Human Cytomegalovirus miR-UL70-3p Downregulates the H2O2-Induced Apoptosis by Targeting the Modulator of Apoptosis-1 (MOAP1)
Abstract
:1. Introduction
2. Results
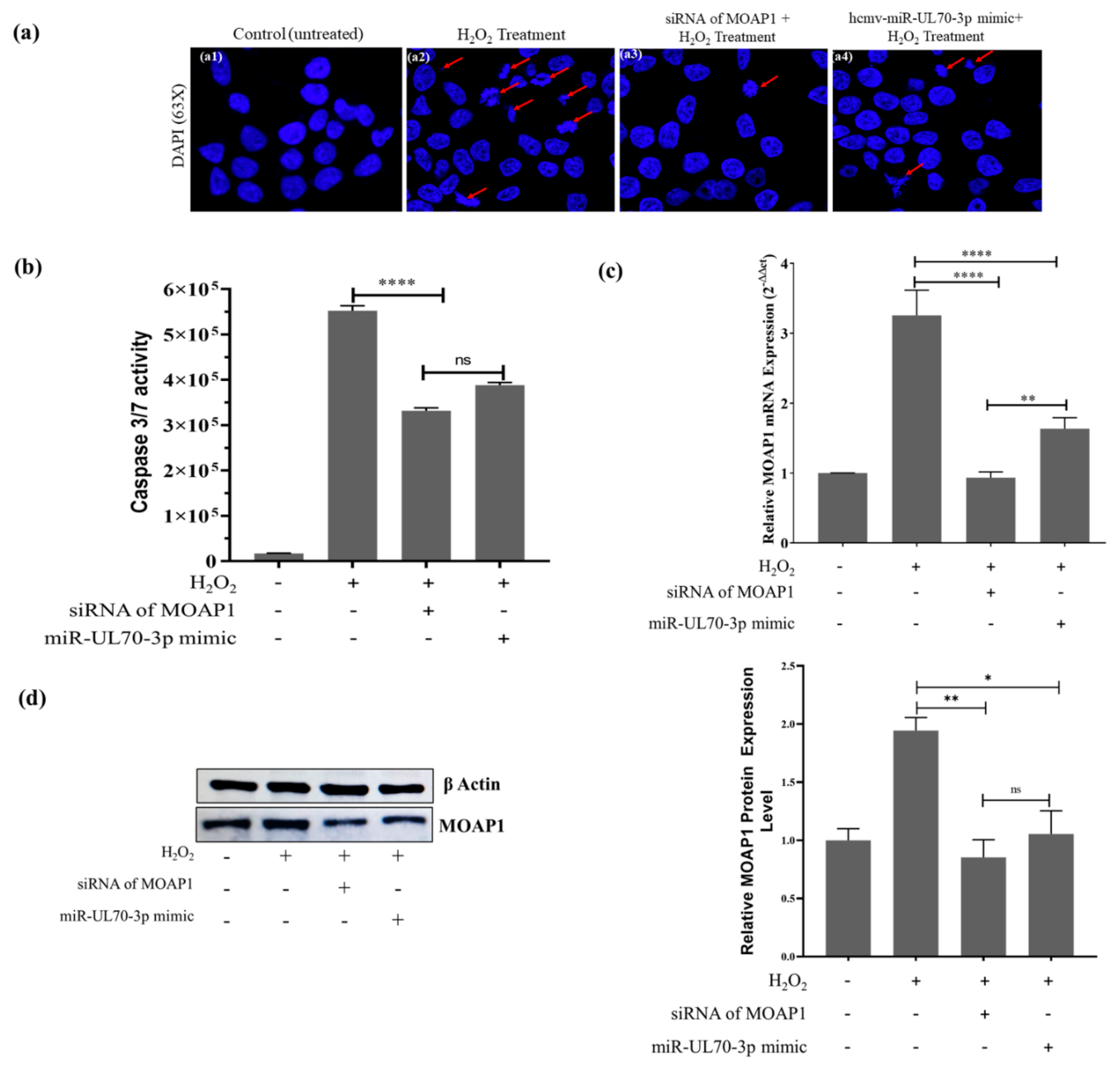
2.1. Effect of hcmv-miR-UL70-3p on H2O2 Induced Apoptosis
2.1.1. DAPI (4’,6-Diamidino-2-phenylindole) Staining
2.1.2. Scanning Electron Microscopy Analysis for Membrane Blebbing
2.1.3. Evaluation of Apoptosis through Flow Cytometry
2.1.4. Measurement of Caspases 3/7 Activity
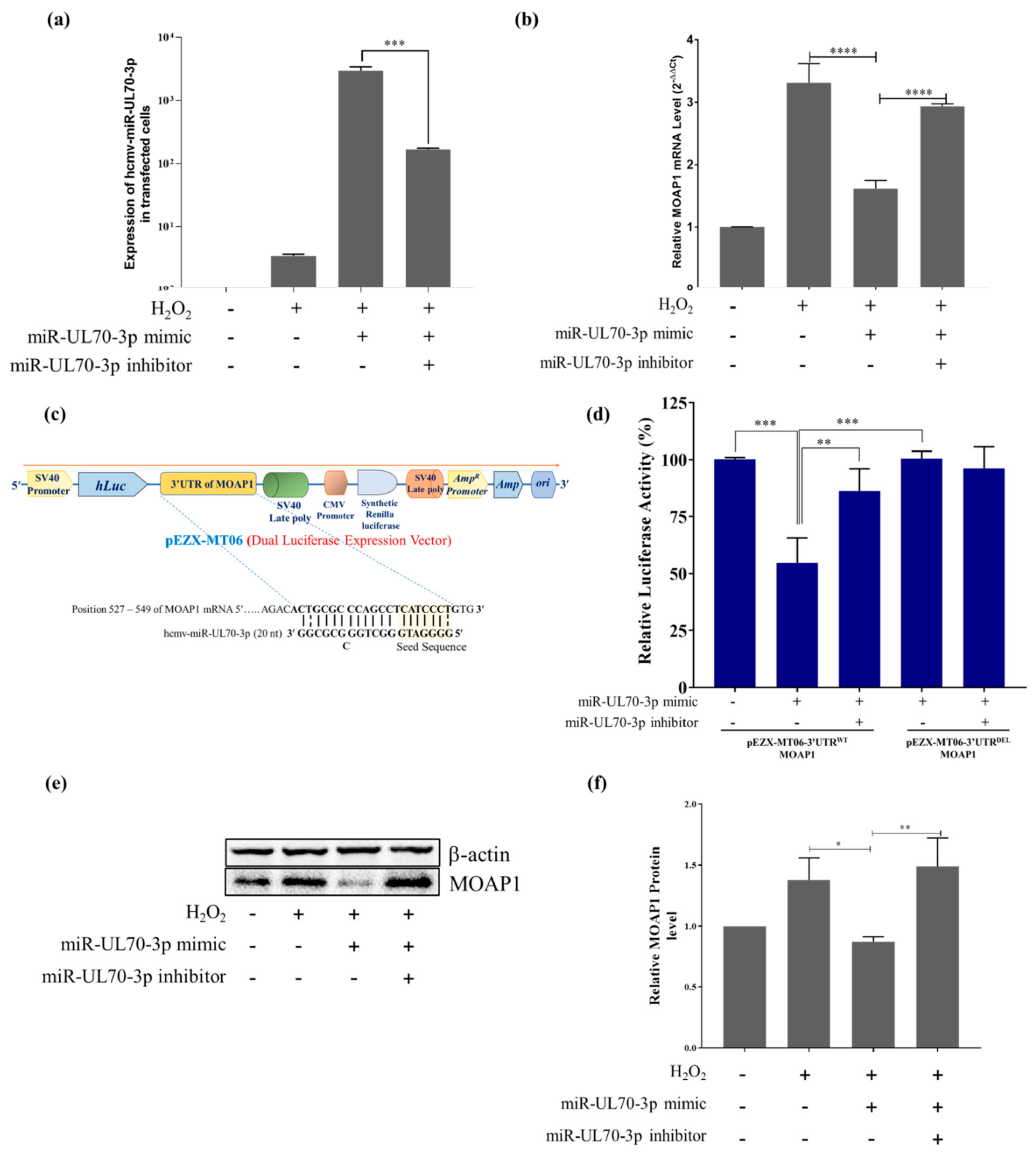
2.2. MOAP1 Is a Direct Target of hcmv-miR-UL70-3p
2.2.1. Hcmv-miR-UL70-3p Regulates the Expression of MOAP1 mRNA
2.2.2. Hcmv-miR-UL70-3p Binds the 3’UTR of MOAP1
2.2.3. Hcmv-miR-UL70-3p Downregulates the MOAP1 Protein Expression
2.3. Hcmv-miR-UL70-3p and siRNA of MOAP1 Effect on the Expression of MOAP1 mRNA and Its Protein
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Apoptosis Induction and Evaluation
4.3. Hcmv-miR-UL70-3p Mimic, Its Inhibitor, siRNA of MOAP1, and UTR Vector Constructs of MOAP1
4.4. 4′,6-Diamidino-2-phenylindole (DAPI) Staining
4.5. Scanning Electron Microscopy
4.6. Analysis of Cell Nuclear Morphology and Membrane Ruffling with ImageJ
4.7. Flow Cytometry
4.8. Caspase 3/7 Assay
4.9. qRT-PCR
4.10. Dual-Luciferase Reporter Assay
4.11. Western Blotting
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuhair, M.; Smit, G.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schottstedt, V.; Blümel, J.; Burger, R.; Drosten, C.; Gröner, A.; Gürtler, L.; Heiden, M.; Hildebrandt, M.; Jansen, B.; Montag-Lessing, T.; et al. Human cytomegalovirus (HCMV)–revised. Transfus. Med. Hemother. 2010, 37, 365–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diggins, N.L.; Skalsky, R.L.; Hancock, M.H. Regulation of Latency and Reactivation by Human Cytomegalovirus miRNAs. Pathogens 2021, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shen, Y.; Shenk, T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 1995, 69, 7960–7970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terhune, S.; Torigoi, E.; Moorman, N.; Silva, M.; Qian, Z.; Shenk, T.; Yu, D. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 2007, 81, 3109–3123. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Bao, Q.; Xuan, B.; Xu, W.; Pan, D.; Li, Q.; Qian, Z. Human cytomegalovirus protein pUL38 prevents premature cell death by binding to ubiquitin-specific protease 24 and regulating iron metabolism. J. Virol. 2018, 92, e00191-18. [Google Scholar] [CrossRef] [Green Version]
- Reeves, M.B.; Davies, A.A.; McSharry, B.P.; Wilkinson, G.W.; Sinclair, J.H. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007, 316, 1345–1348. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Kalejta, R.F.; Kerry, J.; Semmes, O.J.; O’Connor, C.M.; Khan, Z.; Garcia, B.A.; Shenk, T.; Murphy, E. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2012, 109, 9575–9580. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Qi, Y.; Huang, Y.J.; Qi, M.L.; Ma, Y.P.; He, R.; Ji, Y.H.; Sun, Z.R.; Ruan, Q. Identification of immediate early gene X-1 as a cellular target gene of hcmv-mir-UL148D. Int. J. Mol. Med. 2013, 31, 959–966. [Google Scholar] [CrossRef] [Green Version]
- Babu, S.G.; Pandeya, A.; Verma, N.; Shukla, N.; Kumar, R.V.; Saxena, S. Role of HCMV miR-UL70-3p and miR-UL148D in overcoming the cellular apoptosis. Mol. Cell Biochem. 2014, 393, 89–98. [Google Scholar] [CrossRef]
- Guo, X.; Huang, Y.; Qi, Y.; Liu, Z.; Ma, Y.; Shao, Y.; Jiang, S.; Sun, Z.; Ruan, Q. Human cytomegalovirus miR-UL36-5p inhibits apoptosis via downregulation of adenine nucleotide translocator 3 in cultured cells. Arch. Virol. 2015, 160, 2483–2490. [Google Scholar] [CrossRef]
- Kim, S.; Seo, D.; Kim, D.; Hong, Y.; Chang, H.; Baek, D.; Kim, V.N.; Lee, S.; Ahn, K. Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 2015, 17, 838–851. [Google Scholar] [CrossRef] [Green Version]
- Hancock, M.H.; Crawford, L.B.; Perez, W.; Struthers, H.M.; Mitchell, J.; Caposio, P. Human cytomegalovirus UL7, miR-US5-1, and miR-UL112-3p inactivation of FOXO3a protects CD34+ hematopoietic progenitor cells from apoptosis. mSphere 2021, 6, e00986-20. [Google Scholar] [CrossRef]
