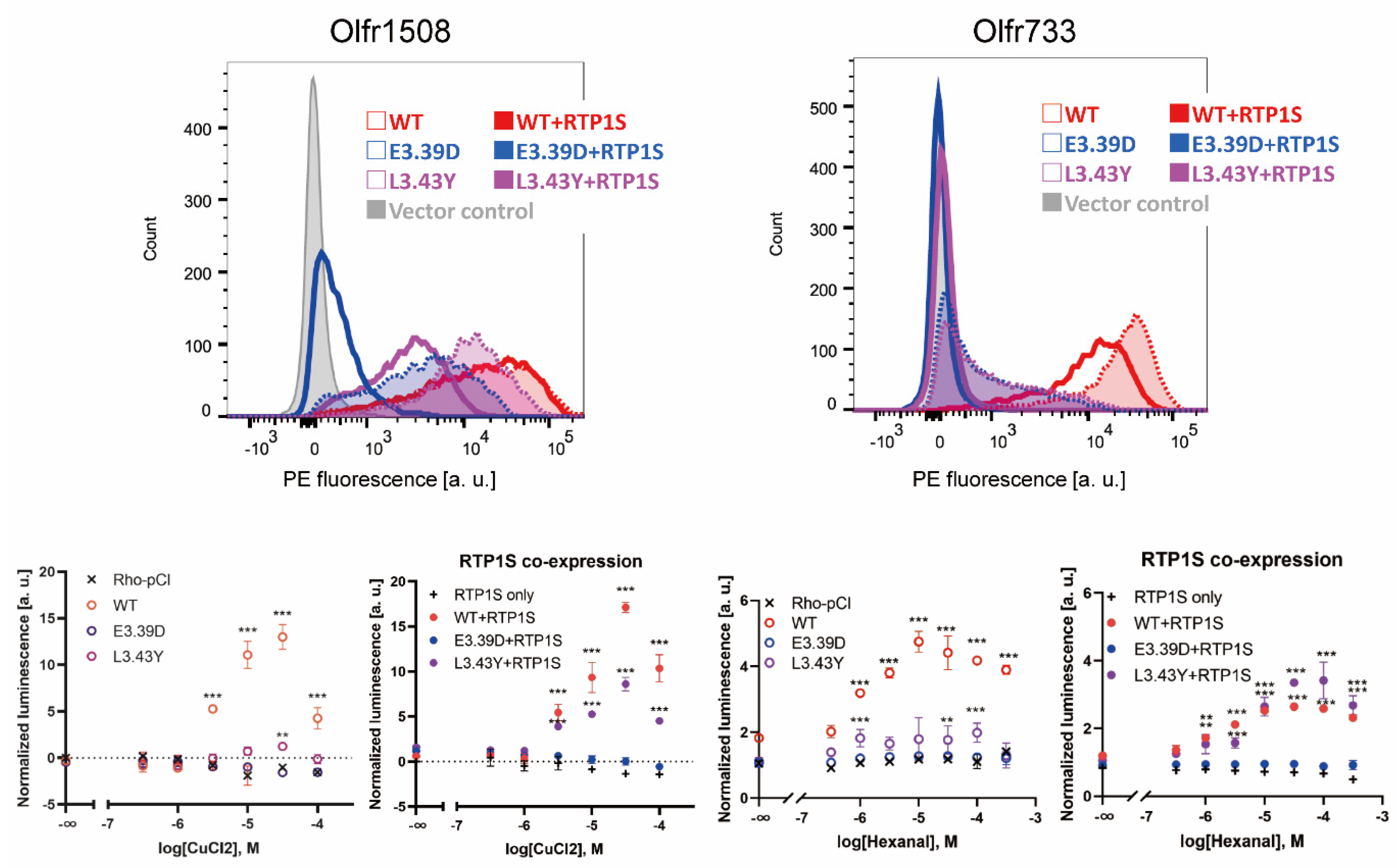
2.2. Improvement of Surface Expression by the 3.39E and 3.43L Mutations in Many ORs
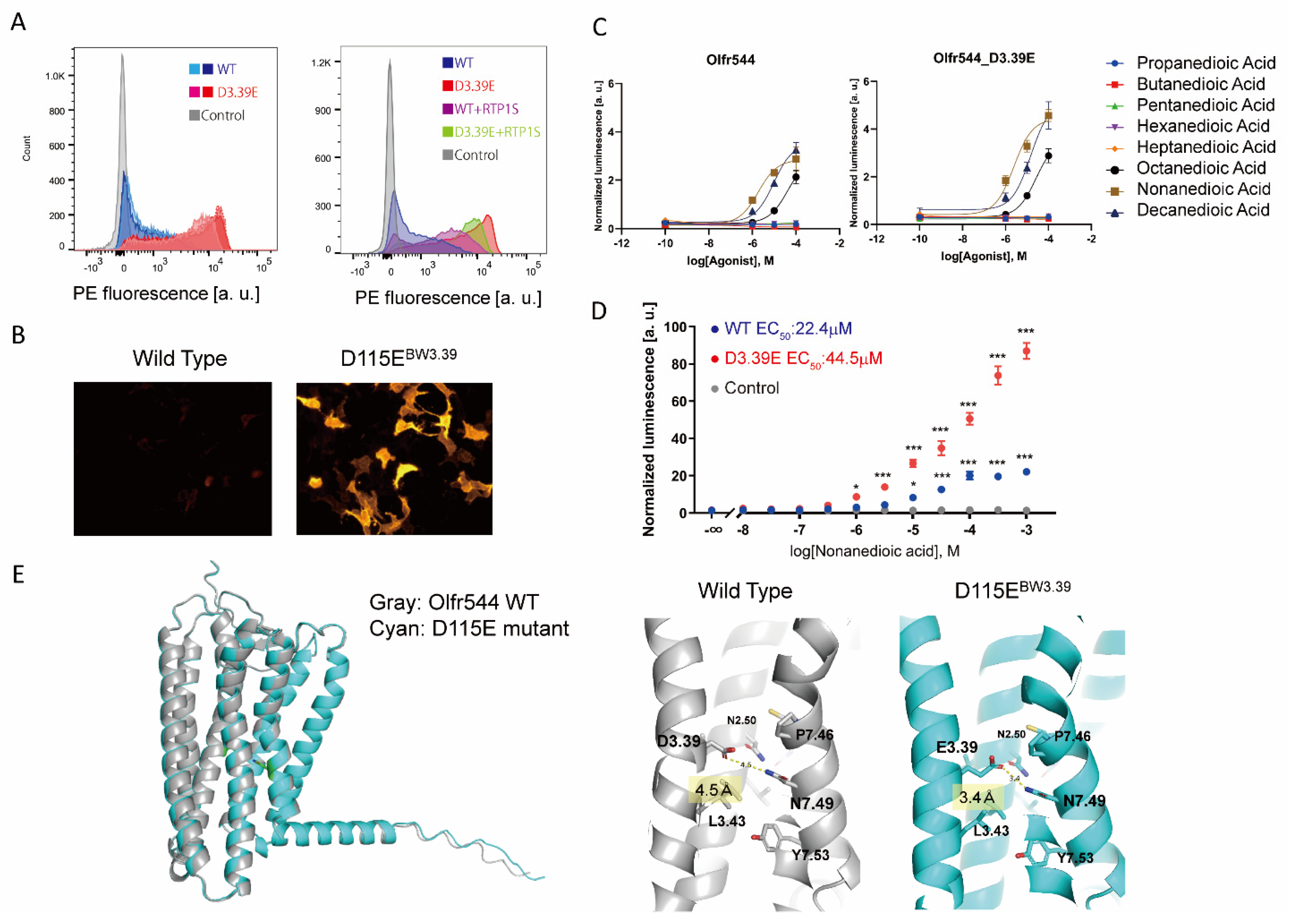
2.3. Olfr544 D3.39E Showed Significantly High Expression without a Change in Ligand Selectivity
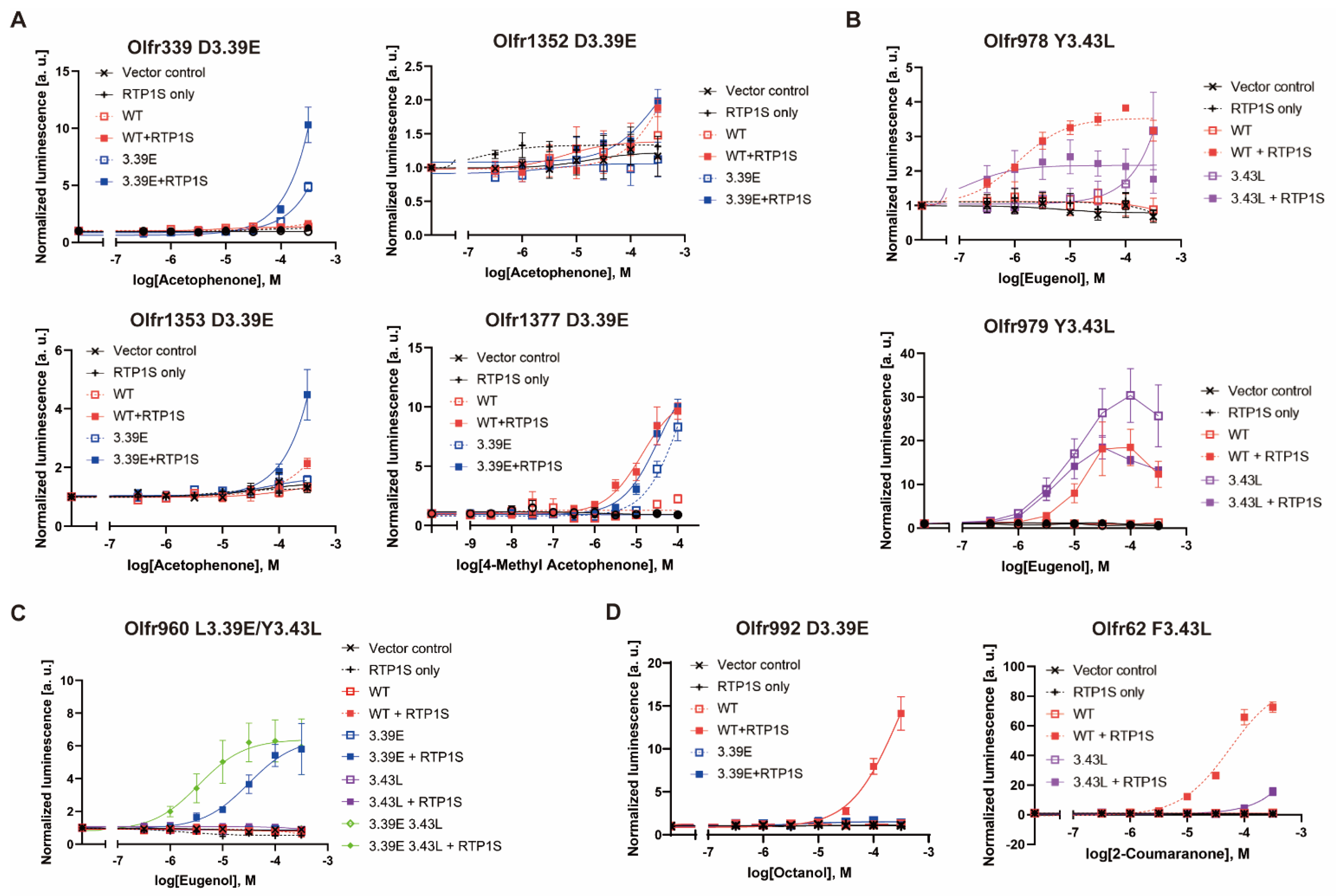
2.4. Agonist Response of NBW3.39E and NBW3.43L Mutant ORs
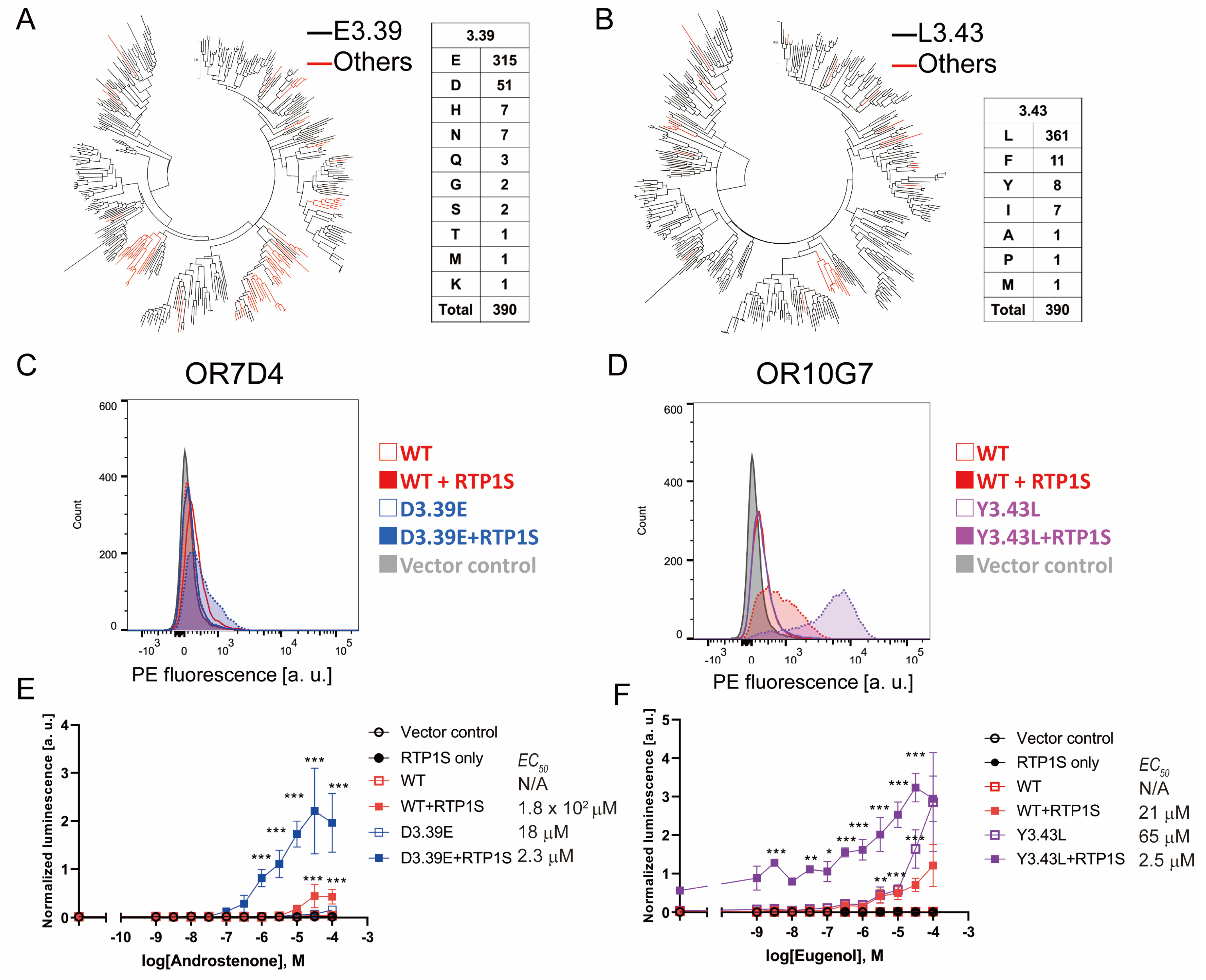
2.5. The Change in Conserved E3.39 and L3.43 Caused a Loss of Function
2.6. NBW 3.39E and NBW 3.43L Mutations Are also Effective in Human Ors
3. Discussion
4. Materials and Methods
4.1. DNA and Vector Preparation
4.2. Cell Culture
4.3. Flow Cytometry Analyses
4.4. Immunocytochemistry
4.5. Dual Luciferase Reporter Gene Assay
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OR | Odorant receptor |
| GPCR | G-protein coupled receptor |
| RTP | Receptor transporting protein |
| BW | Ballesteros-Weinstein number [20] |
| ER | endoplasmic reticulum |
| HEK293T | Human Embryonic Kidney 293T |
| FACS | fluorescence-activated cell sorting |
References
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Lu, M.; Echeverri, F.; Moyer, B.D. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic 2003, 4, 416–433. [Google Scholar] [CrossRef] [PubMed]
- Malnic, B.; Godfrey, P.A.; Buck, L.B. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2584–2589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraiva, L.R.; Kondoh, K.; Ye, X.; Yoon, K.H.; Hernandez, M.; Buck, L.B. Combinatorial effects of odorants on mouse behavior. Proc. Natl. Acad. Sci. USA 2016, 113, E3300–E3306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, I.H.A.; Ibarra-Soria, X.; Fitzgerald, S.; Gonzalez, J.M.; Davidson, C.; Hardy, M.P.; Manthravadi, D.; Van Gerven, L.; Jorissen, M.; Zeng, Z.; et al. Expert curation of the human and mouse olfactory receptor gene repertoires identifies conserved coding regions split across two exons. BMC Genom. 2020, 21, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.Y.; Menuz, K.; Carlson, J.R. Olfactory perception: Receptors, cells, and circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Kida, H.; Fukutani, Y.; Mainland, J.D.; de March, C.A.; Vihani, A.; Li, Y.R.; Chi, Q.; Toyama, A.; Liu, L.; Kameda, M.; et al. Vapor detection and discrimination with a panel of odorant receptors. Nat. Commun. 2018, 9, 4556. [Google Scholar] [CrossRef] [Green Version]
- Krautwurst, D.; Yau, K.W.; Reed, R.R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 1998, 95, 917–926. [Google Scholar] [CrossRef] [Green Version]
- Noe, F.; Geithe, C.; Fiedler, J.; Krautwurst, D. A bi-functional IL-6-HaloTag((R)) as a tool to measure the cell-surface expression of recombinant odorant receptors and to facilitate their activity quantification. J. Biol. Methods 2017, 4, e82. [Google Scholar] [CrossRef] [Green Version]
- Noe, F.; Frey, T.; Fiedler, J.; Geithe, C.; Nowak, B.; Krautwurst, D. IL-6-HaloTag((R)) enables live-cell plasma membrane staining, flow cytometry, functional expression, and de-orphaning of recombinant odorant receptors. J. Biol. Methods 2017, 4, e81. [Google Scholar] [CrossRef] [Green Version]
- Neuhaus, E.M.; Mashukova, A.; Zhang, W.; Barbour, J.; Hatt, H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chem. Senses 2006, 31, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Dannecker, L.E.; Mercadante, A.F.; Malnic, B. Ric-8B promotes functional expression of odorant receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 9310–9314. [Google Scholar] [CrossRef] [Green Version]