- Lau, B.; Poole, E.; Krishna, B.; Sellart, I.; Wills, M.R.; Murphy, E.; Sinclair, J. The Expression of Human Cytomegalovirus MicroRNA MiR-UL148D during Latent Infection in Primary Myeloid Cells Inhibits Activin A-triggered Secretion of IL-6. Sci. Rep. 2014, 6, 31205. [Google Scholar] [CrossRef]
- Hook, L.M.; Grey, F.; Grabski, R.; Tirabassi, R.; Doyle, T.; Hancock, M.; Landais, I.; Jeng, S.; McWeeney, S.; Britt, W.; et al. Cytomegalovirus miRNAs Target Secretory Pathway Genes to Facilitate Formation of the Virion Assembly Compartment and Reduce Cytokine Secretion. Cell Host Microbe 2014, 15, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Qi, Y.; Ma, Y.; He, R.; Ji, Y.; Sun, Z.; Ruan, Q. Down-regulation of human cytomegalovirus UL138, a novel latency-associated determinant, by hcmv-miR-UL36. J. Biosci. 2013, 38, 479–485. [Google Scholar] [CrossRef]
- Chaudhry, M.Z.; Kasmapour, B.; Plaza-Sirventl, C.; Bajagic, M.; Casalegno Garduño, R.; Borkner, L.; Lenac Roviš, T.; Scrima, A.; Jonjic, S.; Schmitz, I.; et al. UL36 Rescues Apoptosis Inhibition and In vivo Replication of a Chimeric MCMV Lacking the M36 Gene. Front. Cell. Infect. Microbiol. 2017, 7, 312. [Google Scholar] [CrossRef] [Green Version]
- Grey, F.; Antoniewicz, A.; Allen, E.; Saugstad, J.; McShea, A.; Carrington, J.C.; Nelson, J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 2005, 79, 12095–12099. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, A.A.; Costa, H.; Landázuri, N.; Lui, W.O.; Hultenby, K.; Rahbar, A.; Yaiw, K.C.; Söderberg-Nauclér, C. Human cytomegalovirus microRNAs are carried by virions and dense bodies and are delivered to target cells. J. Gen. Virol. 2017, 98, 1058–1072. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Shango, J.; Seal, A.; Shukla, D.; Nares, S. Viral miRNAs Alter Host Cell miRNA Profiles and Modulate Innate Immune Responses. Front. Immunol. 2018, 9, 433. [Google Scholar] [CrossRef]
- Shen, Z.Z.; Pan, X.; Miao, L.F.; Ye, H.Q.; Chavanas, S.; Davrinche, C.; McVoy, M.; Luo, M.H. Comprehensive analysis of human cytomegalovirus microRNA expression during lytic and quiescent infection. PLoS ONE 2014, 9, e88531. [Google Scholar] [CrossRef] [Green Version]