- Saito, H.; Kubota, M.; Roberts, R.W.; Chi, Q.; Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 2004, 119, 679–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, H.; Matsunami, H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 2007, 282, 15284–15293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Pan, Y.; Chen, G.Q.; Matsunami, H.; Zhuang, H. Receptor-transporting protein 1 short (RTP1S) mediates translocation and activation of odorant receptors by acting through multiple steps. J. Biol. Chem. 2012, 287, 22287–22294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Su, X.; Pan, Y.; Zhuang, H. Receptor-transporting protein (RTP) family members play divergent roles in the functional expression of odorant receptors. PLoS ONE 2017, 12, e0179067. [Google Scholar] [CrossRef] [Green Version]
- Fukutani, Y.; Tamaki, R.; Inoue, R.; Koshizawa, T.; Sakashita, S.; Ikegami, K.; Ohsawa, I.; Matsunami, H.; Yohda, M. The N-terminal region of RTP1S plays important roles in dimer formation and odorant receptor-trafficking. J. Biol. Chem. 2019, 294, 14661–14673. [Google Scholar] [CrossRef]
- Sharma, R.; Ishimaru, Y.; Davison, I.; Ikegami, K.; Chien, M.S.; You, H.; Chi, Q.; Kubota, M.; Yohda, M.; Ehlers, M.; et al. Olfactory receptor accessory proteins play crucial roles in receptor function and gene choice. Elife 2017, 6, e21895. [Google Scholar] [CrossRef]
- Ikegami, K.; de March, C.A.; Nagai, M.H.; Ghosh, S.; Do, M.; Sharma, R.; Bruguera, E.S.; Lu, Y.E.; Fukutani, Y.; Vaidehi, N.; et al. Structural instability and divergence from conserved residues underlie intracellular retention of mammalian odorant receptors. Proc. Natl. Acad. Sci. USA 2020, 117, 2957–2967. [Google Scholar] [CrossRef]
- Juan, A.; Ballesteros, H.W. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Olender, T.; Nativ, N.; Lancet, D. HORDE: Comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013, 1003, 23–38. [Google Scholar] [PubMed]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-de-Teran, H.; Massink, A.; Rodriguez, D.; Liu, W.; Han, G.W.; Joseph, J.S.; Katritch, I.; Heitman, L.H.; Xia, L.; Ijzerman, A.P.; et al. The role of a sodium ion binding site in the allosteric modulation of the A(2A) adenosine G protein-coupled receptor. Structure 2013, 21, 2175–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katritch, V.; Fenalti, G.; Abola, E.E.; Roth, B.L.; Cherezov, V.; Stevens, R.C. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 2014, 39, 233–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Yasuda, S.; Kajiwara, Y.; Toyoda, Y.; Morimoto, K.; Suno, R.; Iwata, S.; Kobayashi, T.; Murata, T.; Kinoshita, M. Hot-Spot Residues to be Mutated Common in G Protein-Coupled Receptors of Class A: Identification of Thermostabilizing Mutations Followed by Determination of Three-Dimensional Structures for Two Example Receptors. J. Phys. Chem. B 2017, 121, 6341–6350. [Google Scholar] [CrossRef]
- Li, S.; Ahmed, L.; Zhang, R.; Pan, Y.; Matsunami, H.; Burger, J.L.; Block, E.; Batista, V.S.; Zhuang, H. Smelling Sulfur: Copper and Silver Regulate the Response of Human Odorant Receptor OR2T11 to Low-Molecular-Weight Thiols. J. Am. Chem. Soc. 2016, 138, 13281–13288. [Google Scholar] [CrossRef]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef]