- Ulasov, I.V.; Kaverina, N.V.; Ghosh, D.; Baryshnikova, M.A.; Kadagidze, Z.G.; Karseladze, A.I.; Baryshnikov, A.Y.; Cobbs, C.S. CMV70-3P miRNA contributes to the CMV mediated glioma stemness and represents a target for glioma experimental therapy. Oncotarget 2017, 8, 25989–25999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, J.; Wan, C.; Guo, R.; Guo, D. Is Hydrogen Peroxide a Suitable Apoptosis Inducer for All Cell Types? BioMed Res. Int. 2016, 2016, 7343965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Zhang, W.; Liu, Q. Human cytomegalovirus-encoded miR-US25-1 aggravates the oxidised low density lipoprotein-induced apoptosis of endothelial cells. BioMed Res. Int. 2014, 2014, 531979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.; Qi, Y.; Huang, Y.; Liu, Z.; Ma, Y.; Guo, X.; Jiang, S.; Sun, Z.; Ruan, Q. Human cytomegalovirus-encoded miR-US4-1 promotes cell apoptosis and benefits discharge of infectious virus particles by targeting QARS. J. Biosci. 2016, 41, 183–192. [Google Scholar] [CrossRef]
- Shao, Y.; Qi, Y.; Huang, Y.; Liu, Z.; Ma, Y.; Guo, X.; Jiang, S.; Sun, Z.; Ruan, Q. Human cytomegalovirus miR-US4-5p promotes apoptosis via downregulation of p21-activated kinase 2 in cultured cells. Mol. Med. Rep. 2017, 16, 4171–4178. [Google Scholar] [CrossRef]
- Yan, B.; Zhao, J.L. miR-1228 prevents cellular apoptosis through targeting of MOAP1 protein. Apoptosis 2012, 17, 717–724. [Google Scholar] [CrossRef]
- Wu, T.; Chen, W.; Kong, D.; Li, X.; Lu, H.; Liu, S.; Wang, J.; Du, L.; Kong, Q.; Huang, X.; et al. miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis 2015, 36, 925–935. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.O.; Tan, K.M.; Chan, S.L.; Yee, K.S.; Bevort, M.; Ang, K.C.; Yu, V.C. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J. Biol. Chem. 2001, 276, 2802–2807. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.O.; Fu, N.Y.; Sukumaran, S.K.; Chan, S.L.; Kang, J.H.; Poon, K.L.; Chen, B.S.; Yu, V.C. MAP-1 is a mitochondrial effector of Bax. Proc. Natl. Acad. Sci. USA 2005, 102, 14623–14628. [Google Scholar] [CrossRef] [Green Version]
- Baksh, S.; Tommasi, S.; Fenton, S.; Yu, V.C.; Martins, L.M.; Pfeifer, G.P.; Latif, F.; Downward, J.; Neel, B.G. The tumor suppressor RASSF1A and MAP-1 link death receptor signaling to Bax conformational change and cell death. Mol. Cell 2005, 18, 637–650. [Google Scholar] [CrossRef]