- Mainland, J.D.; Li, Y.R.; Zhou, T.; Liu, W.L.; Matsunami, H. Human olfactory receptor responses to odorants. Sci. Data 2015, 2, 150002. [Google Scholar] [CrossRef] [Green Version]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J.D. Odor coding by a Mammalian receptor repertoire. Sci. Signal 2009, 2, ra9. [Google Scholar] [CrossRef] [Green Version]
- Bubnell, J.; Jamet, S.; Tomoiaga, D.; D’Hulst, C.; Krampis, K.; Feinstein, P. In Vitro Mutational and Bioinformatics Analysis of the M71 Odorant Receptor and Its Superfamily. PLoS ONE 2015, 10, e0141712. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, M.; Bauer, J.; Haag, F.; Krautwurst, D. Conserved C-terminal motifs in odorant receptors instruct their cell surface expression and cAMP signaling. FASEB J. 2021, 35, e21274. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chun, E.; Thompson, A.A.; Chubukov, P.; Xu, F.; Katritch, V.; Han, G.W.; Roth, C.B.; Heitman, L.H.; Ap, I.J.; et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 2012, 337, 232–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abaffy, T.; Bain, J.R.; Muehlbauer, M.J.; Spasojevic, I.; Lodha, S.; Bruguera, E.; O’Neal, S.K.; Kim, S.Y.; Matsunami, H. A Testosterone Metabolite 19-Hydroxyandrostenedione Induces Neuroendocrine Trans-Differentiation of Prostate Cancer Cells via an Ectopic Olfactory Receptor. Front. Oncol. 2018, 8, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushdid, C.; de March, C.A.; Topin, J.; Do, M.; Matsunami, H.; Golebiowski, J. Mammalian class I odorant receptors exhibit a conserved vestibular-binding pocket. Cell. Mol. Life Sci. 2019, 76, 995–1004. [Google Scholar] [CrossRef]
- de March, C.A.; Topin, J.; Bruguera, E.; Novikov, G.; Ikegami, K.; Matsunami, H.; Golebiowski, J. Odorant Receptor 7D4 Activation Dynamics. Angew. Chem. Int. Ed. Engl. 2018, 57, 4554–4558. [Google Scholar] [CrossRef]
- Charlier, L.; Topin, J.; de March, C.A.; Lai, P.C.; Crasto, C.J.; Golebiowski, J. Molecular modelling of odorant/olfactory receptor complexes. Methods Mol. Biol. 2013, 1003, 53–65. [Google Scholar]
- Zhuang, H.; Matsunami, H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 2008, 3, 1402–1413. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukutani, Y.; Nakamura, Y.; Muto, N.; Miyanaga, S.; Kanemaki, R.; Ikegami, K.; Noguchi, K.; Ohsawa, I.; Matsunami, H.; Yohda, M. Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors. Int. J. Mol. Sci. 2022, 23, 277. https://doi.org/10.3390/ijms23010277
Fukutani Y, Nakamura Y, Muto N, Miyanaga S, Kanemaki R, Ikegami K, Noguchi K, Ohsawa I, Matsunami H, Yohda M. Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors. International Journal of Molecular Sciences. 2022; 23(1):277. https://doi.org/10.3390/ijms23010277
Chicago/Turabian StyleFukutani, Yosuke, Yuko Nakamura, Nonoko Muto, Shunta Miyanaga, Reina Kanemaki, Kentaro Ikegami, Keiichi Noguchi, Ikuroh Ohsawa, Hiroaki Matsunami, and Masafumi Yohda. 2022. "Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors" International Journal of Molecular Sciences 23, no. 1: 277. https://doi.org/10.3390/ijms23010277
APA StyleFukutani, Y., Nakamura, Y., Muto, N., Miyanaga, S., Kanemaki, R., Ikegami, K., Noguchi, K., Ohsawa, I., Matsunami, H., & Yohda, M. (2022). Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors. International Journal of Molecular Sciences, 23(1), 277. https://doi.org/10.3390/ijms23010277