- Zhang, A.; Hildreth, R.L.; Colberg-Poley, A.M. Human cytomegalovirus inhibits apoptosis by proteasome-mediated degradation of Bax at endoplasmic reticulum-mitochondrion contacts. J. Virol. 2013, 87, 5657–5668. [Google Scholar] [CrossRef] [Green Version]
- Chazotte, B. Labeling nuclear DNA using DAPI. Cold Spring Harb. Protoc. 2011, 87. [Google Scholar] [CrossRef] [Green Version]
- Eidet, J.R.; Pasovic, L.; Maria, R.; Jackson, C.J.; Utheim, T.P. Objective assessment of changes in nuclear morphology and cell distribution following induction of apoptosis. Diagn. Pathol. 2014, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandeya, A.; Khalko, R.K.; Mishra, A.; Singh, N.; Singh, S.; Saha, S.; Yadav, S.; Saxena, S.; Gosipatala, S.B. Human Cytomegalovirus miR-UL70-3p Downregulates the H2O2-Induced Apoptosis by Targeting the Modulator of Apoptosis-1 (MOAP1). Int. J. Mol. Sci. 2022, 23, 18. https://doi.org/10.3390/ijms23010018
Pandeya A, Khalko RK, Mishra A, Singh N, Singh S, Saha S, Yadav S, Saxena S, Gosipatala SB. Human Cytomegalovirus miR-UL70-3p Downregulates the H2O2-Induced Apoptosis by Targeting the Modulator of Apoptosis-1 (MOAP1). International Journal of Molecular Sciences. 2022; 23(1):18. https://doi.org/10.3390/ijms23010018
Chicago/Turabian StylePandeya, Abhishek, Raj Kumar Khalko, Anup Mishra, Nishant Singh, Sukhveer Singh, Sudipta Saha, Sanjay Yadav, Sangeeta Saxena, and Sunil Babu Gosipatala. 2022. "Human Cytomegalovirus miR-UL70-3p Downregulates the H2O2-Induced Apoptosis by Targeting the Modulator of Apoptosis-1 (MOAP1)" International Journal of Molecular Sciences 23, no. 1: 18. https://doi.org/10.3390/ijms23010018
APA StylePandeya, A., Khalko, R. K., Mishra, A., Singh, N., Singh, S., Saha, S., Yadav, S., Saxena, S., & Gosipatala, S. B. (2022). Human Cytomegalovirus miR-UL70-3p Downregulates the H2O2-Induced Apoptosis by Targeting the Modulator of Apoptosis-1 (MOAP1). International Journal of Molecular Sciences, 23(1), 18. https://doi.org/10.3390/ijms23010018





